Abstract
Key message
New molecular markers were developed and mapped to the FHB resistance QTL region in high resolution. Micro-collinearity of the QTL region with rice and Brachypodium was revealed for a better understanding of the genomic region.
Abstract
The wild emmer wheat (Triticum dicoccoides)-derived Fusarium head blight (FHB) resistance quantitative trait locus (QTL) Qfhs.ndsu-3AS previously mapped to the short arm of chromosome 3A (3AS) in a population of recombinant inbred chromosome lines (RICLs). This study aimed to attain a better understanding of the genomic region harboring Qfhs.ndsu-3AS and to improve the utility of the QTL in wheat breeding. Micro-collinearity of the QTL region with rice chromosome 1 and Brachypodium chromosome 2 was identified and used for marker development in saturation mapping. A total of 42 new EST-derived sequence tagged site (STS) and simple sequence repeat (SSR) markers were developed and mapped to the QTL and nearby regions on 3AS. Further comparative analysis revealed a complex collinearity of the 3AS genomic region with their collinear counterparts of rice and Brachypodium. Fine mapping of the QTL region resolved five co-segregating markers (Xwgc1186/Xwgc716/Xwgc1143/Xwgc501/Xwgc1204) into three distinct loci proximal to Xgwm2, a marker previously reported to be closely linked to the QTL. Four other markers (Xwgc1226, Xwgc510, Xwgc1296, and Xwgc1301) mapped farther proximal to the above markers in the QTL region with a higher resolution. Five homozygous recombinants with shortened T. dicoccoides chromosomal segments in the QTL region were recovered by molecular marker analysis and evaluated for FHB resistance. Qfhs.ndsu-3AS was positioned to a 5.2 cM interval flanked by the marker Xwgc501 and Xwgc510. The recombinants containing Qfhs.ndsu-3AS and new markers defining the QTL will facilitate utilization of this resistance source in wheat breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fusarium head blight (FHB), caused mostly by Fusarium graminearum Schwabe [telomorph: Gibberella zeae (Schw.) Petch] in North America, is a destructive fungal disease of wheat. Infection and development of FHB are accelerated by warm and humid conditions (Stack and McMullen 1985; Xu et al. 2008). Outbreak of FHB can cause severe losses of grain yield and quality in both common wheat (Triticum aestivum L., 2n = 6x = 42, genome AABBDD) and durum wheat (T. durum Desf., 2n = 4x = 28, genome AABB) (McMullen et al. 1997). Multiple QTL for FHB resistance have been identified and successfully deployed in common wheat cultivars (Liu and Anderson 2003; Liu et al. 2006, 2008; Buerstmayr et al. 2009; Chu et al. 2011). However, FHB remains a major threat to durum wheat production due to the lack of effective resistance sources in durum (Oliver et al. 2007). Incorporation of FHB resistance genes from common wheat into durum has not been very successful because of the complex inheritance of hexaploid-derived FHB resistance in the durum background (Buerstmayr et al. 2012; Zhu et al. 2012).
Sources of FHB resistance have been found in durum wheat and its tetraploid relatives even though they are not as effective as those in common wheat (Stack et al. 2002; Buerstmayr et al. 2003; Cai et al. 2005; Oliver et al. 2007, 2008; Ghavami et al. 2011; Talas et al. 2011; Ruan et al. 2012; Zhang et al. 2014). Molecular mapping has identified several wild emmer wheat (T. dicoccoides)-derived FHB resistance QTL, including Qfhs.ndsu-3AS on chromosome 3A (Otto et al. 2002), Qfhs.fcu-7AL on 7A (Kumar et al. 2007), and another on 6B (Stack et al. 2006). Also, FHB resistance QTL has been detected in the durum-related tetraploids T. cathlicum (Somers et al. 2006) and T. dicoccum (Buerstmayr et al. 2012; Zhang et al. 2014). In addition, extensive screening of durum accessions for FHB resistance has identified several durum landraces with detectable resistance to the disease over the last few years (Ghavami et al. 2011; Talas et al. 2011; Zhang et al. 2014). Some of these tetraploid-derived FHB resistance QTL mapped to the same genomic regions as those identified in hexaploids, suggesting collinearity of the resistance gene loci in tetraploids and hexaploids (Somers et al. 2006; Buerstmayr et al. 2012).
Qfhs.ndsu-3AS is a wild emmer-derived FHB resistance QTL located on 3AS (Otto et al. 2002; Stack et al. 2002). It is the first FHB resistance QTL identified in tetraploid wheat and confers moderate resistance in the durum background. Saturation mapping positioned Qfhs.ndsu-3AS to a chromosomal interval of 11.5 cM flanked by the molecular markers Xfcp401 and Xfcp397.2 on 3AS (Chen et al. 2007). Wheat chromosomes in homoeologous group 3 are collinear to rice chromosome 1 (R1) and Brachypodium chromosome 2 (B2) (Moore et al. 1995; the International Brachypodium Initiative 2010; Sehgal et al. 2012; Luo et al. 2013). Wheat researchers have been taking advantage of the collinearity with these two grass models to characterize genes in the large and complex polyploid genome of wheat (Liu and Anderson 2003; Foote et al. 2004; Liu et al. 2006; Chen et al. 2007; Kumar et al. 2009). This study aimed to saturate the genomic region harboring Qfhs.ndsu-3AS and improve the map resolution of the QTL region by exploring the micro-collinearity of the QTL region with R1 and B2. This will facilitate understanding of the genomic region harboring the FHB resistance QTL Qfhs.ndsu-3AS and provide effective molecular markers to assist selection of this resistance QTL in wheat breeding.
Materials and methods
Mapping populations
Eighty-three recombinant inbred chromosome lines (RICLs) derived from the cross between durum wheat cultivar ‘Langdon’ (LDN) and disomic LDN-T. dicoccoides Israel-A (ISA) substitution line 3A [LDN(DIC-3A)] (Joppa 1993) were employed for saturation mapping and QTL analysis in this study. RICL#10, having the shortest T. dicoccoides chromosomal fragment harboring Qfhs.ndsu-3AS, was crossed with LDN to generate additional meiotic recombinants within the QTL region for fine mapping of the QTL. A large F2 population (n > 1800) segregating only in the QTL region was developed from that cross. The F2 individuals with T. dicoccoides chromosomal fragments smaller than that in RICL#10 were selected to develop recombinants homozygous at the marker loci within the QTL region. Two other RICLs (RICL#15 and RICL#49) critical in the QTL region were also included in this study. Both RICL#10 and RICL#49 exhibited FHB resistance as the resistant parent LDN(DIC-3A), while RICL#15 exhibited susceptibility to FHB as the susceptible parent LDN (Chen et al. 2007). RICL#10 and the homozygous recombinants developed in this study were evaluated for FHB resistance to identify shortened T. dicoccoides chromosomal fragments harboring Qfhs.ndsu-3AS for fine mapping of the QTL and for germplasm development.
Comparative analysis and molecular marker development
An initial comparative analysis was performed to develop EST (expressed sequence tag)-derived STS and SSR markers for saturation mapping of the Qfhs.ndsu-3AS QTL region based on the collinearity of the QTL region with the genomic regions on rice chromosome R1. The EST sequences of BE517736 and BF484475, from which the STS markers Xfcp402 and Xfcp399 flanking the QTL region were developed (Chen et al. 2007), were used as queries to perform BLASTn against rice and Brachypodium genomic sequences and to identify the collinear counterparts of the QTL region on rice chromosome R1 and Brachypodium chromosome B2 using the J. Craig Venter Institute (JCVI) wheat genome database (http://blast.jcvi.org/euk-blast/index.cgi?project=tae1) and Brachypodium database (http://www.brachypodium.org/gmod/alignment/blast_finders/new), respectively. A threshold of an expected (E) value equal to or less than e−15 was adopted in the BLAST search and comparative analysis. The rice bacterial artificial chromosome (BAC)/P1-derived artificial chromosome (PAC) clones and Brachypodium genomic sequence hits in the BLAST search were used as anchor points to identify the rice and Brachypodium genomic regions collinear to the QTL region. Then the retrieved rice and Brachypodium genomic sequences were used as queries to search for wheat ESTs in the JCVI wheat genome database. The corresponding tentative sequences (TCs) (http://compbio.dfci.harvard.edu/cgi-bin/tgi/est_ann.pl?gudb=wheat) or contigs in GrainGenes 2.0 (http://wheat.pw.usda.gov/GG2/index.shtml) for the identified ESTs were adopted to eliminate redundant EST sequences. The STS and SSR primers were designed from the EST sequences using Primer 3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi).
Genetic mapping and QTL analysis
All EST-derived STS and SSR primers were surveyed for polymorphisms between LDN and LDN (DIC-3A). The polymorphic markers were employed to genotype the RICL population (n = 83) for saturation mapping of the FHB resistance QTL Qfhs.ndsu-3AS. The markers mapped to the QTL region were used to genotype a subset of the F2 population (n = 372) for fine mapping of the QTL. PCR amplification was carried out in a 20-µl mixture containing 40 ng genomic DNA, 0.5 µM each of forward and reverse primers, 1× PCR buffer, 1.5 mM MgCl2, 0.25 mM dNTP and 0.25 U of Taq DNA polymerase. PCR was run as follows: 94 °C for 3 min; 45 cycles of 94 °C for 1 min, 52~60 °C (varied with specific primer pairs) for 1 min and 72 °C for 1.5 min; then with a final 72 °C for 7 min. PCR products were separated on 8 % non-denaturing polyacrylamide gel and visualized by ethidium bromide staining or separated on 5 % denaturing polyacrylamide gel and visualized by silver staining (Liu and Anderson 2003; Chen et al. 2007). Additional molecular marker techniques used in this study included cleaved amplified polymorphism (CAP) (Chen et al. 2003), single-strand conformation polymorphism (SSCP) (Kumar et al. 2006), and heteroduplex analysis (Mohamed et al. 2004). The genetic maps were constructed using Mapmaker 2.0 for Macintosh as described by Lander et al. (1987). All the newly developed molecular markers in this project were designated Xwgc followed by a number.
Qfhs.ndsu-3AS was analyzed against the saturated linkage map of the QTL region in the RICL population using the computer software QGene 4.3.10 (http://www.qgene.org/qgene/index.php). The same sets of phenotypic data as those used in the previous study by Chen et al. (2007) were employed in the composite interval mapping. A permutation test with 1000 permutations was performed to determine the LOD threshold. A LOD threshold of about 2.0 was determined with an experiment-wise error rate of 0.05 in the RICL population.
Immature embryo culture
The immature embryo culture technique was used to accelerate generations of the recombinants for FHB evaluation. Immature embryos of the recombinants at early generations were excised from caryopses and cultured in vitro on MS medium (Cai 1993). Seedlings regenerated from immature embryos were transplanted to the greenhouse for further studies.
FHB disease evaluation and data analysis
The recombinants homozygous at the molecular marker loci within the QTL region were recovered from the LDN x RICL#10 cross by molecular marker analysis. They were grown for evaluation of FHB resistance in randomly arranged 6-inch plastic pots with one plant per pot in 2–3 greenhouse seasons (Fall 2013, Spring 2013, and Summer 2014). Their parents, RICL#10 and LDN, were used as resistant and susceptible controls, respectively. Response of the recombinants and their parents to FHB was evaluated by the point inoculation method as described by Stack et al. (2002). A single floret from a central spikelet was inoculated at first anthesis using 10 µl of spore suspension. The inoculum was prepared by mixing equal number of spores harvested from four isolates of F. graminearum and concentrated to 1 × 105 spores per mL. The temperature in the greenhouse was kept at approximately 25 °C with a 16 h photoperiod during the disease development stage. To facilitate disease development, high humidity was maintained for 72 h with the inoculated spikes covered by a plastic bag. At 21 days post inoculation, the percentage of infected spikelets in a spike was scored. For each pot, the average percentage of all spikes (5–10) was calculated as FHB severity for each individual homozygous recombinant. With regard to the control, the mean value of three pots was recorded as FHB severity.
ANOVA was performed on FHB severity of the homozygous recombinants and their parents. Fisher’s protected LSD was used for mean separation between the genotypes. All statistical analyses were conducted using the Statistical Analysis System version 9.4.
Results
Identification and analysis of micro-collinearity
The wheat ESTs BE517736 (Xfcp402) and BF484475 (Xfcp399) mapped proximally and distally to the Qfhs.ndsu-3AS QTL region defined in the previous study by Chen et al. (2007). The contig sequences of BE517736 (Ta.9622.1.S1_x_at) and BF484475 (Ta.1999.3.S1_a_at) were used as queries of BLAST search against rice genomic sequences. One rice PAC (AP003282) and one rice BAC (AP004225) were hit by the contig ‘Ta.9622.1.S1_x_at’ with the lowest E values of 3.2e−144 and 6.8e−90, respectively. AP003282 is located on the short arm of rice chromosome 1, while AP004225 is on the long arm. Since Qfhs.ndsu-3AS mapped to the short arm of wheat chromosome 3A (3AS), AP003282 was selected as the anchor point of BE517736 on rice chromosome 1. BLAST search with the contig ‘Ta.1999.3.S1_a_at’ identified three PACs (AP003610, AP002969, and AP003727) on rice chromosome 1 with similar E values, i.e., 5.4e−40, 5.8e−40, and 8.0e−40, respectively. Sequence alignment of the three PACs indicated that AP003610 was part of AP003727 and 84 % of AP002969 was included in AP003727. Thus, AP003727 was considered the anchor point of BF484475 on rice chromosome 1. Therefore, the rice genomic region from AP003727 to AP003282 (3724 kb) was considered collinear with the wheat genomic region spanning Qfhs.ndsu-3AS on 3AS. BLAST search against the Brachypodium genome with BE517736 identified a genomic region from 3139,341 to 3141,007 bp on chromosome 2 with an E value of 1.4e−131, while BF484475 identified a genomic region from 2,644,263 to 2,644,440 bp with an E value of 4.7e−53 on the same chromosome. Thus, the Brachypodium genomic region from 2,644,263 to 3,141,007 bp on chromosome 2 was considered collinear with the wheat genomic region spanning Qfhs.ndsu-3AS on 3AS.
Molecular mapping and QTL analysis
A total of 793 pairs of STS primers and 42 pairs of SSR primers were designed from the wheat EST singletons, TCs, and contigs identified based on the micro-collinearity of the Qfhs.ndsu-3AS QTL region with the genomic regions of rice and Brachypodium. Forty-eight pairs of STS primers and three pairs of SSR primers amplified polymorphisms between the two parents of the mapping population. Of these, 42 new STS/SSR markers mapped to a genomic region of 143.3 cM spanning the QTL on 3AS (Fig. 1). The primer sequences of the newly mapped STS and SSR markers on 3AS are listed in Table 1. Twenty-two of the newly developed markers mapped within the QTL region previously defined by the marker Xfcp401 and Xfcp397.2 (Chen et al. 2007). Five co-segregating STS markers, Xwgc501, Xwgc716, Xwgc1143, Xwgc1188, and Xwgc1204, mapped 0.6 cM proximal to Xgwm2, an SSR marker locus identified closely linked to the QTL peak in previous studies (Otto et al. 2002; Chen et al. 2007). Two co-segregating markers (Xwgc774/Xwgc1226) mapped 1.2 cM proximal to the five co-segregating markers within the QTL region. Immediately proximal to Xwgc774/Xwgc1226 were three other co-segregating markers (Xwgc510, Xwgc1296, and Xwgc1301) (Fig. 1). In addition, twelve markers mapped closely distal to the Xbcd1532/Xbarc45 loci. Ten of them, including Xwgc1238, Xwgc1240, Xwgc1248, Xwgc1241, Xwgc1244, Xwgc901, Xwgc1263, Xwgc1254, Xwgc1100, and Xwgc1094, co-segregated in the RICL mapping population. In addition, 11 newly developed markers mapped distal to the previously defined QTL region (i.e., distal to Xfcp401) (Chen et al. 2007; Fig. 1). The distal region defined by these eleven markers on 3AS is collinear with the genomic region encompassing the Fhb1 locus on the short arm of wheat chromosome 3B (Chen et al. 2007; Fig. 1).
Saturated genetic map of wheat 3AS distal region constructed in the RICL population (left), physical map of the distal region (4360 kb) of rice R1S (middle), and distal region of Brachypodium B2S (right) and comparative analysis of these three genomic regions. The hatched region refers to the QTL region in the genetic map (left). The breaks refer to the regions where genetic and physical map distances are not to scale. Rice PACs/BACs: A AP003727, B AP002882, C AP002747, D AP002541, E AP002868, F AP002487, G AP003046, H AP003233, I AP002538, J AP002872, K AP002540, L AP002522, M AP003045, N AP003225, O AP002521, P AP003209, Q AP003301, R AP003339, S AP003282, T AP003215, U AP002523, V AP002903, W AP002524, X AP003118, Y AP003047, Z AP002484
Composite interval mapping of Qfhs.ndsu-3AS against the saturated genetic map delimited the QTL to the chromosomal segment of 6.2 cM flanked by Xbarc45 and Xwgc510 in the RICL population (Figs. 1, 2). Each of the four intervals defined by the markers within the chromosomal segment (i.e., Xbarc45-Xgwm2, Xgwm2-Xwgc501, Xwgc501-Xwgc1226, and Xwgc1226-Xwgc510) explained a phenotypic variation ranging from 43.3 to 46.9 % with a LOD score of over 10.0, which were higher than the intervals outside the chromosomal segment flanked by Xbarc45 and Xwgc510 (Fig. 2).
Nine co-dominant STS markers (Xwgc1188, Xwgc716, Xwgc1143, Xwgc501, Xwgc1204, Xwgc1226, Xwgc1296, Xwgc1301, and Xwgc510) and two SSR markers (Xbarc45 and Xgwm2) mapped to the QTL region (Fig. 1) were used to genotype the F2 mapping population (n = 372). Xwgc774, co-segregating with Xwgc1226, was not included because it is a dominant marker and could affect mapping accuracy in an F2 population (Jiang and Zeng 1997). Xbarc45, a dominant SSR marker, was used to identify homozygous recombinants, but was not used in fine mapping. Xwgc716 and Xwgc1188 still co-segregated and mapped 0.8 cM proximal to Xgwm2 in the F2 population. Xwgc1143 and Xwgc1204 also co-segregated and mapped 0.1 cM proximal to Xwgc716/Xwgc1188 (Fig. 3). Five other STS markers, including Xwgc501, Xwgc1226, Xwgc510, Xwgc1296, and Xwgc1301, mapped farther proximal to the above markers. The map distances from Xwgc1226 to its flanking markers Xwgc501 (distal) and Xwgc510 (proximal) were stretched, whereas Xwgc1296 and Xwgc1301 still co-segregated in the F2 population (Figs. 1, 3).
Identification and evaluation of recombinants for FHB resistance
Genotyping of the F2 population (n = 372) at the 11 SSR and STS marker loci within the QTL region identified three meiotic recombinants (3AS07-39-17, 3AS07-42-26, and 3AS07-53-16). They all contained a shortened T. dicoccoides 3AS fragment in the QTL region (Table 2). 3AS07-39-17 was homozygous at all 11 marker loci in the QTL region and designated 3AS-Rec I (Table 2). Prior to FHB evaluation, this recombinant line was advanced to the F3 and F4 generations to increase homozygosity at other loci. 3AS07-42-26 and 3AS07-53-16 were heterozygous at some of the marker loci within the QTL region (Table 2). They were self-pollinated to produce homozygous recombinants for FHB evaluation and mapping of Qfhs.ndsu-3AS. Two homozygous recombinants with the same genotype at the marker loci were identified from the 10 F3 individuals of 3AS07-42-26 and designated 3AS-Rec II (Table 2). One of these two 3AS-Rec II recombinants was advanced to the F4 generation for FHB evaluation. Three recombinants heterozygous at different marker loci in the QTL region were identified in the F3 generation of 3AS07-53-16. Forty F4 individuals derived from each of these three heterozygous recombinants were genotyped using the 11 markers within the QTL region to recover homozygous recombinants in the QTL region. Eleven individuals were identified as homozygous recombinants from each of the two F4 populations, which were designated 3AS-Rec III and 3AS-Rec IV, respectively (Table 2). Meiotic recombination events were recovered between Xwgc716 and Xwgc1188 and between Xwgc1143 and Xwgc1204 in 3AS-Rec III (F4). A similar recombination event between Xwgc1143 and Xwgc1204 was also recovered in 3AS-Rec IV (F4) (Table 2). Xwgc716 co-segregated with Xwgc1188, and Xwgc1143 co-segregated with Xwgc1204 in the F2 population (Fig. 3). Nine homozygous recombinants were recovered from the third 3AS07-53-16-derived F4 population and designated 3AS-Rec V (Table 2). In total, five types of homozygous recombinants with shortened T. dicoccoides 3AS fragments were generated in the QTL region (Table 2).
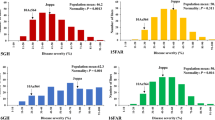
The five types of homozygous recombinants at different generations were evaluated for FHB resistance in three greenhouse seasons (Table 3). Nine F3 individuals of 3AS-Rec I showed FHB severity ranging from 12.0 to 76.9 % in the greenhouse season of Fall 2013. A mean FHB severity of 44.8 % was observed in these nine 3AS-Rec I plants, which was not significantly different from RICL#10, but significantly lower than LDN. Similar FHB severity as 3AS-Rec I was obtained for 3AS-Rec III, IV, and V in Fall 2013. 3AS-Rec II, however, exhibited FHB severity similar to LDN, which was significantly higher than RICL#10 in the greenhouse season of Fall 2013 (Table 3).
The five recombinants selected for FHB evaluation in the first greenhouse season (Fall 2013) were all homozygous at the 11 marker loci within the QTL region. Thus, only one plant of 3AS-Rec I (F3) and one plant from each of 3AS-Rec III, IV, and V (F4) were randomly selected for immature embryo culture to accelerate generation. Ten immature embryo-regenerated F4 plants of 3AS-Rec I had FHB severity similar to LDN in Spring 2014, which was significantly higher than RICL#10. 3AS-Rec II consistently showed a high FHB severity as LDN, while 3AS-Rec III and IV showed FHB severity similar to RICL#10 at the F5 generation in Spring 2014. F5 progeny were not obtained from embryo culture for FHB evaluation in 3AS-Rec V (Table 3).
Seed set on the plants with an FHB severity less than 30 % in 3AS-Rec I (F3), 3AS-Rec III (F4), and 3AS-Rec V (F4) were selected for further FHB evaluation based on seed availability in Summer 2014. All three recombinants at the advanced generations (i.e., F4 for 3AS-Rec I and F5 for 3AS-Rec III and V) exhibited FHB severity similar to RICL#10. FHB severity of RICL#10, however, was not significantly different from LDN in this greenhouse season. FHB severity of the three recombinants and LDN were lower than other two greenhouse seasons (Table 3).
The genotypes of the three critical RICLs (RICL#10, RICL#15, and RICL#49) and five recombinants homozygous at the marker loci within the QTL region are graphically represented in Fig. 4. Both RICL#10 and RICL#49 were resistant to FHB as their resistance parent LDN(DIC-3A), while RICL#15 was susceptible as the susceptible parent LDN (Chen et al. 2007). RICL#10, LDN, and the five recombinants were evaluated for FHB resistance in 2–3 greenhouse seasons (Table 3). The genotypes with FHB severity significantly lower than LDN were considered resistant and designated “R”, whereas those with FHB severity comparable to LDN were considered susceptible and designated “S” (Fig. 4; Table 3; Chen et al. 2007). Qfhs.ndsu-3AS was delimited to the chromosomal interval of 5.2 cM flanked by Xwgc501 and Xwgc510 based on the genotypes of the RICLs and recombinants at the marker loci within the region and their response to FHB (Fig. 4).
Comparative analysis
The chromosomal interval proximal to Xgwm2 harbors three groups of co-segregating markers within the QTL region (Fig. 1). The co-segregating markers, including Xwgc501, Xwgc716, Xwgc1143, Xwgc1188, and Xwgc1204, were 0.6 cM away from Xgwm2. These five EST-derived STS markers identified a collinear region of ~154 kb on the short arm of rice chromosome 1 (R1S) and ~100 kb on the short arm of Brachypodium chromosome 2 (B2S) (Fig. 1). The other two co-segregating markers, Xwgc774 and Xwgc1226, were 1.2 cM proximal to the five co-segregating markers. These two markers identified a collinear region of ~78 and ~50 kb on rice R1S and Brachypodium B2S, respectively. The orientation of the chromosomal interval harboring these two groups of co-segregating markers on 3AS is inverted relative to the corresponding collinear regions on R1S and B2S (Fig. 1). The other three co-segregating markers proximal to Xwgc774/Xwgc1226, Xwgc1296, Xwgc1301, and Xwgc510, detected a collinear region of ~339 kb on R1S and ~330 kb on B2S, both of which are located proximally to the rice and Brachypodium collinear regions identified by the five co-segregating markers (Fig. 1). The chromosomal segment distal to the QTL region, defined by Xwgc1229 and Xgwc500, showed good collinearity with a genomic region of ~340 kb on R1S and ~300 kb on B2S. A group of ten co-segregating markers distal to Xwgc1229 detected two collinear regions on R1S and B2S, respectively. Two of the ten markers Xwgc901 and Xwgc1263, identified a collinear region of ~13 kb on R1S and ~10 kb on B2S. This collinear region on R1S was about 2658 kb away from the terminal end of this chromosome arm. The other eight co-segregating markers detected a collinear region of ~141 kb near the terminal end of R1S. However, the collinear region identified by these eight markers on B2S (~90 kb) was located 2600 kb away from the terminal end of the chromosomal arm and ~150 kb proximal to the collinear region detected by Xwgc901 and Xwgc1263 on B2S. The collinear regions on R1S and B2S were in inverted orientations (Fig. 1). Eleven additional new markers mapped to the chromosomal region farther distal to the QTL region on 3AS. Some of them (Xwgc503/Xwgc811/Xwgc802 and Xwgc803/Xwgc819) co-segregated in the RICL mapping population. These two groups of co-segregating markers and three other markers (Xwgc810, Xwgc826, and Xwgc796) proximal to Xwgc503/Xwgc811/Xwgc802 detected a collinear region of 244 kb on R1S, which was about 910 kb away from the terminal end of the chromosomal arm. The collinear region identified by these eight markers was about 320 kb long on B2S. High collinearity was detected within these genomic regions, in terms of order and chromosomal locations of the marker loci in the respective genomes (Fig. 1). Apart from the inversion of marker loci, deletions of marker loci on the Brachypodium chromosome were also observed. For example, the TC sequences of Xwgc501 and Xwgc1301 have orthologous sequences on rice chromosome R1, but not on Brachypodium chromosome B2 (Fig. 1).
To estimate the physical size of the QTL region, comparative analysis was performed between the refined QTL region and the released ‘Chinese Spring’ (CS) chromosome 3B pseudomolecules (Choulet et al. 2014). Contigs were identified from the CS chromosome 3AS-specific survey sequence database (http://urgi.versailles.inra.fr/blast/blast.php) using the wheat EST singletons, TCs, or contig sequences corresponding to the newly developed markers in the refined QTL region (Sehgal et al. 2012). Two identical T. durum microsatellite sequence clones, Dwm213 and Dwm2, were identified using the 40-bp primer sequence of Xgwm2, an SSR marker closely linked to the Qfhs.ndsu-3AS QTL peak (Chen et al. 2007). Contigs corresponding to the microsatellite sequence of Xgwm2 were identified in the 3AS-specific survey sequence database (https://urgi.versailles.inra.fr/blast/blast.php). The identified 3AS contig sequences were used to search against CS chromosome 3B genomic sequences and hit the pseudomolecule HG670306.1 on 3BS. Eight of the ten markers encompassing Qfhs.ndsu-3AS, including Xgwm2, Xwc1188, Xwgc716, Xwgc1143, Xwgc501, Xwgc1226, Xwgc510, and Xwgc1301, were identified to have anchor points in the 3BS pseudomolecule. Those markers, except Xwgc501 and Xwgc1226, maintained the same order in the QTL region on 3AS and its collinear counterpart on 3BS. The collinear counterparts of Xwgc501 and Xwgc1226 reside toward the distal region of 3BS relative to those of the markers Xwgc1143 and Xwgc1188, and overlap with the 3BS collinear region identified by Xgwm2 and Xwgc716 (Fig. 3). Xwgc501 and Xwgc1226 identified a ~3.4 Mb collinear region (75.7–79.1 Mb) in the pseudomolecule. A ~38.8 Mb genomic region in the pseudomolecule was found to be collinear with the chromosome interval flanked by Xwgc1226 and Xwgc510 (Fig. 3). Two other markers in the QTL region (Xwgc1204 and Xwgc1296) did not detect significant collinearity in the 3BS pseudomolecule.
Discussion
The T. dicoccoides-derived FHB resistance QTL Qfhs.ndsu-3AS previously mapped to the genomic region (11.5 cM) flanked by two target region amplification polymorphism (TRAP) markers Xfcp401 and Xcfp397.2 on 3AS by Chen et al. (2007). Only three markers, including Xbcd1532, Xbarc45, and Xgwm2, mapped within the QTL region (Otto et al. 2002; Chen et al. 2007). In the present study, the Qfhs.ndsu-3AS QTL region defined by Xfcp401 and Xcfp397.2 was saturated by 22 newly developed wheat EST-derived markers, which stretched the genetic distance of this region to 13.6 cM (Fig. 1). Evidently, the micro-collinearity of the wheat genome with rice and Brachypodium provides an effective approach for marker development in saturation and fine mapping of specific wheat genomic regions.
Interval mapping against the saturated genetic map delimited Qfhs.ndsu-3AS to a chromosomal segment of 6.2 cM flanked by Xbarc45 and Xwgc510, which is about half the size of the QTL region defined in the previous study (Fig. 1; Chen et al. 2007). Nine of the thirteen markers mapped to the QTL region are newly developed in this study. Xgwm2, an SSR marker closely linked to the QTL peak in previous studies (Otto et al. 2002; Chen et al. 2007), still resides within the redefined QTL region. All four intervals defined by the markers within the QTL region explained a predominant phenotypic variation of FHB resistance, suggesting these four intervals encompass Qfhs.ndsu-3AS (Fig. 2).
Fine mapping resolved some of the co-segregating markers and stretched map distance between the markers within the QTL region (Figs. 1, 3). The F2 population employed in this study segregated only at loci within the genomic region harboring the QTL, making it ideal for fine mapping of the QTL. Five meiotic recombinants containing shortened T. dicoccoides chromosomal segments within the QTL region defined by Xbarc45 and Xwgc510 were recovered in the F2 and advanced generations of the cross (Fig. 4; Table 2). The recombinant 3AS-Rec I recovered in the F2 generation exhibited a higher FHB severity than the FHB-resistant parent RICL#10 in the F3 generation, but the difference was not statistically significant (Fall 2013). In addition, a significantly higher FHB severity than RICL#10 was observed with 10 randomly selected F4 individuals (Spring 2014). However, the 20 F4 individuals derived from the F3 plants selected with FHB severity less than 30 % showed a resistance level similar to RICL#10 (Summer 2014) (Table 3). Apparently, 3AS-Rec I might be still segregating at Qfhs.ndsu-3AS in the F2–4 generations even though the markers flanking the QTL were under homozygous conditions. Some of the selected 20 F4 individuals, if not all, were likely homozygous at Qfhs.ndsu-3AS as well as the marker loci within the QTL region.
The recombinants 3AS-Rec III, IV, and V were all derived from the same F2 individual (3AS07-53-16) that was heterozygous at six of the 11 marker loci in the QTL region (Table 2). These three recombinants were recovered in the F4 generation. They consistently exhibited a resistance level similar to the FHB-resistant parent RICL#10 across the three seasons (Table 3). Most likely, they were homozygous at Qfhs.ndsu-3AS in addition to the marker loci in the region. The recombinant 3AS-Rec II showed a high FHB severity as the susceptible parent LDN in both seasons, suggesting absence of Qfhs.ndsu-3AS in 3AS-Rec II (Table 3).
The four FHB-resistant recombinants (3AS-Rec I, III, IV, and V) and two FHB-resistant RICLs (#10 and #49) all contained the T. dicoccoides allele at the Xwgc1226 locus, whereas the susceptible recombinant 3AS-Rec II contained the LDN allele at this marker locus. In addition, the four FHB-resistant recombinants had the LDN alleles at the two marker loci (Xwgc501 and Xwgc510) flanking the Xwgc1226 locus, whereas 3AS-Rec II had the T. dicoccoides allele at the Xwgc501 locus. Such a genotype at the Xwgc1226 locus was not observed at other marker loci within the QTL region in the RICLs and recombinants, suggesting Qfhs.ndsu-3AS resides within the chromosomal interval defined by Xwgc501 and Xwgc510 (Fig. 4).
Comparative analysis suggested that the collinear counterparts of Xwgc501 and Xwgc1226 were 3.4 Mb apart on 3BS; and the collinear counterpart of Xwgc510 was 38.8 Mb away from that of Xwgc1226 on 3BS (Fig. 3). Thus, the collinearity-based estimate for the physical size of the QTL region defined by Xwgc501 and Xwgc510 could be around 42 Mb on 3AS. This estimate, however, could fluctuate because of the complex collinearity of Xwgc501 and Xwgc1226 with their counterparts on 3BS in terms of their order relative to other markers in the region (Fig. 3). In addition, this estimate suggests a much higher recombination frequency in the distal segment flanked by Xwgc1226 and Xwgc501 (Mb/cM = 3.4/1.6 = 2.13) than the proximal segment flanked by Xwgc1226 and Xwgc510 (Mb/cM = 38.8/3.6 = 10.78) within this newly defined QTL region. Meiotic recombination hot spots generally cluster within the gene-rich regions (Faris et al. 2000). Thus, Qfhs.ndsu-3AS likely resides closer to the Xwgc501 locus than to Xwgc510 based on the distribution of recombination within the QTL region (Fig. 4). Also, this is consistent with the interval mapping results of the QTL, i.e., a higher LOD score with the Xwgc501-Xwgc1226 interval than the Xwgc1226-Xwgc510 interval. All these information will be useful in further characterization of this genomic region and cloning of Qfhs.ndsu-3AS. The four FHB-resistant recombinants (3AS-Rec I, III, IV, and V) share a shortened T. dicoccoides chromosomal interval that contains the resistance QTL Qfhs.ndsu-3AS. This reduces potential T. dicoccoides 3AS-associated deleterious effects on agronomic traits. They should be breeding-friendly and can be utilized directly in durum wheat breeding for variety development.
Collinearity among the genomes of cereal crops and their models has been extensively investigated (Moore et al. 1995). Wheat homoeologous group 3 chromosomes are collinear to rice chromosome R1 and Brachypodium chromosome B2 (Moore et al. 1995; the International Brachypodium Initiative 2010; Sehgal et al. 2012; Luo et al. 2013). In this study, almost all STS/SSR marker sequences mapped on wheat 3AS were found to have orthologous counterparts on both R1 and B2 (Fig. 1). R1 had a higher collinearity with B2 than with wheat group 3 chromosomes in the distal regions of the short arms in terms of the order of the marker loci (Fig. 1). However, the similarity between the genomic sequences of wheat and Brachypodium was found to be higher than that between wheat and rice in the collinear regions according to the BLAST search results (data not shown). Thus, Brachypodium is more closely related to wheat than rice based on DNA sequence similarities in the genomic regions on the short arms of these three collinear chromosomes. These results are consistent with the observations reported in the previous studies (Catalan and Olmstead 2000; Draper et al. 2001; Foote et al. 2004; the International Brachypodium Initiative 2010; Zhang et al. 2013). However, higher collinearity between wheat and rice than between wheat and Brachypodium has been observed in specific genomic regions (Faris et al. 2008). To date, many genes in wheat have been well characterized through comparative analysis with rice and Brachypodium genomic sequences (Liu et al. 2003, 2006; Bossolini et al. 2007; Faris et al. 2008; Somyong et al. 2011; Zhang et al. 2013). Comparative analysis of the Qfhs.ndsu-3AS QTL and nearby regions on 3AS tremendously enhanced genetic as well as physical mapping of the region in this study. The micro-collinearity revealed in this study will facilitate further understanding of this genomic region.
Conclusions
The genomic region of the wild emmer wheat-derived FHB resistance QTL Qfhs.ndsu-3AS was saturated by 42 newly developed wheat EST-based STS and SSR markers. Qfhs.ndsu-3AS mapped to a shortened wild emmer chromosomal interval in a higher resolution and was redefined by newly developed markers. The QTL region had a complex collinearity with rice and Brachypodium. The micro-collinearity revealed in this study will enhance further understanding and characterization of the QTL and nearby genomic regions on 3AS. The molecular markers defining Qfhs.ndsu-3AS and FHB-resistant recombinants containing the QTL on the reduced wild emmer chromosomal segments will facilitate utilization of this resistance QTL in durum wheat breeding.
Author contribution statement
XC and XZ conceived and designed the experiments in this study. XZ performed this study and drafted the manuscript. XC made major revision of the initial draft. SZ provided inoculum for FHB disease evaluation. SC helped in marker genotyping. YQG participated in Brachypodium sequence analysis for comparative mapping. SK provided part of the phenotyping data for QTL analysis. EE was co-PI of the grant that financially supported this research project. All authors reviewed, edited, and approved the manuscript for publication.
References
Bossolini E, Wicker T, Knobel PA, Keller B (2007) Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J 49:704–717
Buerstmayr H, Stierschneider M, Steiner B, Lemmens M, Griesser M, Nevo E, Fahima T (2003) Variation for resistance to head blight caused by Fusarium graminearum in wild emmer (Triticum dicoccoides) originating from Israel. Euphytica 130:17–23
Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128:1–26
Buerstmayr M, Huber K, Heckmann J, Steiner B, Nelson JC, Buerstmayr H (2012) Mapping of QTL for Fusarium head blight resistance and morphological and development traits in three backcross populations derived from Triticum dicoccum × Triticum durum. Theor Appl Genet 125:1751–1765
Cai X (1993) Production and cytogenetics of hybrid and amphiploid between Triticum tauschiiand Secale cereale. In: Li ZS, Xin ZY (eds) Proceedings of the Eighth International Wheat Genetics Symposium, Beijing, pp 361–363
Cai X, Chen PD, Xu SS, Oliver RE, Chen X (2005) Utilization of alien genes to enhance Fusarium head blight resistance in wheat-A review. Euphytica 142:309–318
Catalan P, Olmstead RG (2000) Phylogenetic reconstruction of the genus Brachypodium P.Beauv. (Poaceae) from combined sequences of chloroplast ndhF gene and nuclear ITS. Plant Syst Evol 220:1–19
Chen X, Soria MA, Yan G, Sun J, Dubcovsky J (2003) Development of sequence tagged site and cleaved amplified polymorphic sequence markers for wheat stripe rust resistance gene Yr5. Crop Sci 43:2058–2064
Chen X, Faris JD, Hu J, Stack RW, Adhikari T, Elias EM, Kianian SF, Cai X (2007) Saturation and comparative mapping of a major Fusarium head blight resistance QTL in tetraploid wheat. Mol Breeding 19:113–124
Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J et al (2014) Structural and functional partitioning of bread wheat chromosome 3B. Science 345:1249721
Chu C, Niu Z, Zhong S, Chao S, Friesen TL, Halley S, Elias EM, Dong Y, Faris JD, Xu SS (2011) Identification and molecular mapping of two QTLs with major effects for resistance to Fusarium head blight in wheat. Theor Appl Genet 123:1107–1119
Draper J, Mur LAJ, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge APM (2001) Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol 127:1539–1555
Faris JD, Haen KM, Gill BS (2000) Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154:823–835
Faris JD, Zhang Z, Fellers JP, Gill BS (2008) Micro-colinearity between rice, Brachypodium, and Triticum monococcum at the wheat domestication locus Q. Funct Integr Genom 8:149–164
Foote TN, Griffiths S, Allouis S, Moore G (2004) Construction and analysis of a BAC library in the grass Brachypodium sylvaticum: its use as a tool to bridge the gap between rice and wheat in elucidating gene content. Funct Integr Genom 4:26–33
Ghavami F, Elias EM, Mamidi S, Ansari O, Sargolzaei M, Adhikari T, Mergoum M, Kianian SF (2011) Mixed model association mapping for Fusarium head blight resistance in Tunisian-derived durum wheat populations. G3 (Genes, Genomes, Genetics) 1:209–218
Jiang C, Zeng Z (1997) Mapping quantitative trait loci with dominant and missing markers in various crosses from two inbred lines. Genetica 101:47–58
Joppa LR (1993) Chromosome engineering in tetraploid wheat. Crop Sci 33:908–913
Kumar D, Gupta N, Ahlawat SPS, Satyanarayana R, Sunder S, Gupta SC (2006) Single strand confirmation polymorphism (SSCP) detection in exon I of the α-lactalbumin gene of Indian Jamunapari milk goats (Capra hircus). Genet Mol Biol 29:287–289
Kumar S, Stack RW, Friesen TL, Faris JD (2007) Identification of a novel Fusarium head blight resistance quantitative trait locus on chromosome 7A in tetraploid wheat. Phytopathology 97:592–597
Kumar S, Mohan A, Balyan HS, Gupta PK (2009) Orthology between genomes of Brachypodium, wheat and rice. BMC Res Notes 2:93
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Liu S, Anderson JA (2003) Targeted molecular mapping of a major wheat QTL for Fusarium head blight resistance using wheat ESTs and synteny with rice. Genome 46:817–823
Liu S, Zhang XL, Pumphrey MO, Stack RW, Gill BS, Anderson JA (2006) Complex microcolinearity among region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct Integr Genom 6:83–89
Liu S, Pumphrey MO, Gill BS, Trick HN, Zhang J, Dolezel J, Chalhoub B, Anderson JA (2008) Toward positional cloning of Fhb1, a major QTL for Fusarium head blight resistance in wheat. Cereal Res Commun 36:195–201
Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y et al (2013) A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci 110:7940–7945
McMullen MP, Jones R, Gallenberg D (1997) Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis 81:1340–1348
Mohamed AM, Bastola DR, Morlock GP, Cooksey RC, Hinrichs SH (2004) Temperature-mediated heteroduplex analysis for detection of pncA mutations associated with pyrazinamide resistance and differentiation between Mycobacterium tuberculosis and Mycobacterium bovis by denaturing high- performance liquid chromatography. J Clinical Microbiology 42:1016–1023
Moore G, Devols KM, Wang Z, Gale MD (1995) Cereal genome evolution: grasses line up and form a circle. Curr Biol 5:737–739
Oliver RE, Stack RW, Miller JD, Cai X (2007) Reaction of wild emmer wheat accessions to Fusarium head blight. Crop Sci 47:893–899
Oliver RE, Cai X, Friesen TL, Halley S, Stack RW, Xu SS (2008) Evaluation of Fusarium head blight resistance in tetraploid wheat (Triticum turgidum L.). Crop Sci 48:213–222
Otto CD, Kianian SF, Elias EM, Stack RW, Joppa LR (2002) Genetic dissection of a major Fusarium head blight QTL in tetraploid wheat. Plant Mol Biol 48:625–632
Ruan Y, Comeau A, Langevin F, Hucl P, Clarke JM, Brule-Babel A, Pozniak CJ (2012) Identification of novel QTL for resistance to Fusarium head blight in a tetroploid wheat population. Genome 55:853–864
Sehgal SK, Li W, Rabinowicz PD, Chan A, Simkova H, Dolezel J, Gill BS (2012) Chromosome arm-specific BAC end sequences permit comparative analysis of homoeologous chromosomes and genomes of polyploid wheat. BMC Plant Biol 12:64
Somers DJ, Fedak G, Clarke J, Cao W (2006) Mapping of FHB resistance QTLs in tetraploid wheat. Genome 49:1586–1593
Somyong S, Munkvold JD, Tanaka J, Benscher D, Sorrells ME (2011) Comparative genetic analysis of a wheat seed dormancy QTL with rice and Brachypodium identifies candidate genes for ABA perception and calcium signaling. Funct Integr Genom 11:479–490
Stack RW, Faris JD (2006) Identification of a Fusarium head blight resistance QTL on chromosome 6B in tetraploid wheat. In: Abstracts of Plant and Animal Genome XIV Conference. San Diego, p 285
Stack RW, McMullen MP (1985) Head blighting potential of Fusarium species associated with spring wheat heads. Can J Plant Pathol 7:79–82
Stack RW, Elias EM, Mitchell FJ, Miller JD, Joppa LR (2002) Fusarium head blight reaction of Langdon durum- Triticum dicoccoides chromosome substitution lines. Crop Sci 42:637–642
Talas F, Longin F, Miedaner T (2011) Sources of resistance to Fusarium head blight within Syrian durum wheat landraces. Plant Breed 130:398–400
The International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
Xu XM, Nicholson P, Thomsett MA, Simpson D, Cooke BM, Doohan FM, Brennan J, Monaghan S, Moretti A, Mule G, Hornok L, Beki E, Tatnell J, Ritieni A, Edwards SG (2008) Relationship between the fungal complex causing Fusarium head blight of wheat and environmental conditions. Phytopathology 98:69–78
Zhang X, Han D, Zeng Q, Duan Y, Yuan F, Shi J, Wang Q, Wu J, Huang L, Kang Z (2013) Fine mapping of wheat stripe rust resistance gene Yr26 based on collinearity of wheat with Brachypodium distachyon and Rice. PLoS One 8:e57885. doi:10.1371/journal.pone.0057885
Zhang Q, Axtman JE, Faris JD, Chao S, Zhang Z, Friesen TL, Zhong S, Cai X, Elias ME, Xu SS (2014) Identification and molecular mapping of quantitative trait loci for Fusarium head blight resistance in emmer and durum wheat using a single nucleotide polymorphism-based linkage map. Mol Breed 34:1677–1687
Zhu X, Zhong S, Xu SS, Cai X (2012) Fusarium head blight reactions of Langdon durum D-genome disomic substitution lines. In: Canty S, Clark A, Anderson-Scully A, Van Sanford D (eds) Proceedings of the 2012 National Fusarium Head Blight Forum. Orlando, USA, p 110
Acknowledgments
The authors would like to acknowledge all members in Cai’s lab and Dr. Chenggen Chu for their help in this research. Also, we thank Drs. Lili Qi and Rebekah Oliver for their critical review of the manuscript. This material is based upon work supported by the US Department of Agriculture, under Agreement No. 59-0790-8-068. This is a cooperative project with the US Wheat and Barley Scab Initiative. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the US Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
This study complies with the current laws of USA.
Additional information
Communicated by A. Zhang.
Rights and permissions
About this article
Cite this article
Zhu, X., Zhong, S., Chao, S. et al. Toward a better understanding of the genomic region harboring Fusarium head blight resistance QTL Qfhs.ndsu-3AS in durum wheat. Theor Appl Genet 129, 31–43 (2016). https://doi.org/10.1007/s00122-015-2606-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2606-x