Abstract
Interspecific crosses contribute significantly to plant evolution enabling gene exchanges between species. The efficiency of interspecific crosses depends on the similarity between the implicated genomes as high levels of genome similarity are required to ensure appropriate chromosome pairing and genetic recombination. Brassica napus (AACC) is an allopolyploid, resulting from natural hybridization between Brassica rapa (AA) and Brassica oleracea (CC), both being diploid species derived from a common ancestor. To study the relationships between genomes of these Brassica species, we have determined simultaneously the pairing and recombination pattern of A and C chromosomes during meiosis of AAC triploid hybrids, which result from the interspecific cross between natural B. napus and B. rapa. Different AAC triploid hybrids and their progenies have been analysed using cytogenetic, BAC-FISH, and molecular techniques. In 71% of the pollen mother cells, homologous A chromosomes paired regularly, and usually one chromosome of each pair was transmitted to the progeny. C chromosomes remained mainly univalent, but were involved in homoeologous pairing in 21.5% of the cells, and 13% of the transmitted C chromosomes were either recombined or broken. The rate of transmission of C chromosomes depended on the identity of the particular chromosome and on the way the hybrid was crossed, as the male or as the female parent, to B. napus or to B. rapa. Gene transfers in triploid hybrids are favoured between A genomes of B. rapa and B. napus, but also occur between A and C genomes though at lower rates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interspecific hybridization is common in plants and has played a crucial role in the evolution of plant species by generating new ecotypes or new species and by allowing gene exchanges across species boundaries. Nowadays, interspecific hybridization is still used in plant breeding to expand the genetic basis of crops and to introduce genes of agronomic importance from wild species into crop germplasm (Frankel et al. 1995). Interspecific hybridization has also received close attention over the past ten years in connection with the development and cultivation of transgenic crops because it could lead to modification of diversity within wild crop relatives through crop-to-wild gene flow. This process has the potential to generate new races of weeds that are more invasive and better adapted to man-made habitats (Ellstrand et al. 1999). At the whole genome scale, the success of hybridization and gene transfer depends on the level of genetic and structural relatedness between the genomes of crops and wild plants. More frequent introgressions are expected when crops and their wild relatives share higher levels of similarity: e.g. Beta vulgaris × B. maritima (Boudry et al. 1993) or Raphanus sativus × R. raphanistrum (Panestos and Baker 1967).
The Brassica genus includes several diploid and tetraploid cultivated species organized around three genomes named A, B, and C (U 1935). Comparative mapping of the diploid species, Brassica rapa (AA), B. nigra (BB), and Brassica oleracea (CC) indicates that genomes of these species are probably derived from a common ancestor through duplications (Lagercrantz and Lydiate 1996; Truco et al. 1996; Parkin et al. 2003, 2005). The closest similarity lies between the A (B. rapa, 2n = 20) and C (B. oleracea, 2n = 18) genomes (Teutonico and Osborn 1994; Truco et al. 1996; Parkin et al. 2003), which are still able to pair (Inomata 1980; Attia and Röbbelen 1986) and which have been reunited in the tetraploid genome of Brassica napus (AACC) (U 1935). A high level of homology remains between the genomes of B. oleracea and B. rapa and those of B. napus, allowing regular pairing and recombination between them (Parkin et al. 1995).
The AAC hybrids, resulting from crosses between B. napus and its progenitor B. rapa, are ideal structures to simultaneously assess homologous pairing and recombination occurring between A genomes of B. rapa and B. napus, and homoeologous pairing and recombination occurring between A and C genomes or within C chromosomes. In particular, these AAC hybrids have been used for production of monosomic addition lines and for gene flow assessment after backcross to B. rapa or selfing. The frequency of C chromosome transmission and the frequency of homoeologous recombination during meiosis of these hybrids have been recurrently discussed.
The transmission of C chromosomes from AAC hybrids has been observed in several studies and contrasting results were obtained. Whereas Mikkelsen et al. (1996) showed that C chromosomes were transmitted to 50% of the backcross progeny, which corresponded to the theoretical distribution of univalent chromosomes between gametes at meiosis, other studies revealed that C chromosomes were transmitted at lower and variable rates, depending upon the C chromosomes involved (Chen et al. 1990; MacGrath and Quiros 1990; Metz et al. 1997). The frequency of homoeologous recombination has been less well studied compared to C chromosome transmission. Multivalents were observed at meiosis of triploid hybrids, indicating that homoeologous pairing, including auto- and allosyndesis, occurred. However, the frequency of multivalents depended on genotypes (Heneen et al. 1995) and the occurrence of “homeologous recombined chromosomes” was not assessed in the first backcross generation on the same material. Using other genotypes, Mikkelsen et al. (1996) found a low rate of deleted or recombined C chromosomes in the first backcross generation. In all these studies, only a few C chromosomes were observed. Accordingly, no clear information exists about the frequency and distribution of homoeologous recombination events on the whole C genome.
Finally, meioses of different genotypes of AAC hybrids were characterized in several studies and supported the contrasting conclusions, which may be explained by genotype effect. However, none of these studies simultaneously investigated the meiotic behaviour and the rate of recombination in the AAC hybrids; so, no relationship can be established between meiotic behaviour, number of chromosomes transmitted, and recombination. In this paper, we determine simultaneously how A and C chromosomes pair and recombine during meiosis of AAC triploid hybrids, resulting from a direct cross between natural B. napus and B. rapa and to analyse what is transmitted from the triploid to its progenies This study is based on the analysis of AAC triploid meioses through meiotic observations and comparisons of chromosome segregation followed by cytogenetic and molecular analyses in different progenies derived from these hybrids.
Materials and methods
Plant materials
The AAC triploid hybrids were produced from crosses between B. rapa plants of cultivar Chicon, as the female, and two B. napus cultivars, Darmor and Yudal, as the male parent. These triploid hybrids CD (Chicon × Darmor) and CY (Chicon × Yudal) were then used to generate progeny by carrying out further crosses, either selfing or backcrosses to recurrent parents (Fig. 1).
Scheme of plant material production for AAC hybrids and their progeny (D Darmor, Y Yudal, C Chicon). The number of plants analysed from each progeny is indicated at the bottom of the figure. a Triploid hybrids Chicon × Darmor (CD) were either self-pollinated (ten F1 plants), crossed as female to Chicon (four F1 plants), to Yudal (one F1 plant) or to Darmor (three F1 plants), or as male to Darmor (four F1 plants). b Three F1 triploid hybrids Chicon × Yudal (CY) were crossed to Yudal as female and as male
Immunolocalization at prophase
Anti ASY1 polyclonal antibodies of Arabidopsis thaliana have been previously described (Armstrong et al. 2002) and were used at a dilution of 1:500. Anti ZYP1 polyclonal antibodies of A. thaliana (Higgins et al. 2005) were also used at a dilution of 1:500. Preparation of prophase stage spreads and fluorescence immunolocalization was performed according to published methods (Armstrong et al. 2002). All observations were made using a Leica DM RXA microscope and photographs taken with a CoolSNAP HQ (Roper, Guluth, GA, USA) camera driven by Open LAB 3.1.5 software. All images were further processed with Open LAB 3.1.5 or Adobe Photoshop 7.0.
Meiotic observation of F1 hybrids and BC progenies
Samples of young floral buds were fixed in Carnoy’s solution (alcohol: chloroform: acetic acid, 6V: 3V: 1V) for 24 h at room temperature and stored until use in 50% ethanol at 4°C. Anthers were then squashed and stained with in 1% aceto-carmine. Pairing configurations were observed on pollen mother cells (PMCs) at Metaphase I.
FISH at meiosis of F1 with genome specific BAC clones
The chromosome preparation was the same as for meiotic observations. Regression was performed with 50% fresh acetic acid and, after freezing the slide in liquid nitrogen, the cover slip was removed with a razor blade and stored at −20°C.
Two BAC clones, one from B. oleracea, BoB014O06 (Howell et al. 2002), and one from B. napus, BAC 54B2, provided by B. Chalhoub (URGV Evry, France; unpublished data, available upon request to Genoplante), were labelled by random priming with biotin-14-dUTP (Invitrogen, life technologies, Paisley, UK) and digoxigenin-11-dUTP (Roche diagnostics, Mannheim, Germany), respectively. The clone BAC 54B2 hybridizes to three A genome chromosomes in B. napus (O. Coriton, unpublished results). The clone BoB014O06 hybridizes to the C chromosomes and allows visualization of the C genome B. napus; the characterization of C specific repeated sequence well spread on the C genome carried by this BAC is in progress (O. Coriton and E. Howell, unpublished results).
Chromosome preparations were incubated in Rnase A (100 ng/μl) and pepsin (0.05%) in 10 mmol HCl, fixed with paraformaldehyde (1%), dehydrated in an ethanol series (70, 90, and 100%), and air-dried. The hybridization mixture consisted of 50% deionized formamide, 10% dextran sulfate, 2 × SSC, 1% SDS, and labelled probes (100 ng per slide). Chromosome preparations and pre-denatured (92°C for 6 min) probes were denatured at 85°C for 10 min. In situ hybridization was carried out overnight in a moist chamber at 37°C. After hybridization, slides were washed for 5 min in 50% formamide in 2 × SSC at 42°C, followed by several washes in 4 × SSC-Tween. Biotinylated probes were immunodetected by fluorescein isothiocyanate (FITC) conjugated avidin antibodies (Vector Laboratories, Burlingame, USA). Digoxigenin-labelled probes were detected with anti-digoxigenin antibody conjugated with rhodamine (Roche Diagnostics). The chromosomes were mounted and conterstained in Vectashield (Vector Laboratories) containing 2.5 μg/ml 4′, 6-diamidino-2-phenylindole (DAPI). Fluorescence images were captured using a CoolSnap HQ camera (Photometrics, Tucson, AZ, USA) on an Axioplan 2 microscope (Zeiss, Oberkochen, Germany) and analysed using MetaVue™ (Universal Imaging corporation, Downington, PA, USA).
Fertility assessment
Male fertility was assessed as the percentage of pollen stained by a 1% aceto-carmine solution. Two flowers and at least 600 pollen grains were analysed per plant. Seed set was assessed by counting the number of seeds per hand-pollinated flower.
Estimation of chromosome number by flow cytometry
Flow cytometry analyses were performed for the selfed and backcrossed progenies using leaf tissue for assessing their chromosome number as described by Eber et al. (1997).
Molecular analyses
Plant DNA was extracted as described by Doyle and Doyle (1990). Molecular analyses were carried out in 77 plants of CD × Chicon progenies with specific markers for each C chromosome and in the progeny of CD × Yudal with specific markers for each A chromosome, according to the map established by Foisset et al. (1996) and Delourme et al. (in preparation) (Table 1). RAPD (Foisset et al. 1996), SINE (Prieto et al. 2005), and PFM were used. PFM are Physical Functional Markers developed from Arabidopsis or Brassica sequences by B. Chalhoub (URGV Evry, France; unpublished data, primers are available upon request to Genoplante). Some markers were not mapped on the reference map, but their position was deduced from linkage analyses in the studied progeny between these unmapped markers and positioned markers.
Statistical analyses
Meiotic behaviour of F1 plants and their progenies and male fertility were analysed using chi-square tests, and seed sets of the different plants were compared through the Kruskal–Wallis nonparametric test. Comparison of the segregation of chromosome number observed or calculated in the different progenies were analysed using chi-square tests.
The correlation factor between flow cytometry values and observed chromosome number was calculated according to meiotic observations from a sample of 74 plants of different progenies.
The theoretical frequency (F i ) of triploid gametes with chromosome number i was defined by a binomial distribution of the nine C chromosomes : \( F_{i} = C^{{i - 10}}_{9} {\left( {1/2} \right)}^{9} \times 100. \).
Theoretical frequency of C chromosome transmission was calculated in CD × C progeny from the average number of chromosomes in the plant according to a hypothesis of equal probability of transmission for each chromosome: \( f_{{{\text{female}}}} = (N - 20)/9, \) where N is the average chromosome number in CD × C progeny.
Results
Characterization of the triploid hybrids
Meiotic behaviour
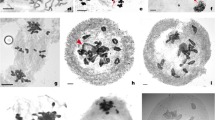
The extent of chromosome pairing was initially investigated during meiotic prophase I by antibodies that recognize the meiotic proteins ASY1, associating with the chromosome axis, and ZYP1, involved in synaptonemal complex (SC) formation; this latter indicates the occurrence of chromosome pairing. In B. napus, used as a control, all chromosomes were paired and formed synapsed bivalents at pachytene, as both proteins were co-localized along each bivalent (Fig. 2a–d). In both CD and CY hybrids, while some chromosomes were paired and formed well synapsed bivalents (overlapped staining with anti-ASY1 and anti-ZYP1 antibodies), some chromosomes remained unsynapsed at pachytene (lack of ZYP1 signal; Fig. 2e–h) and persisted unsynapsed until diplotene (Fig. 2i–l) when both ASY1 and ZYP1 signals progressively disappeared.
Meiotic observation of AAC hybrids. Immunolocalization of Asy1 and Zyp1 meiotic proteins in Brassica napus (a–d) and in AAC hybrids (e–l). m PMCs at metaphase I showing ten bivalents and nine univalents. n–o FISH analysis of metaphase I PMCs: FISH was carried out using two BACs that identify three pairs of chromosomes A (red) and all the C chromosomes (green), respectively. n Nine C univalent chromosomes, three A bivalents marked in red, and seven unmarked A bivalents (in blue DAPI); o seven C univalent chromosomes, one autosyndetic C-C bivalent, three A bivalents marked in red, and seven unmarked A bivalents
At metaphase I, all 15 CD and CY F1 hybrids displayed a similar meiotic behaviour (Table 2). The most frequently occurring configuration of the 29 chromosomes, 9I + 10II, was observed in ca. 71% of the analysed PMCs (Fig. 2m). The percentage of chromosomes paired (from 67.8 to 70.7%) was not significantly different from the theoretical value [20/29 chromosomes paired as bivalents (ll), i.e. 69.0%], suggesting that B. rapa homologues pair preferentially. However, pairing did not occur exclusively between homologues as 5.3 and 3.3% of the PMCs formed trivalents (lll) or tetravalents (lV) in the CD and CY hybrids, respectively. Ca. 17% of PMCs showed seven univalents (l) and 11 bivalents, and 7% of PMCs displayed less than ten bivalents and no multivalents.
The BAC FISH analyses confirmed that nine C chromosomes, painted green by BoB014O06, largely remained at a univalent stage (Fig. 2n), but other configurations, such as autosyndesis between C chromosomes (Fig. 2o), also occurred. The average meiotic behaviour of CD hybrids, deduced from the observation of 25 PMCs by BAC FISH, was: 8.52 IC + 0.36 IA + 9.76 IIAA + 0.2 IICC + 0.04 IIAC + 0.04 IIIAAC.
Fertility
The male fertility of hybrids established through pollen analysis was high (Table 2) but significant differences were observed between the different CD hybrids and between the different CY hybrids. On average, the male fertility of the CY hybrids (61.8%) was lower than that of CD hybrids (78.1%).
Female fertility was also very variable as the mean number of seeds produced per pollinated flower ranged from 0.4 to 8.5 among the different CD and CY hybrids. Interestingly, these hybrids produced on average the same number of seeds per pollinated flower whether they had been pollinated by the oilseed rape (Darmor or Yudal) or by B. rapa (Chicon) recurrent parent. Likewise, no significant difference was detected between reciprocal crosses of CD to its recurrent parent Darmor or between reciprocal crosses of CY to its recurrent parent Yudal. Finally, seed set was significantly lower when the AAC hybrids were selfed (0.4–2.2 seeds produced per pollinated flower) than when backcrossed to any of its parents (1.0–8.5 seeds produced per pollinated flower).
Characterization of the progenies
Chromosome number
The chromosome number of all BC-plants was estimated by flow cytometry. In a subset of 74 plants, encompassing the whole range of cytometric values, the true chromosome number was also determined cytologically. Regression analysis between the cytometric values and the true chromosome numbers (Fig. 3) indicated that flow cytometry gave an excellent prediction of the chromosome number (R² = 0.99). The maximal difference between the observed and estimated values for the chromosome number was only one chromosome.
For every BC-plant, the number of chromosomes transmitted from the AAC hybrid was estimated by substracting the gametic chromosome number of the recurrent parent (either 10 for B. rapa or 19 for B. napus) from the estimated total number of chromosomes of the BC-plant.
As expected, most BC-plants had inherited between 10 and 19 chromosomes from their AAC parent (Table 3), but a few (2.4%) inherited more than 19 chromosomes, differing from the expected binomial distribution. Of the CDxC plants, 2.8% had inherited nine chromosomes.
Chi-square analyses indicated that none of the observed distributions of gametic structure corresponded to expectations (α = 5%). The number of chromosomes transmitted to the progeny was different when the triploid hybrid was used as female versus as male (Fig. 4). When the triploid hybrid was used as female, gametes with extreme chromosome number were more represented than expected. When the triploid hybrid was used as male, gametes with 17, 18, or 19 chromosomes were highly over-represented (50.9 or 49.3% of gametes had more than 17 chromosomes in D × CD and Y × CY crosses, respectively). Neither the origin of the hybrid (CY or CD) nor that of the recurrent parent (B. napus, Darmor or Yudal, or B. rapa) had a significant effect (α = 5%) on the chromosome number of gametes when the hybrid was crossed as female.
The average number of chromosomes transmitted by the triploid was 14.2 in the CD × C cross, 14.9 in the CD × D cross, 14.6 in the CD × Y cross, 16.1 in the D × CD cross, 15.2 in the CY × Y cross, and 15.9 in the Y × CY cross.
The distribution of chromosome numbers in the CD-S progeny, obtained after selfing the CD hybrids, was different from the theoretical distribution expected if both male and female gametes fitted a binomial distribution of chromosome numbers (P = 0.5). The observed distribution of chromosome numbers fitted the product of the observed distribution of chromosome number in male and female gametes, which were deduced from the BC progenies (Fig. 5).
Molecular analyses to deduce A and C chromosome transmission
The transmission of A and C chromosomes was assessed with molecular markers present in hybrid parents and absent in the recurrent parent.
Transmission of A-chromosomes from triploid hybrids
The transmission of A chromosomes from CD hybrids was studied in 76 CD × Y plants, which were genotyped with markers present in Darmor and/or in Chicon, but absent in Yudal (Table 4). Provided homologous A-chromosomes paired and segregated regularly in CD hybrids, markers present both in Chicon and Darmor would be transmitted to the whole progeny while markers present only in Chicon or in Darmor to half of the progeny in the absence of segregation distortion.
Transmission of 6 A-chromosome pairs (DY1b/N1, DY4/N3, DY5/N9, DY9/N8, DY10/N7, and DY16/N10) was investigated with at least one marker present in both Darmor and Chicon. Molecular markers of DY1b (N1), DY4 (N3), DY9 (N8), and DY10 (N7) were detected in the whole progeny. The molecular marker for DY16 (N10) was detected in 75 of the 76 plants of the progeny. Transmission of DY5 (N9) was studied using two different markers present in both Darmor and Chicon; one of these markers was present in all plants of the progeny, while the second one was only detected in 74 of 76 plants of the progeny.
Transmission of the four other A-chromosome pairs (DY6/N6, DY11/N2, DY14/N4, and DY19/N5) was studied using markers, which were only present in one of the parents; so, the expected transmission frequency of these markers in progeny was 50%. Molecular markers of these four linkage groups were transmitted with the expected frequency.
C chromosome transmission and recombination events
C chromosome transmission was assessed in CD × C progeny instead of in CD × Y progeny, because in this progeny, all the markers specific to C genome can be used since the recurrent parent transmits only A chromosomes.
Transmission of C chromosomes from the CD hybrids was assessed in a subset of CD × C progeny (77 plants), using two to five molecular markers per chromosome. Transmission frequency of each marker was compared to an expected value, which was calculated by assuming an equal transmission of every C chromosome. In the surveyed subset of CD × C progeny, which did not represent the whole variability of the CD × C progeny, the average chromosome number transmitted from CD hybrid was 13.2; so, the theoretical frequency of transmission for each C chromosome was (13.2–10)/9, i.e. 35.8%. No significant difference was observed between observed transmission frequency of markers and the theoretical one (α = 5%), except for one marker on DY2 and the three markers on DY18, which were transmitted at a higher than expected rate (Fig. 6).
Transmission of the C chromosomes was assessed through co-segregation analyses of markers from each linkage group in CD × C progeny. Most of C chromosomes were transmitted as whole chromosomes, and only 13.2% of transmitted C chromosomes were incomplete (i.e. some markers of the chromosome were detected in a plant while others were not). Each C chromosome was transmitted as a whole chromosome to 23.3–46.8% of plants, and as an incomplete chromosome to 1.3–13% of plants, depending upon the chromosome (Fig. 6). Significant differences of transmission were detected, with DY8 being less frequently transmitted and DY18 being more frequently transmitted than the average. We also observed that DY15 was more frequently transmitted as incomplete than the other linkage groups, as it was transmitted as incomplete in 13% of plants in the progeny.
In the population, 49 of 77 plants presented only whole C chromosomes. Among these 49 plants, 12 were observed at metaphase I. For ten of these, ten bivalents were observed and the number of univalents corresponded to the number of C chromosomes detected through molecular analyses. In the two remaining plants, one supernumerary C chromosome was detected: in one of these two plants, four C chromosomes were detected through molecular analyses, but 23 chromosomes forming 10II + 3I were observed at meiosis. In the second plant, six C chromosomes were detected, whereas only 25 chromosomes were observed at meiosis, forming either 5I + 10II (7PMC/12) or 7I + 9II (4PMC/12) or 4I + 9II + 1III (1PMC/12).
Incomplete C chromosomes were detected in 28 of the 77 plants. Most of these plants (22) contained one incomplete C chromosome and six plants contained more than one incomplete chromosome. Ten of the plants containing one incomplete C chromosome were observed at metaphase I: in three plants, ten bivalents were consistently observed and the number of univalents corresponded to the total number of detected C chromosomes (i.e. whole plus incomplete). In four plants, ten bivalents were also consistently observed, the number of univalents corresponded to the number of detected whole chromosomes, but incomplete chromosomes were not observed. The last three plants presented different meiotic configurations: one of these plants, having three whole and one incomplete C chromosome, showed at meiosis either 4I + 10II (6PMC/12), or 1I + 10II + 1III (3PMC/12) or 2I + 11II (3PMC/12). The second plant, having one whole and one incomplete C chromosome, showed either 2I + 10II + 1IV (1PMC/11) or 2I + 12II (9PMC/11) or 1I + 11II + 1III (1PMC/11). The last plant, having two whole and one incomplete C chromosomes, showed at meiosis either 3I + 10II (5PMC/12) or 3I + 8II + 1III (1PMC/7) or 1I + 11II (6PMC/7).
Discussion
The results of this study suggest that pairing and recombination occur preferentially between homologous chromosomes in triploid AAC hybrids, but also between homoeologues at a lower rate. This study also reveals that the different C chromosomes can be transmitted by the triploid hybrid at different rates. These results depend, however, on the fertility of the triploid hybrid and on the viability of the different gametes and embryos according to their structure.
In this study, the fertility of interspecific hybrids between B. rapa and B. napus was found to be highly variable, as mentioned in previous reports (MacKay 1977; Quiros et al. 1987; Lee and Namai 1992; Mikkelsen et al. 1996; Lu and Kato 2001). Whereas the seed set was quite similar to that observed by Mikkelsen et al. (1996), MacKay (1977) and Lu and Kato (2001), the male fertility was higher in our crosses (from 58.5 to 84.5%) than that reported by others (MacKay 1977; Quiros et al. 1987;Lee and Namai 1992; Heneen et al. 1995; Mikkelsen et al. 1996; Lu and Kato 2001). This variability may indicate that male fertility is a property of the parental genotypes rather than that of the genomic AAC structure. In addition, our study showed that CD and CY hybrids had different male fertilities in spite of similar meiotic behaviours. This may point to the presence of genetic factors affecting fertility with no obvious meiotic effects (Ramsey and Schemske 2002) and this shows that PrBn, the genetic factor that control homoeologous pairing at the haploid stage (Jenczewszki et al. 2003), has no effect on the meiotic behaviour of triploid hybrids.
The structure of gametes produced by triploid hybrids and transmitted to the progenies did not fit the expected binomial distribution and depended on the recurrent parent when triploid hybrids were used as the male parent. First, gametes with extreme chromosome numbers (n = 10–12 and 17–19) occurred at higher rates than predicted by binomial distribution, which was in agreement with previous studies (MacKay 1977; Namai 1987; Lee and Namai 1992; Lu and Kato 2001). This skewed distribution indicates that either gametes with n = 13–16 are less viable and fertile (i.e. gametophytic selection) or that they generate embryos that are counter-selected (i.e. sporophytic selection). Second, our results compared to previous studies showed that the recurrent parent affected the structure of gametes only when the triploid hybrid was crossed as the male parent. Indeed, when B. napus was used as the female parent, the mean number of chromosomes transmitted by the hybrid in our study varied between 15.9 and 16.1 according to the genotype with 50.2% of gametes containing more than 17 chromosomes as found in previous data (MacKay 1977). By contrast, when B. rapa was used as the female parent, 13.1 and 13.8 chromosomes were transmitted on average with 12–15% of gametes having more than 17 chromosomes (Namai 1987; Lu and Kato 2001). Several hypotheses could explain the differences of distribution of chromosome number between female and male gametes according to the recurrent parent. First, one may imagine that gametic competition between male gametes is higher than between ovules with the advantage of male gametes having a higher chromosome number. Secondly, it is well known that interspecific crosses are more fertile when the female gamete contains more chromosomes than the male gamete (Johnston et al. 1980).
The transmission of A and C chromosomes was observed in progenies from the triploid hybrids used as the female parent.
In AAC hybrids, the A genomes are in a disomic stage and one may therefore expect that regular homologous pairing should occur almost systematically between the A chromosomes of B. rapa and B. napus (MacNaughton 1973; Heneen et al. 1995). Our observations confirmed these expectations since on average 19.5 A chromosomes among the 20% per cell are involved in AA bivalents. Our molecular analyses carried out in CD × Y progeny also confirm that one chromosome of every A-homologous pair is mainly transmitted to the progeny of AAC hybrids.
Rare abnormal transmission of A chromosome specific markers was detected in three plants and may have originated from two mechanisms. First, unequal segregation of A chromosome can be due to mispairing of homologous chromosomes and then lead to unbalanced gametes at the origin of nullisomic or trisomic plants for one A chromosome. This scenario, which leads to the nontransmission or triplication of an entire chromosome, is likely to occur since 20% of the cells contained one or two A univalents at Metaphase I that should partition randomly at Anaphase I. It may explain the absence of the marker of DY16 (N10) in one plant of the progeny even if genotyping other markers of this chromosome is necessary to conclude that the whole chromosome is lacking in this plant. Trisomic plants containing three A chromosomes of one homology group probably occurred at low rate in our material even if we never observed backcross plants with nine bivalents and one trivalent at metaphase I. Such trisomic plants were, however, detected in previous studies at the subsequent backcross generations (Chen et al. 1992; Cheng et al. 1994; Heneen and Jorgensen 2001). Secondly, homoeologous recombination could explain the nontransmission of an A chromosome fragment, which was substituted by its C homoeologous region. This could explain why only a fragment of DY5 (N9) was detected in two plants but cannot account for the nontransmission or triplication of an entire chromosome. Homoeologous recombination will be more precisely discussed with regard to the results obtained on C genome.
The C chromosomes are in a monosomic stage in the AAC hybrid and we showed that they are transmitted less regularly than the A chromosomes. Our results, therefore, contradict the prediction of Tomiuk et al. (2000) who proposed that a gene carried by the C genome should be transmitted to the next generation with an equal probability than a gene carried by the A genome. First, we clearly showed, using molecular markers well spread on the nine C chromosomes of the B. napus genetic map, that C chromosomes were transmitted to less than 50% of the progeny when the triploid hybrid was crossed to B. rapa. This result is in agreement with those of Lee and Namai (1992), Lu and Kato (2001), and MacGrath and Quiros (1990) but in disagreement with the observations of Mikkelsen et al. (1996), who showed that C chromosomes were transmitted to 50% of plants. Secondly, our transmission analyses showed that C chromosomes have different probabilities to be transferred to the progeny of the triploid hybrid. Indeed, DY18 (N15) and DY2 (N12) tend to be more frequently transmitted, whereas, DY8 (N18) is less transmitted than expected. This result supports several previous studies on transmission of one to seven C chromosomes (Fantes and MacKay 1979; Chen et al. 1990; MacGrath and Quiros 1990; Metz et al. 1997). In all these studies (Fantes and MacKay 1979; Chen et al. 1990; MacGrath and Quiros 1990; Chen et al. 1992; Metz et al. 1997) including ours, these analyses were carried out on small progenies (between 24 and 77 plants) and, therefore, only achieved a low-statistical power for detecting differences, and hence the significance of the differences of transmission rate was probably underestimated. Moreover, absence of common markers between studies did not allow comparison of the different transmission rates. Notwithstanding these two points, different hypotheses are proposed to explain differences of transmission rates: either the transmission frequency of each C chromosome may depend on the size of the chromosome (Chen et al. 1990) or the transmission frequency of each C chromosome is affected by an effect of the chromosome on the competitiveness or viability of the gametes or of the embryo.
Our study provided genetic evidence for chromosomal rearrangements affecting C chromosomes. Indeed, we have detected 28 plants in the CD × C progeny (36.4% of the progeny) that contain incomplete C chromosomes. According to the cytological observations we made on the meiosis of the triploid AAC hybrids, it is likely that most of these rearrangements were mediated by recombination between A and C chromosomes (allosyndesis) or between two C chromosomes (autosyndesis). Similar results were reported from monosomic addition lines by Haterok et al. (2005). This assertion is also supported by our molecular analyses; indeed, in four of the ten plants that contained one incomplete C chromosome, the number of univalents corresponded to the number of whole-C chromosomes detected with molecular markers, therefore, demonstating that the incomplete C chromosome had been introgressed onto another A or C chromosome.
Autosyndesis is expected to occur in at least 17% of the PMCs that showed 7 univalents and 11 bivalents. Furthermore, bivalents formed by two C chromosomes were showed by BAC FISH. This rate of autosyndetic pairing is among the highest rates ever reported among different genotypes of haploid B. oleracea (Armstrong and Keller 1982). A higher rate has only been reported once by Attia et al. (1987) who observed that 30% of PMCs of AAC hybrids contain more than ten bivalents. Differences in the rate of autosyndesis may suggest that pairing and recombination between C chromosomes is genotype dependent. Such events of recombination should produce chromosomes formed by parts of different C chromosomes, and by our molecular analyses we observed simultaneously two or more incomplete C chromosomes in 7.8% of plants, which is close to the rate expected from our cytogenetic observation (8.5%).
The detected C chromosome rearrangements may also be due to homoeologous recombination between A and C chromosomes. Indeed, 4.4% of the PMCs contained multivalents and one allosyndetic bivalent was observed by BAC FISH in one on 25 cells. As already discussed, occurrence of incomplete A chromosomes in two or three plants provides genetic evidence that recombination had occurred between A and C chromosomes. Our results suggest that homoeologous recombination had occurred in a minimum of 2.6–3.9% of PMCs. Extending this estimates to the whole genome scale, homoeologous recombination should occur in at least 4.3–6.5% of PMCs, which is in agreement with the frequency of homoeologous pairing observed at meiosis. Interestingly, rearrangement events on C chromosomes were detected at variable rates, ranging from 1.3 to 13%, according to the C chromosome. One chromosome, DY15 (N19), was significantly more frequently rearranged than the other C chromosomes. This variation was not only due to marker density or position on chromosome, because, for each chromosome except DY2 (N12) and DY8 (N9), the markers we used were located on the two chromosome arms and covered a large part of the chromosome. According to previous studies (Parkin et al. 2003), chromosome DY15 (N19) presents homoeologous regions to two A chromosomes, DY5 and DY16 (N9 and N10, respectively). Udall et al. (2005) have recently observed that recombination occurs quite frequently between N19 (DY15) and N9 (DY5) in euploid lines. Although it is not possible to assess the extent to which our results are in agreement with those of Udall et al. (2005) because some of the markers we used have not been mapped, they suggest that DY15 (N19) and its homeologue(s) are among the least divergent homeologous chromosomes. In turn, this interpretation raises the question of the “low rate” of homeologous recombination observed for DY1a (N11) in our study; whereas, this chromosome was shown to recombine with its homeologues at a high rate in euploids (Parkin et al. 1995; Udall et al. 2005).
One may finally notice that incomplete C chromosomes were observed at a higher rate than expected from the meiotic behaviour, suggesting that some of the rearrangements we have detected may have originated from mechanisms other than recombination. It is possible that some of these incomplete chromosomes could sometimes correspond to telosomes or isochromosomes resulting from centromere breakage of univalents, as described in wheat (Friebe et al. 2005). One may therefore speculate that DY15 (N19), which is one of the largest metacentric C chromosomes (Howell et al. 2002), may be rearranged at higher rates because of its physical size and the position of its centromere. Breakage events may also explain why in three plants containing one incomplete C chromosome, one additional chromosome than the number of whole chromosome expected from molecular analyses was observed. In these plants, the incomplete chromosome should be independent from the other chromosomes and could be a broken C chromosome. This hypothesis was supported by molecular analyses that revealed that only markers of one chromosome arm were detected in these plants.
In conclusion, the analysis of meiosis of triploid AAC hybrids revealed that the transmission frequency of C chromosomes varies between C chromosomes, that it depends on the triploid hybrid being crossed as the male or as the female parent and whether it is crossed to B. rapa or to B. napus. For gene flow assessment, we showed that a gene carried by a C chromosome is less likely to be transferred in B. rapa background than a gene carried by a A chromosome, and within the C genome there is variability in likelihood of transfer. Introgression of genes from C to A chromosomes, which is mediated by homoeologous recombination, occurred at low rates in the AAC hybrid but can nonetheless be enhanced in advanced backcross generations (Chen et al. 1992). Our study also confirmed that A chromosomes of B. rapa and of B. napus are similar enough to pair. Triploid hybrids could then be used as well as synthetic B. napus to transfer genes from B. rapa to B. napus and reciprocally. Recombination rates between A homologous chromosomes have yet to be assessed before concluding the efficiency of such crosses.
References
Armstrong KC, Keller WA (1982) Chromosome pairing in haploids of Brassica oleracea. Can J Genet Cytol 24:735–739
Armstrong SJ, Caryl AP, Jones GH, Franklin FCH (2002) Asy1, a protein required for melotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115:3645–3655
Attia T, Busso C, Röbbelen G (1987) Digenomic triploids for an assessment of chromosome relationships in the cultivated diploid Brassica species. Genome 29:326–330
Attia T, Röbbelen G (1986) Cytogenetic relationship within cultivated Brassica analyzed in amphihaploids from the three diploid ancestors. Can J Genet Cytol 28:323–329
Boudry P, Morchen M, Saumitoulaprade P, Vernet P, Vandijk H (1993) The origin and evolution of weed beets—consequences for the breeding and release of herbicide-resistant transgenic sugar-beets. Theor Appl Genet 87:471–478
Chen BY, Heneen WK, Simonsen V (1990) Genetics of isozyme loci in Brassica campestris L. and in the progeny of a trigenomic hybrid between B. napus L. and B. campestris L. Genome 33:433–440
Chen BY, Simonsen V, Lannerherrera C, Heneen WK (1992) A Brassica campestris-alboglabra addition line and its use for gene-mapping, intergenomic gene-transfer and generation of trisomics. Theor Appl Genet 84:592–599
Cheng BF, Heneen WK, Chen BY (1994) Meiotic studies on a Brassica campestris-alboglabra monosomic addition line and derived Brassica campestris primary trisomics. Genome 37:584–589
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Eber F, Letanneur JC, Chèvre AM (1997) Chromosome number of oilseed rape (Brassica napus)-wild radish (Raphanus raphanistrum) spontaneous hybrids and of their progeny estimated by flow cytometry. Cruciferae Newsl 19:17–18
Ellstrand NC, Prentice HC, Hancock JF (1999) Gene flow and introgression from domesticated plants into their wild relatives. Annu Rev Ecol Syst 30:539–563
Fantes JA, MacKay GR (1979) The production of disomic addition lines of Brassica campestris. Eucarpia Cruciferae Newsl 4:36–37
Foisset N, Delourme R, Barret P, Hubert N, Landry BS, Renard M (1996) Molecular-mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a doubled haploid progeny. Theor Appl Genet 93:1017–1025
Frankel OM, Brown AHD, Burdon JJ (1995) The conservation of plant biodiversity, vol, edn. Cambridge University Press, Cambridge
Friebe B, Zhang P, Linc G, Gill BS (2005) Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenet Genome Res 109:293–297
Hasterok R, Wolny E, Kulak S, Zdziechiewicz A, Maluszynska J, Heneen WK (2005) Molecular cytogenetic analysis of Brassica rapa-Brassica oleracea var. alboglabra monosomic addition lines. Theor Appl Genet 111:196–205
Heneen WK, Chen BY, Cheng BF, Jonsson A, Simonsen V, Jorgensen RB, Davik J (1995) Characterization of the A and C genomes of Brassica campestris and B. alboglabra. Hereditas 123:251–267
Heneen WK, Jorgensen RB (2001) Cytology, RAPD, and seed colour of progeny plants from Brassica rapa-alboglabra aneuploids and development of monosomic addition lines. Genome 44:1007–1021
Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19:2488–2500
Howell EC, Barker GC, Jones GH, Kearsey MJ, King GJ, Kop EP, Ryder CD, Teakle GR, Vicente JG, Armstrong SJ (2002) Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics 161:1225–1234
Inomata N (1980) Hybrid progenies of the cross, Brassica campestris x B. oleracea. I. Cytogenetical studies on F1 hybrids. Jap J Genet 55:189–202
Jenczewski E, Eber F, Grimaud A, Huet S, Lucas MO, Monod H, Chèvre AM (2003) PrBn, a major gene controlling homoeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164:645–653
Johnston SA, Den Nijs TPM, Peloquin SJ, Hanneman RE (1980) The significance of genic balance to endosperm development in interspecific crosses. Theor Appl Genet 57:5–9
Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144:1903–1910
Lee KH, Namai H (1992) Pollen fertility and seed set percentage after backcrossing of sesquidiploids (AAC genomes) derived from interspecific hybrid between Brassica campestris L (AA) and Brassica oleracea L (CC) and frequency-distribution of aneuploids in the progenies. Jap J Breed 42:43–53
Lu CM, Kato M (2001) Fertilization fitness and relation to chromosome number in interspecific progeny between Brassica napus and B. rapa: a comparative study using natural and resynthesized B. napus. Breed Sci 51:73–81
MacGrath JM, Quiros CF (1990) Generation of alien chromosome addition lines from synthetic Brassica napus: morphology, cytology, fertility and chromosome transmission. Genome 33:374–383
MacKay GR (1977) The introgression of S alleles into forage rape, Brassica napus L. from turnip, Brassica campestris L. ssp. rapifera. Euphytica 26:511–519
MacNaughton IH (1973) Brassica napocampestris L. (2n = 58). 1. Synthesis, cytology, fertility and general considerations. Euphytica 22:301–309
Metz PLJ, Jacobsen E, Nap JP, Pereira A, Stiekema WJ (1997) The impact on biosafety of the phosphinothricin-tolerance transgene in inter-specific B. rapa x B. napus hybrids and their successive backcrosses. Theor Appl Genet 95:442–450
Mikkelsen TR, Jensen J, Jorgensen RB (1996) Inheritance of oilseed rape (Brassica napus) RAPD markers in a backcross progeny with Brassica campestris. Theor Appl Genet 92:492–497
Namai H (1987) Inducing cytogenetical alterations by means of interspecific and intergeneric hybridization in Brassica crops. Gamma Field Symp 26:41–89
Panestos CA, Baker H (1967) The origin of variation in « wild » Raphanus sativus in California. Genetica 38:243–274
Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781
Parkin IAP, Sharpe AG, Keith DJ, Lydiate DJ (1995) Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38:1122–1131
Parkin IAP, Sharpe AG, Lydiate DJ (2003) Patterns of genome duplication within the Brassica napus genome. Genome 46:291–303
Prieto JL, Pouilly N, Jenczewski E, Deragon JM, Chevre AM (2005) Development of crop-specific transposable element (SINE) markers for studying gene flow from oilseed rape to wild radish. Theor Appl Genet 111:446–455
Quiros CF, Ochoa O, Kianan SF, Douches D (1987) Analysis of the Brassica oleracea genome by the generation of B. campestris-oleracea chromosome addition lines: characterization by isozymes and rDNA genes. Theor Appl Genet 74:758–766
Ramsey J, Schemske DW (2002) Neopolyploidy in flowering plants. Annu Rev Ecol Syst 33:589–639
Teutonico RA, Osborn TC (1994) Mapping of RFLP and qualitative trait loci in Brassica rapa and comparison to the linkage maps of Brassica napus, Brassica oleracea, and Arabidopsis thaliana. Theor Appl Genet 89:885–894
Tomiuk J, Hauser TP, Bagger-Jorgensen R (2000) A- or C-chromosomes, does it matter for the transfer of transgenes from Brassica napus. Theor Appl Genet 100:750–754
Truco MJ, Hu J, Sadowski J, Quiros CF (1996) Inter- and intra-genomic homology of the Brassica genomes: implications for their origin and evolution. Theor Appl Genet 93:1225–1233
U N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Udall JA, Quijada PA, Osborn TC (2005) Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics 169:967–979
Acknowledgments
Our colleagues of OUEST-Génopole® (Le Rheu, France) are gratefully acknowledged for their technical assistance. We thank B. Chalhoub and H. Belcram (URGV Evry, France) for providing PFM and the BAC 54B2, T. Ksiazczyk and J. Maluszynska (University of Silesia, Katowice, Poland) for providing a stab of BoBO14O06, R. Delourme (INRA Rennes, France) for Brassica mapping data and review of the paper, and M. Barbetti (University of Western Australia) for his critical reading of the manuscript. This work was funded by INRA, CETIOM and ANR Biodiversité.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Friebe.
Rights and permissions
About this article
Cite this article
Leflon, M., Eber, F., Letanneur, J.C. et al. Pairing and recombination at meiosis of Brassica rapa (AA) × Brassica napus (AACC) hybrids. Theor Appl Genet 113, 1467–1480 (2006). https://doi.org/10.1007/s00122-006-0393-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0393-0