Abstract
Key message
Provide evidence that the Brassica B genome chromosome B3 carries blackleg resistance gene, and also the B genome chromosomes were inherited several generations along with B. napus chromosomes.
Abstract
Blackleg disease caused by fungus Leptosphaeria maculans causes significant yield losses in Brassica napus. Brassica carinata possesses excellent resistance to this disease. To introgress blackleg resistance, crosses between B. napus cv. Westar and B. carinata were done. The interspecific-hybrids were backcrossed twice to Westar and self-pollinated three times to produce BC2S3 families. Doubled haploid lines (DH1) were produced from one blackleg resistant family. SSR markers were used to study the association between B genome chromosome(s) and blackleg resistance. The entire B3 chromosome of B. carinata was associated with blackleg resistance in DH1. A second DH population (DH2) was produced from F1s of resistant DH1 lines crossed to blackleg susceptible B. napus cv. Polo where resistance was found to be associated with SSR markers from the middle to bottom of the B3 and top of the B8 chromosomes. The results demonstrated that the B3 chromosome carried gene(s) for blackleg resistance. Genomic in situ hybridization (GISH) and GISH-like analysis of the DH2 lines revealed that susceptible lines, in addition to B. napus chromosomes, possessed one pair of B genome chromosomes (2n = 40), while resistant lines had either one (2n = 40) or two pairs (2n = 42) of B chromosomes. The molecular and GISH data suggested that the B chromosome in the susceptible lines was B7, while it was difficult to confirm the identity of the B chromosomes in the resistant lines. Also, B chromosomes were found to be inherited over several generations along with B. napus chromosomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oilseed rape or canola (Brassica napus) contributes about 15 % to the world’s total supply of vegetable oils (Rahman et al. 2013). It is cultivated extensively in North America, Europe, China and Australia. In 2012, Canada was the top producing country (15.4 MMT), followed by China (14.0 MMT), India (6.8 MMT), France (5.5 MMT) and Germany (4.8 MMT) (FAOSTAT). Revenue to the major producing countries from canola is projected to increase even further in the coming years due to soaring demands for canola on the world market. However, B. napus is affected by many diseases among which blackleg or stem canker caused by the fungus Leptosphaeria maculans Desm. (Ces. & de Not) is of major concern mainly because of the significant yield losses associated with the disease worldwide (for reviews, see Gugel and Petrie 1992; West et al. 2001; Howlett 2004; Fitt et al. 2006). Yield loses of up to 50 % have been reported in infected B. napus fields (Gugel and Petrie 1992) while total crop failure have also been reported in areas where highly susceptible cultivars are grown or when there is breakdown of a major resistance gene in a susceptible background (Li et al. 2003; Rouxel et al. 2003; Sprague et al. 2006). Breeding for resistance to blackleg offers by far the least expensive, the most environmentally friendly and the best practical means of controlling the disease (Rimmer and Buchwaldt 1995; for reviews, see Fernando et al. 2007; Kutcher et al. 2011).
Genetic mapping studies on the resistance of B. napus to L. maculans have identified six endogenous race-specific blackleg resistance (Rlm) genes in this crop. The loci controlling these genes have been mapped onto the A7 (≈N7) (Rlm1, Rlm3, Rlm4, Rlm7 and Rlm9) and A10 (≈N10) (Rlm2) linkage groups of the A-genome (Ansan-Melayah et al. 1998; Balesdent et al. 2001, 2002; Delourme et al. 2004, 2006; Rimmer 2006). Other blackleg resistance genes identified in B. napus cultivars and thought to be allelic forms of the genes located on the A7 linkage group or members of the previously mapped gene cluster include LEM1 in B. napus cv. ‘Major’ (Ferreira et al. 1995), LmFr1 (Dion et al. 1995) and CLmR1 (Mayerhofer et al. 2005) in cv. ‘Cresor’, LmR1 in cv. ‘Shiralee’ (Mayerhofer et al. 1997), cRLMm in cv. ‘Maluka’ (Rimmer et al. 1999) and cRLMrb/aRLMrb in line ‘RB87-62’ (Rimmer et al. 1999). Four other disease resistant genes LepR1 on A2 (≈N2), LepR2/LepR3 on A10 and LepR4 on A6 (≈N6) thought to be different from any of the aforementioned genes (Yu et al. 2005, 2008, 2013) as well as Rlm8 and Rlm11 which have not been genetically mapped (Balesdent et al. 2002, 2013) have been introgressed into B. napus cultivars from B. rapa. Thus, to date a total of 12 race-specific resistance genes to L. maculans that originate from the A genome of B. napus and B. rapa have been identified.
Canadian canola breeders, including the University of Alberta, have been utilizing LmR1 and cRLMm resistance present on the A7 linkage group of the Australian canola (B. napus) cvs. ‘Shiralee’ and ‘Maluka’, respectively, to develop L. maculans resistant canola cultivars for the Prairie Provinces where pathogenicity group 2 (PG2) is most prevalent. The two resistant genes were initially thought to be Rlm4 (see review by Delourme et al. 2006; Hayward et al. 2012), but recent data indicate they could be Rlm3 since the two are thought to be allelic versions of the same gene and are not present together in the same cultivar. However, more virulent pathotypes of the fungus, e.g. pathogenicity groups PG3, PG4 and PGT, have emerged in the Canadian Praires rendering the resistance ineffective (Fernando and Chen 2003; Kutcher et al. 2007). There is, therefore, the threat of single gene resistance breakdown in B. napus cultivars in Canada as was reported in Australia (Li et al. 2003; Sprague et al. 2006) and France (Brun et al. 2000; Rouxel et al. 2003).
Brassica species that contain the B genome, namely B. nigra (BB, 2n = 16), B. carinata (BBCC, 2n = 34) and B. juncea (AABB, 2n = 36), are known to carry excellent resistance to blackleg that is effective throughout the life of the plant (Roy 1984; Sjödin and Glimelius 1988; Rimmer and van den Berg 1992). Therefore, to produce B. napus cultivars that are resistant to blackleg disease many workers have attempted to use these B genome containing Brassica species as donors in either sexual crosses (Roy 1978, 1984; Sacristan and Gerdemann 1986; Zhu et al. 1993; Struss et al. 1996; Chèvre et al. 1996, 1997; Plieske et al. 1998; Dixelius 1999; Dixelius and Wahlberg 1999; Navabi et al. 2010, 2011) or somatic-hybridization (Sjodin and Glimelius 1989a, b; Dixelius and Glimelius 1995, Waara and Glimelius 1995; Glimelius 1999).
For the introgression of B genome resistance into B. napus several pre- and post- fertilization barriers need to be overcome. In spite of these challenges, Chèvre et al. (1997) successfully introgressed a B genome resistance gene, Rlm6, from B. juncea into B. napus. It is important to incorporate additional resistance genes from the Brassica B genome not only to provide resistance to specific pathotypes but also for resistance against emerging new forms of the pathogen as well as to be used in combination with quantitative resistance in this crop. For this, knowledge of the location of the B genome resistance genes is important. A recent paper (Brun et al. 2010) showed that one of the best strategies to minimize the devastating effects of blackleg disease on B. napus production is to combine dominant genes and polygenic resistance to prevent breakdown of major genes. Therefore, the introduction of new major resistance genes from B. carinata into B. napus is important for the diversification of resistance genes for the optimal management of blackleg disease in this crop.
In this work, we used different generations from a B. napus × B. carinata cross progenies and simple sequence repeat (SSR) markers to show the association of a B genome chromosome of B. carinata and blackleg disease resistance in the B. napus background. In addition, we studied the behaviour of different B genome chromosomes through genomic in situ hybridization (GISH) and GISH-like techniques.
Materials and methods
Plant materials
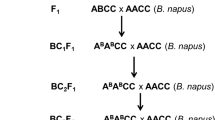
The parental materials used for this interspecific cross were Brassica napus cv. ‘Westar’ and B. carinata accession #98-14513. Westar is a registered cultivar in Canada and is highly susceptible to blackleg while B. carinata line #98-14513 is highly resistant to this disease. Seed of this line was obtained from the germplasm collection of the Canola Breeding and Research Program at the University of Alberta (U of A). Crosses between Westar × B. carinata were done using Westar as female, and the interspecific hybrids were backcrossed twice to Westar and subsequently were self-pollinated three times to produce BC2S3 families (Fig. 1). In each generation selection was made for cotyledon resistance to L. maculans PG2 isolate #UA3356. Thirty BC2S3 families were produced from the B. napus cv. Westar × B. carinata interspecific cross. Of these, four plants from the family #02-17006 which exhibited resistance to L. maculans PG2 isolate #UA3356 at the cotyledon stage were used for the production of DH lines through bulk culture of microspore and the DH population was designated Popl#3 which for simplicity, hereafter will be referred to as DH1 (Fig. 1). The microspore culture technique applied for the production of DH lines was as described by Kebede et al. (2010). To study the inheritance of blackleg resistance and transmission of B genome chromosomes in a different genetic background, we crossed two resistant DH1 lines carrying B genome chromosomes to the oil-rich blackleg susceptible B. napus cv. ‘Polo’ and produced DH lines from the F1 plants of each of these two crosses (Fig. 1). These two DH populations were designated as Polo-Popl#31151 and Polo-Popl#31152, and collectively as Polo-Popl#3, which for simplicity, hereafter will be referred to as DH2 population (Fig. 1).
Interspecific crossing for the introgression of Leptosphaeria maculans resistance from B. carinata into B. napus. The DH1 lines were produced from four cotyledon resistant plants of the BC2S3 family #02-17006 while the DH2 lines were produced from F1 plants obtained by crossing two resistant DH1 lines with Polo
Leptosphaeria maculans culture and inoculum preparation
Leptosphaeria maculans PG2 isolate #UA3356 originally collected from a canola field in Alberta (Canada) and maintained by the Canola Breeding and Research Program at the U of A was used for all inoculation experiments. The isolate was cultured on V8 vegetable juice agar plates containing 40 mg/L Rose Bengal and 100 mg/L streptomycin sulphate (Dhingra and Sinclair 1985). The culture was maintained under 12-h photoperiod at 21 °C in a NuAire™ incubator (Fernbrrok Lane, MN, USA). After 16 days, each plate was flooded with 5 mL sterile distilled water and the culture surface was gently scraped with a sterile metal rod to manually dislodge the pycnidiospores. The pycnidiospore suspensions were then filtered through sterile cheesecloth. The concentration of the spore suspensions were determined using a haemocytometer and adjusted to 1 × 107 pycnidiospores/mL. The prepared inoculum was used immediately or stored at +4 °C for not more than 24 h until use.
Inoculation tests for blackleg resistance
Screening for reaction to L. maculans isolate #UA3356 was carried out by inoculating four to eight seedlings of each DH line and each inoculation experiment was replicated two to four times. To do this, 7-day-old seedlings with fully expanded cotyledons were wounded on each lobe and inoculated with L. maculans spore suspension as described by Sjödin and Glimelius (1988). Disease symptoms were rated 10 to 13 days after inoculation for cotyledon reaction. The plants were left to grow without any further inoculation and evaluated for stem canker at maturity by cutting stems of the inoculated plants. The severity of the disease symptoms at the cotyledon stage was scored based on the 0–9 scale devised by Delwiche (1980), while the severity of the symptoms at the adult stage was determined based on the 0–5 scale proposed by Western Canada Canola/Rapeseed Recommending Committee (WCC/RRC). Based on mean scores for cotyledon infection, the plants were grouped as resistant (score ≤1), intermediate (score 1 to ≤4) and susceptible (score >4); while based on mean scores for internal necrosis at adult stage, the plants were grouped as resistant (score ≤1), intermediate (score 1 to ≤2) and susceptible (score >2). In these tests, B. carinata was used as the resistant control while B. napus cultivars Westar and Polo were used as the susceptible controls.
DNA extraction for molecular marker analysis
DNA was extracted from the parental materials (B. carinata and the B. napus cultivars Westar and Polo) and from the DH lines using the GENELUTE Plant Genomic DNA Kit (Sigma-Aldrich, St. Louis, CA, USA) according to the manufacturer’s instructions. The quality of the DNA was evaluated by spectrometry using the 260/280 nm absorbance ratio method and the DNA concentration estimated at 260 nm with an ND-2000c Nanodrop spectrophotometer (Nanodrop Technologies, Inc., Wilmington, DE, USA). DNA was stored at −80 °C until use.
Identification of SSR marker(s) linked to blackleg disease resistance
Bulk segregant analysis as described by Michelmore et al. (1991) was performed using DNA from the resistant parent (B. carinata), the susceptible parents (B. napus cvs. Westar and Polo), as well as resistant and susceptible bulks of DH1 and by the use of 465 SSR markers. The SSR markers comprised 87, 70 and 273 markers from the Brassica A, B and C genomes, respectively, obtained from Dr. Isobel Parkin [Agriculture and Agri-Food Canada (AAFC), Saskatoon Research Centre, Saskatoon, Canada] through a material transfer agreement. In addition, 35 B genome SSR markers obtained from Ramchiary et al. (2007) were also used in the bulk segregant analysis. Putative polymorphic markers were evaluated on the parents as well as on five resistant and five susceptible DH1 lines. Once the linkage association of a marker and disease resistance had been established, fine-scale mapping was done using all DH1 and DH2 lines. Linkage group designation of the B genome chromosomes was done using B1–B8 notation proposed by Lagercrantz and Lydiate (1996) and accepted by the Multinational Brassica Genome Project (MBGP) Steering Committee Meeting of January 2010, while linkage group assignments of the A and C genome chromosomes were done using A1–A10 and C1–C9, respectively.
PCR amplification
Each PCR reaction was performed in a total volume of 12 μL containing 1 × Taq buffer, 2.0 mM MgCl2, 200 μM dNTPs, 0.4 μM forward primer modified at the 5′-end with an M13 tail, 0.4 μM reverse primer, 0.2 μM fluorescently labelled M13 primer, 1.25 U Taq polymerase (Promega Corp., Madison, WI, USA) and 3 to 10 ng of genomic DNA. Amplifications were carried out in an MJ Research PTC-200 DNA Engine Thermal Cycler (Bio-Rad laboratories, Hercules, CA, USA) with initial denaturation step at 94 °C for 5 min, followed by 35 cycles at 94 °C for 1 min, optimum annealing temperature of primer for 1 min, and at 72 °C for 1.5 min and a final extension at 72 °C for 30 min. An aliquot of the PCR products was separated by capillary electrophoresis on an ABI PRISM 3730 × l DNA analyzer (Applied Biosystems). Amplified PCR products from promising markers were also separated on 6 % denaturing polyacrylamide gels (Bio-Rad laboratories, Hercules, CA, USA). To confirm the reproducibility of the molecular data we repeated PCR, genotyping and gel electrophoresis in about 10 % of the individual samples.
Meiotic observation in the Polo-Popl#3-DH2 lines
For the study of meiotic behaviour in the interspecific hybrids, immature flower buds were collected from two susceptible and seven resistant DH2 lines. Meiotic chromosome preparations were carried out as described by Leflon et al. (2006) and Mason et al. (2010), that is, the buds were fixed in Carnoy’s solution (ethanol: chloroform: acetic acid, 6: 3: 1) for 24 h at room temperature and stored in 50 % ethanol at 4 °C. Anthers were squashed and stained in a drop of 1 % acetocarmine solution. The slides were examined under a phase contrast microscope and chromosome pairing was observed in 10–24 pollen mother cells (PMCs) per plant at metaphase I (MI).
GISH and GISH-like at mitosis with genome-specific probes
Total genomic DNA used for genomic in situ hybridization (GISH) analyses were extracted from young leaves of B. napus using the CTAB method (Rogers and Bendich 1988). The genomic DNA samples were autoclaved to give fragments of 100–300 bp and used as a blocking agent. The bacterial artificial chromosome (BAC) clone BoB014O06 from B. oleracea (Howell et al. 2002) was used as a probe to identify all C genome chromosomes in the interspecific hybrids using GISH-like technique, while B. nigra genomic DNA was used to probe the Brassica B genome of B. carinata in the interspecific hybrids using GISH (Mason et al. 2010) techniques. Bacterial artificial chromosome BoB014O06 clone and the genomic DNA of B. nigra were labelled by random priming with Alexa488-5-dUTP (Molecular probes) and by nick translation with biotin-14-dUTP (Invitrogen), respectively. A 1:50 ratio of B. nigra probe to B. napus blocking DNA was used to prepare the hybridization mixture. Preparation of slides was as described by Leflon et al. (2006) and Mason et al. (2010). The biotinylated probes were immunodetected by Texas Red avidin DCS (Vector Laboratories).
Statistical analysis
Fisher’s exact test was done to investigate independence for resistance and susceptibility among the crosses and Chi-square test for goodness of fit was done to determine segregation pattern for the phenotypic classes.
Results
Evaluation of DH lines for resistance to blackleg PG2 isolate #UA3356
Sixty DH1 lines, obtained from the BC2S3 family #02-17006, were evaluated for resistance to L. maculans PG2 isolate #UA3356. At the cotyledon stage, 48 % of the lines were found to be resistant, 7 % intermediate and 45 % susceptible (Table 1). The resistant parent B. carinata line always had disease score ≤1; therefore, a Chi-square test was done by grouping the DH lines with disease score ≤1 into one class (resistant) and all other DH lines into a separate class (intermediate and susceptible). The distribution of the DH1 lines for cotyledon resistance followed a 1:1 segregation ratio for resistance to susceptibility (χ 2 = 0.067, P = 0.7957). Assessment of adult plant resistance could not be carried out in 8 % of the DH1 lines because the plants died before maturity while among the remaining DH1 lines 33 % were found to be resistant, 20 % intermediate and 47 % susceptible (Table 1). Thirty-one per cent of the cotyledon resistant DH1 lines became intermediate resistant at the adult stage while 33 % of the cotyledon intermediate resistant lines became susceptible and 33 % became resistant. This indicates that in spite of the specific resistance gene the fungus was able to continue growth in some plants. On the other hand, almost all (96 %) of the cotyledon susceptible DH1 lines remained susceptible at the adult stage while about 4 % of the susceptible DH1 lines became intermediate resistant at the adult stage).
In the case of DH2, 8 DH lines of Polo-Popl#31151 and 44 DH lines of Polo-Popl#31152 were evaluated for resistance to L. maculans PG2 isolate #UA3356. Cotyledon stage phenotypic evaluation of DH2 lines from the Polo-Popl#31151 and Polo-Popl#31152 showed that a total of 48 % of the lines were resistant, 6 % intermediate and 46 % susceptible to the isolate (Table 1). At the adult stage, 40 and 28 % of the cotyledon resistant DH2 lines became susceptible and intermediate resistant, respectively, while 32 % of the lines were found to remain resistant. In addition, all cotyledon intermediate DH2 lines became susceptible at the adult stage. Thus, the fungus was able to grow in some cotyledon resistant and intermediate resistant DH2 lines. Fisher’s exact test showed that segregation pattern for cotyledon or adult resistance was not different in the DH2 lines derived from the two crosses. As a result, Chi-square test for fit to 1:1 segregation for resistant to susceptible phenotype was done on pooled data of the DH2 lines of the two crosses. The results fitted well with 1:1 segregation for cotyledon resistance (χ 2 = 0.077, P = 0.781) but deviated significantly for adult resistance (χ 2 = 26.741, P ≤ 0.0001) (Table 1). Also, pooled data of DH1 and DH2 also fitted well to a 1:1 segregation for resistance and susceptibility at the cotyledon stage (χ 2 = 0.143, P = 0.705), but not at the adult stage (χ 2 = 28.271, P ≤ 0.0001) (Table 1). This suggests that cotyledon resistance to L. maculans PG2 isolate #UA3356 in DH1 and DH2 populations is under monogenic control. Also, the fact that none of the cotyledon susceptible plants became resistant at the adult stage suggests that the same gene conferring cotyledon resistance is also involved in the control of adult plant resistance.
Genotypic analysis of SSR markers used in the bulk segregant analysis
Analysis of the PCR amplified products of the 87 A genome SSR markers screened showed that 28 produced polymorphic bands, 54 monomorphic bands while 5 failed to amplify genomic DNA in the resistant (B. carinata) and susceptible (B. napus cvs. Westar and Polo) parents. Among the 273 C genome SSR markers screened, 186 produced polymorphic bands, 61 monomorphic bands while 26 did not amplify genomic DNA in the samples. Comparison of the electrophoretic pattern of PCR-amplified products of the polymorphic SSR markers from the A and C genome (28 + 186 = 214) used in the bulk segregant analysis showed that alleles of about 90 % of the markers detected in the B. napus parents (but not in B. carinata) were also detected in the resistant and susceptible bulks of DH1 and DH2. The remaining 10 % of the polymorphic SSR markers from the A and C genomes detected alleles which were shared by both B. carinata and the B. napus parents, and these alleles were also detected in the DH1 and DH2 bulks. In addition, these markers also amplified alleles from other loci which were unique to B. carinata; however, these alleles were not detected in the DH1 and DH2 bulks. Thus, about 10 % of the polymorphic A and C markers may also have amplified the B genome. Based on the molecular marker data, all ten A (A1–A10) and nine C (C1–C9) genome chromosomes were inherited in the interspecific hybrids.
Of the 105 B genome SSR markers screened, 37 markers detected polymorphism between the parents, 62 gave monomorphic bands, while six failed to amplify genomic DNA in any of the samples. Fourteen out of the 37 polymorphic markers, comprising four, one, two, three and four markers from the B1, B2, B4, B5 and B6 chromosomes, respectively, did not detect any alleles in DH1 and DH2. This indicates that not all B genome chromosomes were inherited in the progenies of the B. napus × B. carinata interspecific cross. However, one marker (SSR #sJ4633) from the terminal region of the B1 chromosome amplified genomic DNA in about 13 and 56 % of both the resistant and susceptible DH1 and DH2 lines, respectively—apparently due to retention of a very small segment of the B1 chromosome in some of the lines (Supplementary Material Table S1). On the other hand, 22 out of the 37 polymorphic SSR markers comprising nine, six and seven from the B3, B7 and B8 chromosomes, respectively, were found to amplify B. carinata alleles in the DH1 and DH2 populations. This suggests that the B3, B7 and B8 chromosomes and a small fragment of the B1 were retained in the interspecific progenies. It should be noted that selection for cotyledon resistance was carried out in each generation during the development of the BC2S3 family #02-17006; this might have contributed to the retention of the extra B chromosome(s) in the DH1 lines.
Linkage association between B genome chromosomes and blackleg resistance
None of the 28 and 186 polymorphic SSR markers from the A and C genomes identified in the bulk segregant analysis was found to be associated with blackleg resistance. Also, the single SSR marker (sJ4633) from the tip of the B1 chromosome as well as the four (sB1822, sJ7046, sB1752 and sJ8469a) B7-specific SSR markers covering about 90 % of the chromosome did not show any linkage association with blackleg resistance in both DH1 and DH2 (Supplementary Table S1 and Figure S2). On the other hand, some of the polymorphic B3 and B8 SSR markers (see below) showed linkage association with cotyledon resistance and hence were selected for mapping of the two DH populations. Sixty DH1 lines were genotyped with one, nine, six and seven markers from the B1, B3, B7 and B8 chromosomes (Supplementary Material Table S1). In the case of the DH2 population, 8 and 46 lines from the two crosses, Polo-Popl#31151 and Polo-Popl#31152, respectively, were genotyped with the same B genome markers used to screen the DH1 population (Supplementary Material Table S1).
In the case of the B3 chromosome, five (sB0860A, sJ34121, sB3751, sA0343a and sB2596) of the nine SSR markers were B-genome-specific and did not amplify Westar or Polo, while the remaining four markers (sJ0954, sJ7166, sB1728 and sB5162) were not specific to the B genome, but produced a clear band in B. carinata and the resistant DH1 lines. Eight of the nine SSR markers from the top (sJ0954, sJ7166 and sB0860A), middle (sB1728, sB3751 and sB5162) and bottom (sA0343a and sB2596) of the B3 chromosome were found to co-segregate with cotyledon resistance in DH1 population (Supplementary Material Figure S1). The eight markers included the telomeric markers (sJ0954 and sB2596) from both ends of the chromosome and span a region of about 222 cM based on the B3 linkage map of Navabi et al. (2010). The remaining marker sJ34121, which is specific to the B genome, amplified genomic DNA in only 22 % of the resistant DH1 lines. Therefore, the SSR marker sJ34121, unlike the other eight B3 chromosome markers, did not co-segregate with cotyledon resistance. This may be due to undetected recombination event(s) that occurred during the generation of interspecific hybrid progenies. Based on these eight SSR markers, it can be inferred that the entire B3 chromosome was present in the resistant DH1 lines and was associated with cotyledon resistance. The co-segregation of three of the eight B3 SSR markers with cotyledon resistance in DH1 population is shown in Fig. 2, while the other five markers are shown in Supplementary Material Figure S1. In the DH2 lines, five (sB1728, sB3751, sB5162, sA0343a and sB2596) of the nine markers from middle to bottom of B3 were found to co-segregate with cotyledon resistance. On the other hand, three markers (sJ0954, sJ7166 and sB0860A) from the top of the B3 chromosome failed to detect the B-genome-specific alleles of B. carinata in the resistant DH2 lines, while the adjacent marker sJ34121 gave monomorphic bands in both resistant and susceptible DH2 (Supplementary Material Figure S1). Thus, based on the marker data, it can be inferred that the top of the B3 chromosome was lost in the resistant DH2 lines, and the presence of the middle to lower segment of this chromosome was enough to confer cotyledon resistance (Supplementary Material Table S1 and Figure S1).
Polyacrylamide gel electrophoretic profiles showing amplified PCR products of resistant and susceptible DH2 lines and the parents B. carinata (C), Polo (P), and Westar (W) with three of the eight B3 chromosome SSR markers (sB1728 (top gel), sB3751 (middle gel), and sB5162 (bottom gel)) that co-segregated with blackleg disease resistance. In all gels the resistant bands (indicated by arrows) are present in the resistant DH lines as well as in B. carinata, but absent in the susceptible DH lines and the susceptible parents, B. napus cv. Westar and Polo. See Supplementary Material Figure S3 for gel images showing co-segregation of the other B3 markers with blackleg resistance in DH1 and DH2 lines
In the case of the B8 chromosome, four (sJ8370a, sJ6846, sJ3302R and sJ03104) of the seven SSR markers from the middle of the chromosome were found to be B-genome-specific. The telomeric marker (sJ7434) from the top of the chromosome was not B-genome-specific but produced a clear band in B. carinata. The two markers from the bottom (sA0499F and the telomeric marker sJ1827a) amplified both B. carinata and B napus giving a similar profile and thus were also not B-genome-specific (Supplementary Material Figure S3). Thus, based on the five B-genome-specific markers (from top to middle) that amplified genomic DNA in the some or all DH1 lines as well as the two markers from the bottom (sA0499F and sJ1827a) that amplified both B. carinata and B. napus, it can be inferred that the whole B8 chromosome (ca. 453 cM, Navabi et al. 2010) was present in both resistant and susceptible lines, and none of these SSR markers were found to be linked with blackleg resistance. Unlike the DH1 lines, cotyledon resistance in the DH2 lines was found to co-segregate with three SSR markers (sJ7434, sJ8370a and sJ6846) from the top of the B8 chromosome. The two remaining B-genome-specific B8 markers (sJ3302R and sJ03104) were present in resistant and susceptible plants and so were not associated with blackleg resistance. Thus, in DH2, the top segment of the B8 chromosome was found to co-segregate with cotyledon resistance while the middle to bottom segment of the chromosome did not co-segregate with the resistance (Supplementary Material Table S1 and Figure S3).
Linkage analysis suggested that SSR markers from the top of the B8 and middle to bottom of the B3 chromosome co-segregated in the DH2 population, and these two chromosome segments jointly co-segregated with cotyledon resistance.
Chromosome behaviour and identification of B chromosomes using GISH and GISH-like technique
Cytogenetic analysis was carried out on two susceptible DH2 lines and seven resistant DH2 lines. Chromosome behaviour at meiosis in pollen mother cells (PMCs) of the DH2 lines showed that the two susceptible lines had 40 chromosomes while four and three of the resistant lines had 40 and 42 chromosomes, respectively (Fig. 4). In PMCs, we observed mostly bivalents (19.5–20.0 per cell in 2n = 40 plants; 20.9 per cell in a 2n = 42 plant), and very few univalent (0.21–0.70 per cell) and multivalent (0.04–0.10 per cell). Investigation of mitotic chromosomes using GISH and GISH-like techniques showed that the 40 chromosomes in the susceptible DH2 lines consisted of two B genome chromosomes (red signals identified by the B. nigra probe fluorescently detected with Texas red avidin), 18 C genome chromosomes (green signals identified by the BoB0104O06 BAC clone fluorescently labelled with Alexa-488) and 20 A genome chromosomes (counter stained blue with DAPI) (Fig. 5a–c; see also Supplementary Material Figure S4). On the other hand, GISH/GISH-like analysis on the seven resistant DH2 lines showed two different patterns: four lines with a pair of B chromosomes (2n = 40) (Fig. 5d–f; see also Supplementary Material Figure S5) and three lines with two pairs of B chromosomes (2n = 42) (Fig. 5g–i; see also Supplementary Material Figure S6). The A and C genome chromosome content in these lines were 20 and 18, respectively, the same as the susceptible lines. Thus, GISH/GISH-like analysis detected one pair of B genome chromosomes in the susceptible DH2 lines, while the resistant DH2 lines possessed either one or two pairs of B chromosomes.
Discussion
In the present study, based on B. napus × B. carinata interspecific hybridization, we provided conclusive evidence that the B genome chromosome B3 of B. carinata carries a blackleg resistance gene. SSR markers spanning the entire B3 chromosome were found to co-segregate with cotyledon resistance in DH1 while five markers covering the middle to bottom segments of the B3 chromosome were found to co-segregate with the resistance in DH2. On the other hand, none of the 360 SSR markers (87 and 273 markers, respectively) from the Brassica A and C genomes used in this study was found to co-segregate with blackleg disease resistance and hence blackleg resistance in our interspecific hybrids progenies is not from endogenous resistance genes in B. napus.
The Brassica B genome is known to carry multiple resistance genes (Zhu et al. 1993). Identification and mapping of these genes has been of interest to researchers and breeders for stable introgression into B. napus. The Rlm genes so far identified to be responsible for blackleg resistance in the B genome include Rlm5 and Rlm6 of B. juncea (Balesdent et al. 2002) and Rlm10 of B. nigra (Chèvre et al. 1996, 1997). Chèvre et al. (1996, 1997) used RAPD markers to demonstrate that cotyledon resistance in B. nigra was located on the B4 chromosome while cotyledon resistance in B. juncea is located on the B8 chromosome of their genetic linkage map. In an RFLP marker study of offspring from asymmetric hybrids between B. napus and the three B genome species B. nigra, B. juncea and B. carinata, Dixelius and Wahlberg (1999) identified four markers co-segregating with adult plant resistance and these were mapped to three different linkage groups of the B genome. A dominant resistant gene (LMJR1) and a recessive resistance gene (LMJR2) associated with blackleg resistance was mapped to the B7 and B3 chromosomes of B. juncea (Christianson et al. 2006) which corresponds to linkage group G3 and G8 chromosomes of B. nigra, respectively, based on the consensus linkage map of Lagercrantz and Lydiate (1995) and Axelsson et al. (2000). It is not possible to compare the chromosome location of the resistant genes reported in most of these studies with the present study due to the lack of common markers or linkage map. The F1 plants from our crosses showed no segregation and were all completely resistant to L. maculans isolate #UA3356. Therefore, the gene on the B3 (middle to bottom) segment that we identified to be linked to L. maculans resistance was dominant and hence could be different from the recessive gene on the B3 chromosome identified by Christianson et al. (2006).
Molecular evidence suggests that the three Brassica genomes evolved from a smaller ancestral genome through extensive chromosome duplication and rearrangements (Song et al. 1988; Warwick and Black 1991; Lagercrantz and Lydiate 1996; Lagercrantz 1998; Lysak et al. 2005). Among the three genomes, the B genome apparently diverged greatly from the A and C genomes resulting in little homoeologous pairing between B genome chromosomes and either of the related genomes (Attia and Röbbelen 1986a, b; Attia et al. 1987; Busso et al. 1987; Mason et al. 2010, 2011). Therefore, the transfer of blackleg disease resistant gene(s) from the B genome chromosomes of B. carinata (BBCC, 2n = 34) into B. napus genome (AACC, 2n = 38) is a difficult task. However, Panjabi et al. (2008) based on consensus block arrangements of the Brassica A (A1–A10), B (B1–B8) and C (C1–C9) genomes demonstrated that some degree of homoeology between B genome and A/C genomes exists. Therefore, intergenomic transfer of genes from the B to the A or C genome can also occur. Indeed, Chèvre et al. (1997) reported stable introgression of blackleg resistance genes from the B genome of B. juncea into B. napus. Using RFLP and RAPD markers, Plieske et al. (1998) demonstrated that the B genome resistance genes of B. nigra, B. juncea and B. carinata were introgressed at the same location of the B. napus genome—apparently due to these three species harbouring the same copy of the resistance gene and sufficient colinearity of the genomic region containing the resistance gene between the B genome and B. napus. However, in the present study two cycles of DH production failed to introgress B genome resistance against L. maculans PG2 isolate #UA3356 from B. carinata into the B. napus genome. Conventional breeding techniques such as backcrossing and pedigree breeding may be applied for introgression of the resistance gene into the B. napus genome. These techniques have proved to be effective for introgression of genes from allied species such as Rfo gene from radish into B. napus (Delourme et al. 1991). Also, we are unable to determine if the B genome resistance gene reported in the present study is the same as those identified by Plieske et al. (1998) due to lack of common markers.
Through working with interspecific progenies derived from B. nigra × B. napus, B. rapa × B. carinata and B. oleracea × B. juncea crosses, Struss et al. (1991) found that B genome chromosomes transmit into progeny predominantly through the ovule. In contrast, Chèvre et al. (1991) developed B. napus-B. nigra disomic chromosome addition lines for six of the eight B. nigra chromosomes. Transmission rate of these alien chromosomes in self-pollinated progeny varied from 50 % to 100 %. This suggests that B genome chromosomes in B. napus background can transmit to the progeny through both ovule and pollen, and the rate of transmission can vary depending on the chromosome. In the present study we also found that the B3 chromosome carrying blackleg disease resistance gene was inherited over several generations through both male and female gametes. In addition, almost normal transmission of the B3 chromosome through microspores was found as evidenced from 1:1 segregation for this chromosome and blackleg disease resistance in the DH population.
In the present study, while applying selection pressure for blackleg resistance at each generation, we found that four chromosomes B1, B3 B7 and B8, were retained as intact or broken chromosomes while the other four B genome chromosomes B2, B4, B5 and B6 were eliminated in the interspecific progenies. Retention of the chromosome B3 may be the consequence of selection for cotyledon resistance as this chromosome is associated with resistance phenotype. Navabi et al. (2010) working with the same B. napus × B. carinata interspecific progeny and a population derived from another B. napus × B. carinata interspecific cross (Navabi et al. 2011) also reported that many of the B genome chromosomes (B1, B3, B6, B7 and B8) were retained as either intact or broken chromosomes in interspecific progeny. This is contrary to the general consensus that the B genome chromosomes are very distinct from the A and C chromosomes of B. napus and are lost in early generations of interspecific hybrids (Dixelius 1999; Li et al. 2004; Schelfhout et al. 2006).
Cytogenetic analysis was carried out in DH2 and not DH1 lines to investigate the intriguing relationship between the B3, B7 and B8 chromosomes as revealed by the molecular marker work. In the case of the DH1 lines the resistance was clearly attributable to the presence of only the B3 chromosome since all the markers that co-segregated with the resistance were on this chromosome; while only the middle to bottom of this chromosome was present in the resistant DH2 lines (Fig. 3; Supplementary Figure S1). Also, the top of the B8 chromosome was found only in the resistant DH2 lines, and this chromosome segment showed linkage association with the middle to bottom of the B3 chromosome. This suggests that a translocation between the top of the B8 and middle to bottom of the B3 chromosomes might have occurred in the DH2 population.
Summary of genotypic analysis of B genome SSR markers used to study the inheritance of B. carinata derived resistance to L maculans PG2 isolate #UA3356 in DH1 and DH2 lines derived from B. napus × B. carinata interspecific cross. a The SSR markers from the entire B3 chromosome were present in the resistant (R) DH1 lines while markers from only the middle to lower segment were found in resistant DH2 lines. The SSR markers from the B3 chromosome were absent in susceptible (S) DH1 and DH2 lines. b The SSR markers from the whole B7 chromosome were found in both resistant and susceptible DH1 and DH2 lines. c The SSR markers from the entire B8 chromosome were found in the resistant and susceptible DH1 lines as well as resistant DH2 lines, but markers from only middle to bottom were found in the susceptible DH2 lines. *Represents the B genome markers that co-segregate with cotyledon resistance to L maculans PG2 isolate #UA3356 in DH2, β represents the markers that amplify both B. napus and B. carinata but possess alleles that are distinct to the B-genome, + represents the markers that amplify only B. carinata but are monomorphic among the resistant and susceptible lines and Θ represents the markers that amplify both B. napus and B. carinata to give monomorphic bands
The meiotic analysis of PMCs from DH2 lines showed that both susceptible and resistant lines possessed more than 38 chromosomes (Figs. 4, 5). The 40 chromosomes observed in susceptible DH2 lines suggest the presence of 20 An chromosomes from B. napus, 18 Cn/c chromosomes from B. napus or B. carinata and two Bc chromosomes from B. carinata. Our molecular marker data suggested that the susceptible lines carried the whole B7 chromosome and a fragment from the middle to bottom of the B8 chromosome (Fig. 3; Supplementary Material Table S1). GISH/GISH-like analysis might have detected only the B7 chromosome, but not the B8 fragment which could be too short to be visualized. In the case of the resistant DH2 lines our molecular marker data suggested that all resistant lines carried a large segment (middle to bottom) of B3, the whole of B7 and the middle to bottom of the B8 chromosome. However, the GISH/GISH-like analysis of the resistant DH2 lines revealed the presence of one or two pairs of B chromosomes. It is difficult to ascertain the identity of the chromosome(s) detected using GISH/GISH-like techniques in the resistant lines. One of the B chromosomes was apparently B7, the same chromosome detected in the susceptible DH2 lines. The other chromosome detected in the resistant lines with 2n = 42 could reflect either the B3 or B8. However, because the susceptible lines lacked the B3 chromosome and this chromosome co-segregated with cotyledon resistance, it can be inferred that GISH detected B3 in these lines. For the resistant lines with 2n = 40 chromosomes, the chromosome detected by GISH could be either B3 or B7, or even B8. The reason why cytological analysis detected only 40 or 42 chromosomes in the resistant DH2 lines when our molecular marker data suggested that these lines carry the B3, B7 and B8 chromosomes, as whole or broken chromosomes, as well as the 2n = 40 chromosomes detected in the susceptible lines, when our marker data suggested the presence of the B7 and B8 chromosomes is difficult to explain—despite the GISH technique generally being able to detect the A, B and C chromosomes (Navabi et al. 2010, 2011; Mason et al. 2010, 2011). Some chromosomal translocation between the B genome chromosomes or between the B and A/C genome chromosomes might have occurred in these lines. In the latter case, the detection by GISH is size dependent and only experiments with B specific BAC probes of each genomic region could efficiently be used to detect the rearrangements occurring in the plants.
Chromosome associations at metaphase I of meiosis in pollen mother cells (PMCs). The susceptible lines were observed to have 20 bivalents a susceptible DH2 line 1152-025 while resistant DH2 plants had either b 20 bivalents (resistant DH2 line 1152-027) or c 21 bivalents (resistant DH2 line 1152-046)
In summary, based on molecular cytogenetic analysis of B. napus × B. carinata interspecific hybrid progenies through the use of SSR markers and GISH/GISH-like chromosome painting techniques, we demonstrated that the B genome chromosome B3 of B. carinata carries a gene conferring resistance at cotyledon stage to L. maculans isolate #UA3356. The fact that the B7 and B8 chromosomes were retained in the resistant lines makes it impossible for us to determine whether the inheritance of the B3 chromosome alone is sufficient to confer blackleg disease resistance. The development of DH lines that contain individual B genome chromosomes is needed to understand the mechanism of the resistance. We also demonstrated that a few B genome chromosomes were inherited along with B. napus chromosomes for several generations of the interspecific hybrid progenies, as well as transmitted normally through microspores during the production of DH lines. This is contrary to the general consensus that B genome chromosomes are very distinct from the A and C chromosomes of B. napus and are easily lost in early generations of interspecific hybrids.
References
Ansan-Melayah D, Balesdent MH, Delourme R, Pilet ML, Tanguy X, Renard M, Rouxel T (1998) Genes for race-specific resistance against blackleg disease in Brassica napus L. Plant Breeding 117:373–378
Attia T, Röbbelen G (1986a) Cytogenetic relationship within cultivated Brassica analyzed in amphihaploids from the three diploid ancestors. Can J Genet Cytol 28(3):323–329
Attia T, Röbbelen G (1986b) Meiotic pairing in haploids and amphidiploids of spontaneous versus synthetic origin in rape, Brassica napus L. Can J Genet Cytol 28(3):330–334
Attia T, Busso C, Röbbelen G (1987) Digenomic triploids for an assessment of chromosome relationships in the cultivated diploid Brassica species. Genome 29(2):326–330
Axelsson T, Bowman CM, Sharpe AG, Lydiate DJ, Lagercrantz U (2000) Amphidiploid Brassica juncea contains conserved progenitor genomes. Genome 43:679–688
Balesdent MH, Attard H, Ansan-Melayah D, Delourme D, Renard R, Rouxel M (2001) Genetic control and host range of avirulence toward Brassica napus cultivars Quinta and Jet Neuf in Leptosphaeria maculans. Phytopathology 91:70–76
Balesdent MH, Attard A, Kuhn AL, Rouxel T (2002) New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans. Phytopathology 92:1122–1133
Balesdent MH, Fudal I, Ollivier B, Bally P, Grandaubert J, Eber F, Chevre AM, Leflon M, Rouxel T (2013) The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa. New Phytol 198:887–898. doi:10.1111/nph.12178
Brun H, Levivier S, Somda I, Ruer D, Renard M, Chevre AM (2000) A field method for evaluating the potential durability of new resistance sources: application to the Leptosphaeria maculans–Brassica napus pathosystem. Phytopathology 90:961–966
Brun H et al (2010) Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol 185(1):285–299. doi:10.1111/j.1469-8137.2009.03049.x
Busso C, Attia T, Röbbelen G (1987) Trigenomic combinations for the analysis of meiotic control in the cultivated Brassica species. Genome 29:331–333
Chèvre AM, This P, Eber E, Deschamps M, Renard M, Delseny M, Quiros CE (1991) Characterization of disomic addition lines Brassica napus-Brassica nigra by isozyme, fatty acid, and RFLP markers. Theor Appl Genet 81(43):49
Chèvre AM, Ever F, This P, Barret P, Tanguy X, Brun H, Delseny M, Renard M (1996) Characterization of Brassica nigra chromosomes and of blackleg resistance in B. napus-B. nigra addition lines. Plant Breeding 115:113–118. doi:10.1111/j.1439-0523.1996.tb00884.x
Chèvre AM, Barret P, Eber F, Dupuy P, Brun PH, Tanguy X, Renard M (1997) Selection of stable Brassica napus-B. Juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 1. Identification of molecular markers, chromosomal and genomic origin of the introgression. Theor Appl Genet 95:1104–1111
Christianson JA, Rimmer SR, Good AG, Lydiate DJ (2006) Mapping genes for resistance to Leptosphaeria maculans in Brassica juncea. Genome 49:30–41. doi:10.1139/G05-085
Delourme R, Eber F, Renard M (1991) Radish cytoplasmic male sterility in rapeseed: breeding restorer lines with a good female fertility. In: Proceedings of 8th Int. Rapeseed, Congress Vol 5, Saskatoon, Canada, pp 1506–1510
Delourme R, Pilet-Nayel MN, Archipiano M, Horvais R, Tangui X, Rouxel T, Brun H, Renard M, Balesdent MH (2004) A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus. Phytopathology 94:578–583
Delourme R, Chevre AM, Brun H, Rouxel T, Balesdent MH, Dias JS, Salisbury P, Renard M, Rimmer SR (2006) Major gene and polygenic resistance to Leptosphaeria maculans in oilseed rape (Brassica napus). Eur J of Plant Path 114:41–52
Delwiche PA (1980) Aspects of blackleg (Leptosphaeria maculans) resistance to rapeseed Brassica napus. PhD Thesis, University of Wisconsin, Madison
Dhingra OD, Sinclair JB (1985) Basic plant pathology methods. CRC Press Inc, Boca Raton, p 355
Dion Y, Gugel RK, Rakow GFW, Séguin-Swartz G, Landry BS (1995) RFLP mapping of resistance to the blackleg disease [causal agent, Leptosphaeria maculans (Desm.) Ces. et de Not.] in canola (Brassica napus L.). Theor Appl Genet 91:1190–1194
Dixelius C (1999) Inheritance of the resistance to Leptosphaeria maculans of Brassica nigra and B. juncea in near-isogenic lines of B. napus. Plant Breeding 118:151–156
Dixelius C, Glimelius K (1995) The use of somatic hybridization for interspecies transfer of plant disease resistance. In: Reuveni R (ed) Novel approaches to integrated pest management. Lewis Publications, Michigan, pp 75–93
Dixelius C, Wahlberg S (1999) Resistance to Leptosphaeria maculans is conserved in a specific region of the Brassica B genome. Theor Appl Genet 99:368–372
Fernando WGD, Chen Y (2003) First report on the presence of Leptosphaeria maculans pathogenicity group-3, the causal agent of blackleg of canola in Manitoba. Plant Dis 87:1268
Fernando WGD, Chen Y, Ghanbarnia K (2007) Breeding for blackleg resistance: the biology and epidemiology. Adv Bot Res 45:271–311
Ferreira ME, Rimmer SR, Williams PH, Osborn TC (1995) Mapping loci controlling Brassica napus resistance to Leptosphaeria maculans under different screening conditions. Phytopathology 85(2):213–217
Fitt BDL, Brun H, Barbetti MJ, Rimmer SR (2006) World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur J Plant Pathol 114:3–15. doi:10.1007/s10658-005-2233-5
Glimelius K (1999) Somatic hybridization. In: Gomez-Campo C (ed) Biology of Brassica cenospecies. Elsevier, New York, pp 107–148
Gugel RK, Petrie GA (1992) History, occurrence, impact, and control of blackleg of rapeseed. Can J Plant Pathol 14:36–45. doi:10.1080/07060669209500904
Hayward A, McLanders J, Campbell E, Edwards D, Batley J (2012) Genomic advances will herald new insights into the Brassica: Leptosphaeria maculans pathosystem. Plant Biol 14(Suppl. 1):1–10. doi:10.1111/j.1438-8677.2011.00481.x
Howell EC, Barker G, Jones GH, Kearsey MJ, King GJ et al (2002) Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics 161:1225–1234
Howlett BJ (2004) Current knowledge of the interaction between Brassica napus and Leptosphaeria maculans. Can J Plant Pathol 26:245–252
Kebede B, Thiagarajah MR, Zimmer C, Rahman MH (2010) Improvement of open-pollinated spring rapeseed (Brassica napus L.) through introgression of genetic diversity from winter rapeseed. Crop Sci 50:1236–1243
Kutcher HR, Keri M, McLaren DL, Rimmer SR (2007) Pathogenic variability of Leptosphaeria maculans in western Canada. Can J Plant Pathol 29:388–393
Kutcher HR, Fernando WGD, Turkington TK, McLaren DL (2011) Best Management Practices for Blackleg Disease of Canola. Prairie Soils Crops J 4:122–134
Lagercrantz U (1998) Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150:1217–1228
Lagercrantz U, Lydiate DJ (1995) RFLP mapping in Brassica nigra indicates differing recombination rates in male and female meioses. Genome 38:255–264
Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144:1903–1910
Leflon M, Eber F, Letanneur JC et al (2006) Pairing and recombination at meiosis of Brassica rapa (AA) × Brassica napus (AACC) hybrids. Theor Appl Genet 113:1467–1480
Li H, Sivasithamparam K, Barbetti MJ (2003) Breakdown of a Brassica rapa subsp sylvestris single dominant blackleg resistance gene in Brassica napus rapeseed by Leptosphaeria maculans field isolates in Australia. Plant Dis 87(6):752
Li M, Qian W, Meng J, Li Z (2004) Construction of novel Brassica napus genotypes through chromosomal substitution and elimination using interploid species hybridization. Chromosome Res 12:417–426
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15:516–525
Mason AS, Huteau V, Eber F, Coriton O, Yan G, Nelson MN, Cowling WA, Chevre AM (2010) Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chromosome Res 18:655–666
Mason AS, Nelson MN, Castello MC, Yan G, Cowling WA (2011) Genotypic effects on the frequency of homoeologous and homologous recombination in Brassica napus × B. carinata hybrids. Theor Appl Genet 122(3):543–553
Mayerhofer R, Bansal VK, Thiagarajah MR, Stringam GR, Good AG (1997) Molecular mapping of resistance to Leptosphaeria maculans in Australian cultivars of Brassica napus. Genome 40:294–301
Mayerhofer R, Wilde K, Mayerhofer M, Lydiate D, Bansal VK, Good AG, Parkin AP (2005) Complexities of chromosome landing in a highly duplicated genome: toward map-based cloning of a gene controlling blackleg resistance in Brassica napus. Genetics 171:1977–1988
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Navabi ZK, Parkin IA, Pires JC, Xiong Z, Thiagarajah MR, Good AG, Rahman MH (2010) Introgression of B-genome chromosomes in a doubled haploid population of Brassica napus × B. carinata. Genome 53(8):619–629. doi:10.1139/G10-039
Navabi ZK, Stead KE, Pires JC, Xiong Z, Sharpe AG, Parkin IAP, Rahman MH, Good AG (2011) Analysis of B-genome chromosome introgression in interspecific hybrids of Brassica napus × B. carinata. Genetics 187:659–673. doi:10.1534/genetics.110.124925
Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, Gupta V et al (2008) Comparative mapping of Brassica juncea and Arabidopsis thaliana using intron polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genom 9(1):113. doi:10.1186/1471-2164-9-113
Plieske J, Struss D, Robbelen G (1998) Inheritance of resistance derived from the B genome of Brassica against Phoma lingam in rapeseed and the development of molecular markers. Theor Appl Genet 97:929–936
Rahman H, Harwood J, Weselake R (2013) Increasing seed oil content in Brassica species through breeding and biotechnology. Lipid Tech 25:182–185
Ramchiary N, Padmaja KL, Sharma S, Gupta V, Sodhi YS, Mukhopadhyay A, Arumugam N, Pental D, Pradhan AK (2007) Mapping of yield influencing QTL in Brassica juncea: implications for breeding of a major oilseed crop of dryland areas. Theor Appl Genet 115:807–817. doi:10.1007/s00122-007-0610-5
Rimmer SR (2006) Resistance genes to Leptosphaeria maculans in Brassica napus. Can J Plant Pathology 28:288–297
Rimmer SR, Buchwaldt L (1995) Diseases. In: Kimber D, McGregor DI (eds) Brassica oilseeds production and utilization. CAB International, Wallingford, pp 111–140
Rimmer SR, van den Berg CGJ (1992) Resistance of oilseed Brassica spp. to blackleg caused by Leptosphaeria maculans. Can J Plant Pathol 14:56–66. doi:10.1080/07060669209500906
Rimmer SR, Borhan MH, Zhu B, Somers D (1999) Mapping resistance genes in Brassica napus to Leptosphaeria maculans. “New horizons for an old crop.” In: Proceedings of the 10th International Rapeseed Congress, Canberra, Australia, 26–29 Sept 1999
Rogers SO, Bendich AJ (1988) Extraction of DNA from plant tissues. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual. Kluwer Academic Publications, Dordrecht, pp 1–10
Rouxel T, Penaud A, Pinochet X, Brun H, Gout L, Delourme R, Schmit J, Balesdent MH (2003) A ten-year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene in oilseed rape. Eur J Plant Pathol 109:871–881
Roy NN (1978) A study on disease variation in the population of an interspecific cross of Brassica juncea L. × B. napus. L. Euphytica 27:145–149
Roy NN (1984) Interspecific transfer of Brassica juncea type high blackleg resistance to Brassica napus. Euphytica 33(2):295–303
Sacristan MD, Gerdemann M (1986) Different behavior of Brassica juncea and B. carinata as sources of Phoma lingam resistance in experiments of interspecific transfer to B. napus. Plant Breed 97:304–314
Schelfhout CJ, Snowdon R, Cowling WA, Wroth JM (2006) Tracing B-genome chromatin in Brassica napus × B. juncea interspecific progeny. Genome 49(11):1490–1497. doi:10.1139/G06-103
Sjodin C, Glimelius K (1989a) Transfer of resistance against P. lingam to Brassica napus L. by asymmetric somatic hybridization and toxin selection. Theor Appl Genet 78:513–520
Sjodin C, Glimelius K (1989b) Brassica naponigra, a somatic hybrid resistant to Phoma lingam. Theor Appl Genet 77:651–656
Sjödin C, Glimelius K (1988) Screening for resistance to blackleg Phoma lingarn (Tode ex Fr.) Desm. within Brassicaceae. J Phytopathol 123:322–332
Song KM, Osborn TC, Williams PH (1988) Brassica taxonomy based on nuclear restriction fragment length polymorphism (RFLPs). 1. Genome evolution of diploid and amphidiploid species. Theor Appl Genet 75:784–794
Sprague SJ, Balesdent MH, Brun H, Hayden HL, Marcroft SJ, Pinochet X, Rouxel T, Howlett BJ (2006) Major gene resistance in Brassica napus (oilseed rape) is overcome by changes in virulence of populations of Leptosphaeria maculans in France and Australia. Eur J Plant Pathol 114:33–40. doi:10.1007/s10658-005-3683-5
Struss D, Bellin U, Robbelen G (1991) Development of B-genome chromosome addition lines of B. napus using different interspecific Brassica hybrids. Plant Breeding 106(3):209–214. doi:10.1111/j.1439-0523.1991.tb00503.x
Struss D, Quiros CF, Plieske J, Röbbelen G (1996) Construction of Brassica B genome synteny groups based on chromosomes extracted from three different sources by phenotypic, isozyme and molecular markers. Theor Appl Genet 93:1026–1032
Waara S, Glimelius K (1995) The potential of somatic hybridization in crop breeding. Euphytica 85:217–233
Warwick SI, Black LD (1991) Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae)–Chloroplast genome and cytodeme congruence. Theor Appl Genet 82:81–92
West JS, Kharbanda PD, Barbetti MJ, Fitt BDL (2001) Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol 50:10–27. doi:10.1046/j.1365-3059.2001.00546.x
Yu F, Lydiate DJ, Rimmer SR (2005) Identification of two novel genes for blackleg resistance in Brassica napus. Theor Appl Genet 110:969–979
Yu F, Lydiate DJ, Rimmer SR (2008) Identification and mapping of a third blackleg resistance locus in Brassica napus derived from B. rapa subsp. sylvestris. Genome 51:64–72
Yu F, Gugel RK, Kutcher HR, Peng G, Rimmer SR (2013) Identification and mapping of a novel blackleg resistance locus LepR4 in the progenies from Brassica napus x B. rapa subsp. Sylvestris. Theor Appl Genet 126(2):307–315
Zhu JS, Struss D, Robbelen G (1993) Studies on resistance to Phoma lingam in Brassica napus-Brassica nigra addition lines. Plant Breeding 111:192–197. doi:10.1111/j.1439-0523.1993.tb00629.x
Acknowledgments
We thank Alberta Agricultural Research Institute (AARI), Alberta Crop Industry Development Fund (ACIDF), Alberta Canola Producers Commission (ACPC), Agriculture and Food Council of Alberta and Canola Agronomic Research Program (Alberta Canola Producers Commission, Manitoba Canola Growers Association, SaskCanola and the Canola Council of Canada) for financial support to this project. We also thank Mohan Thiagarajah, Marnie Newell, Julie Bernier, Zahidur Rahman, An Vo, and many others who have contributed in various aspects of the project.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments herein comply with the current laws of Canada.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. G. F. Visser.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2014_2298_MOESM1_ESM.doc
Polyacrylamide gels showing the whole B3 chromosome co-segregate with cotyledon resistance in DH1 lines. In case of DH2, the SSR markers sJ0954 and sJ7166 gave the B. napus profile while sB0860A failed to amplify genomic DNA in both resistant and susceptible lines. This suggests that the top of the B3 chromosome was lost or deleted in the DH2 lines. Thus, only the middle to bottom segment (from sB1728 to sB2596, represented by dotted oval) of the B3 chromosome co-segregated with blackleg cotyledon resistance in DH2 (DOC 691 kb)
122_2014_2298_MOESM2_ESM.doc
Polyacrylamide gels showing that the two SSR markers (sB23190a and sJ8126b) from the top of the B7 chromosome amplified genomic DNA in both resistant and susceptible DH1 and DH2 lines to give B. napus profile while the remaining four (sB1822, sJ7046, sB1752 and sJ8469a) markers from middle to bottom were B-genome-specific and amplified the resistant and susceptible DH1 and DH2 lines as well. Therefore, either the whole or middle to bottom part of the B7 chromosome was present in both DH1 and DH2 lines. None of the SSR markers from the B7 chromosome were found to be linked to blackleg resistance in these two DH populations (DOC 667 kb)
122_2014_2298_MOESM3_ESM.doc
Polyacrylamide gels showing SSR markers, either specific to B. carinata or amplified both B. napus and B. carinata, spanning the entire B8 chromosome was present in both resistant and susceptible DH1 lines. In case of the DH2 lines, three markers from the top segment of the B8 chromosome (markers sJ7434 to sJ6846) co-segregated with blackleg cotyledon resistance in this population (but not in DH1); two SSR markers (sJ3302R and sJ03104) from the middle of the chromosome gave B-genome-specific bands and two markers (sA0499F and sJ1827a) from the bottom of the chromosome gave B. napus/B. carinata profile. This suggests that the whole B8 chromosome was present in the resistant and susceptible DH1 lines. In DH2, chromosome segment from the middle to bottom of B8 was present in both the resistant and susceptible lines, while top of this chromosome co-segregated with cotyledon resistance (top of B8 co-segregated with middle to bottom of B3) (DOC 784 kb)
122_2014_2298_MOESM4_ESM.doc
(a-f) FISH-BAC/GISH labelling of mitotic chromosome spreads in susceptible DH2 lines with 20 bivalents. Chromosomes painted red are from the B genome. Based on the molecular and cytogenetic data we speculate that the extra B chromosome in the susceptible DH2 lines is B7 (a-c) Susceptible DH line 1152-025 (2n=40) and (d-f) 1152-033 (2n=40). Bars: 5μm (DOC 549 kb)
122_2014_2298_MOESM5_ESM.doc
(a-i) FISH-BAC/GISH labelling of mitotic chromosome spreads in resistant DH2 lines with 20 bivalents. Chromosomes painted red are from the B genome. Based on the molecular and cytogenetic data we speculate the extra B genome chromosome in the resistant DH2 lines with 2n = 40 could be the B3, B7 or B8 chromosomes. (a-c) Resistant DH line 1152-027 (d-f) 1152-028, and (g-i) 1152-030 Bars: 5μm (DOC 701 kb)
122_2014_2298_MOESM6_ESM.doc
(a-i) FISH-BAC/GISH labelling of mitotic chromosome spreads in resistant DH2 lines with 21 bivalents. Chromosomes painted red are from the B genome. Based on the molecular and cytogenetic data we speculate the two B genome chromosomes in the resistant DH2 lines with 2n = 42 are the B7 and B3 chromosomes (a-c) Resistant DH line 1152-031, (d-f) 1152-046 and (g-i) 1152-040. Bars: 5μm (DOC 590 kb)
Rights and permissions
About this article
Cite this article
Fredua-Agyeman, R., Coriton, O., Huteau, V. et al. Molecular cytogenetic identification of B genome chromosomes linked to blackleg disease resistance in Brassica napus × B. carinata interspecific hybrids. Theor Appl Genet 127, 1305–1318 (2014). https://doi.org/10.1007/s00122-014-2298-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2298-7