Abstract
Cytoplasmic male sterility (CMS), which is a maternally inherited trait and controlled by novel chimeric genes in the mitochondrial genome, plays a pivotal role in the production of hybrid seed. In cotton, no PCR-based marker has been developed to discriminate CMS-D8 (from Gossypium trilobum) from its normal Upland cotton (AD1, Gossypium hirsutum) cytoplasm. The objective of the current study was to develop PCR-based single nucleotide polymorphic (SNP) markers from mitochondrial genes for the CMS-D8 cytoplasm. DNA sequence variation in mitochondrial genes involved in the oxidative phosphorylation chain including ATP synthase subunit 1, 4, 6, 8 and 9, and cytochrome c oxidase 1, 2 and 3 subunits were identified by comparing CMS-D8, its isogenic maintainer and restorer lines on the same nuclear genetic background. An allelic specific PCR (AS-PCR) was utilized for SNP typing by incorporating artificial mismatched nucleotides into the third or fourth base from the 3′ terminus in both the specific and nonspecific primers. The result indicated that the method modifying allele-specific primers was successful in obtaining eight SNP markers out of eight SNPs using eight primer pairs to discriminate two alleles between AD1 and CMS-D8 cytoplasms. Two of the SNPs for atp1 and cox1 could also be used in combination to discriminate between CMS-D8 and CMS-D2 cytoplasms. Additionally, a PCR-based marker from a nine nucleotide insertion–deletion (InDel) sequence (AATTGTTTT) at the 59–67 bp positions from the start codon of atp6, which is present in the CMS and restorer lines with the D8 cytoplasm but absent in the maintainer line with the AD1 cytoplasm, was also developed. A SNP marker for two nucleotide substitutions (AA in AD1 cytoplasm to CT in CMS-D8 cytoplasm) in the intron (1,506 bp) of cox2 gene was also developed. These PCR-based SNP markers should be useful in discriminating CMS-D8 and AD1 cytoplasms, or those with CMS-D2 cytoplasm as a rapid, simple, inexpensive, and reliable genotyping tool to assist hybrid cotton breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The acquisition of a new cytoplasmic male sterile (CMS) line and the introduction of its characteristics into breeding programs are very important to the development of commercial hybrid cultivars. As an initial key step in hybrid breeding, identification of cytoplasmic types is usually done by a time-consuming progeny test. However, molecular markers capable of distinguishing mitochondrial genotypes (mitotypes) at the DNA level enable breeders to save time and effort, and PCR-based DNA markers have been developed for this purpose in many crops.
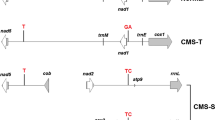
In Upland cotton (Gossypium hirsutum, AD1), two CMS systems, CMS-D2 and CMS-D8, were developed by introducing cytoplasm from two wild diploid species Gossypium harknessii (D2-2) and Gossypium trilobum (D8), respectively (Meyer 1975; Stewart 1992). Two nuclear encoded Rf genes were also introduced from the two diploid species for the two different CMS cytoplasms, i.e., Rf 1 from CMS-D2-2 (Weaver and Weaver 1977) and Rf 2 from CMS-D8 (Wang et al. 2007, 2009; Zhang and Stewart 2001a, b, 2004). Using seven mtDNA gene probes and five restriction enzymes, Wang et al. (2010) demonstrated that restriction fragment length polymorphic (RFLP) markers using atp1 and atp6 genes as probes can distinguish the three cytoplasms, i.e., CMS-D2, CMS-D8 and AD1 cytoplasm. Wu et al. (2011) further used 10 mitochondrial gene-specific probes (atpA, atp6, atp9, cob, cox1, cox2, cox3, nad3, nad6 and nad9) to perform a RFLP analysis and identified three probes (atpA, cox3, and nad6) that showed polymorphic patterns to discriminate the CMS-D2 and its isogenic maintainer line with the AD1 cytoplasm. After further comparing the sequences of the three genes between the two cytoplasms, they identified 1–2 single nucleotide replacement within the three genes. However, they were unable to develop any single nucleotide polymorphic (SNP) markers except for a sequence characterized amplified region (SCAR) marker based on an insertion downstream of the atpA gene. Most recently, Zhang et al. (2012) reported that both CMS line P30A (most likely CMS-D2 cytoplasm) and its maintainer with normal AD1 cytoplasm contained an intact and a truncated copy of atpA but with sequence variation, which allowed the development of a SCAR and a simple sequence repeat (SSR) marker from the two copies, respectively. Up to now, no PCR-based markers have been reported for the CMS-D8 system.
The SNP marker system has become the marker system of choice in recent years, as they are most abundant in a genome. Of many SNP typing methods (Sobrino et al. 2005), the allele-specific PCR method utilizes allele-specific PCR primers, which correspond to the site of the SNPs. In this method, one allele-specific primer is perfectly compatible with one allele, and another primer mismatches with the DNA template at the 3′ end (Hayashi et al. 2004). Since the allele-specific primer preferentially amplifies the corresponding allele, this phenomenon causes PCR products with the nonspecific allele primer to be obtained with much lower amplification efficiency than properly matched termini. This PCR system with DNA polymerase results in SNPs straightforwardly typed by the presence or absence of PCR-amplified products on an ordinary agarose gel electrophoresis (Hayashi et al. 2004). Although the allele-specific PCR method has not been extensively utilized because single allele mismatch at the 3′ end of the primer is usually not satisfactory to completely discriminate two alleles, allele-specific PCR development has been recently conducted by modifying primer sequences to increase specificity for allele discrimination (Kwok et al. 1990; Drenkard et al. 2000; Hayashi et al. 2004).
In this study, we followed the PCR-based allele discrimination strategies developed by Hayashi et al. (2004) to distinguish the two different cytoplasms (CMS-D8 and AD1) using mitochondrial genes in cotton. SNPs in mitochondrial genes involved in the oxidative phosphorylation chain including genes encoding ATP synthase subunit 1, 4 and 8 (atp1, atp4 and atp8) and cytochrome c oxidase 1, 2 and 3 subunits (cox1, cox2 and cox3) were identified in CMS-D8, and maintainer lines on the same nuclear genetic background, 8518. SNPs and InDel in mitochondrial genes were employed to discriminate the different cytoplasmic types by a simple PCR with allele-specific modified primers and PCR products were confirmed by the regular agarose-based electrophoresis.
Materials and methods
Plant materials and total DNA extraction
Cotton plants from three lines, 8518 (a B line with normal fertile AD1 cytoplasm and homozygous non-functional recessive fertility restorer genes rf 2 rf 2 ), D8CMS8518 (an A line with CMS-D8 cytoplasm and homozygous non-functional recessive fertility restorer genes rf 2 rf 2 ), and D8R8518 (an R line for CMS-D8 with the CMS-D8 cytoplasm and homozygous functional dominant fertility restorer genes Rf 2 Rf 2 ), were grown in the greenhouse, New Mexico State University, Las Cruces, NM, USA. One folded or newly unfolded young leaf each from 5 to 10 plants from each genotype was collected and pooled in a 1.5 ml tube and stored at −80 °C until use. Total DNA was isolated using a quick CTAB method (Zhang and Stewart 2000). The presence and integrity of genomic DNA was confirmed by 1.0 % agarose gel and quantified using DU 530 UV/VIS spectrophotometer (Beckman Coulter, Brea, CA, USA).
PCR, cloning, sequencing and multiple sequence alignment
Mitochondrial gene sequences including atp1, atp4, atp6, atp8, atp9, cox1, cox2 and cox3 were amplified using a high-fidelity DNA polymerase by primers for each gene (Table 1; Fig. 1). The PCR products were purified using the QIAquick PCR Purification kit (Qiagen, Valencia, CA, USA) and cloned using pGEM-T Easy vector system (Promega, Madison, WI, USA). Eight clones from each gene fragment were sequenced by ElimBiopharm (Hayward, CA, USA; http://www.elimbio.com/dna_sequencing.htm). Multiple sequence alignment (http://www.ebi.ac.uk/Tools/msa/clustalw2/) using ClustalW was utilized to estimate the percentage of identity match sequences between clones from each gene and among sequences from the three genotypes (8518, D8CMS8518, and D8R8518). The translated protein sequences for these mitochondrial genes, i.e., atp1, 4, 6, 8 and 9, and cox1, 2 and 3 were also compared. Sequence variations among the three cotton genotypes were identified by conducting ClustalW and manually checking sequence differences among the genotypes (Supplementary Figs. 1 and 2). Here, the DNA sequences for atp1 and cox1 from D2RB418, an R line for CMS-D2 with CMS-D2 cytoplasm and homozygous functional dominant fertility restorer genes Rf 1 Rf 1 , were obtained from a separate study using different primers (Wang 2008) and used in the alignment for a comparative analysis to confirm the discriminatory power of SNPs among the three cytoplasms (AD1, D2 and D8).
Primer designing
SNPs and insertion–deletions (InDels) were detected by comparing consensus sequences for the same gene among the four genotypes. Primers for confirmation of the SNPs and InDels were then designed by the utilization of Primer3 (Integrated DNA Technologies; http://www.idtdna.com). The allele and InDel specific primers were used for the amplification of specific mitochondrial genes, i.e. atp1, atp4, atp6 and atp8, and cox1, cox2 and cox3 by the use of regular PCR for the development of allele-specific markers and InDel markers to discriminate the two different cytoplasms (AD1 and D8) or the three cytoplasms (AD1, D8 and D2). The primer information is listed in Table 2. Both allele-specific and nonspecific forward primers have an artificial mismatch within four bases of the 3′ terminus. Reverse primers are commonly designed for both allele-specific and nonspecific primers. As a comparison, primer sequences used in primer extension are listed in Supplementary Table 1.
PCR amplification and gel electrophoresis
To verify the SNPs and InDels, DNA from 8518 (with fertile AD1 cytoplasm) and D8CMS8518 (with sterile CMS-D8 cytoplasm) was used without using D8R81518 since both D8CMS8518 and D8R8518 share the same CMS-D8 cytoplasm and had the same DNA sequences when the eight mtDNA genes were sequenced. SNPs in 8518, D8CMS8518, D8R8518 and D2RB418 (Wang 2008) were also identified and employed for confirmation of the sequence variations. A total of 25 μl PCR reaction contained 2.5 ng of genomic DNA, 0.125 U Taq polymerase (Promega, Madison, WI, USA), 5 μl of 5 × PCR buffer, 25 mM of MgCl2, 10 mM of dNTPs, and 11.875 μl of deionized water. The primers used in the PCR reaction were 2.5 μl of both forward gene-specific primer (5 μM) and reverse primer (5 μM). The PCR amplification program consisted of 30 s, 94 °C denaturing step; a 30 s, 52–60 °C annealing step; a 1 min, 72 °C elongation step; in 27–30 cycles. Finally, 7 min, 72 °C final elongation step was conducted. The PCR products were separated by electrophoresis using 2.0 % agarose gels in 1 × TBE buffer.
Results
Sequence differences between AD1 and D8 cytoplasms
An assembled DNA, cDNA and protein contig sequences based on eight clones sequenced from each mtDNA gene and each genotype were submitted to NCBI under accession ID KC149532 to KC149552. Genomic DNA (gDNA) and protein sequence alignments among the three isogenic genotypes (D8CMS8518, D8R8518, and 8518) are shown in Supplementary Figs. 1 and 2. The gDNA sequence alignments among those with D2RB418 in only atp1, atp6 and cox1, cox2 and cox3 are also included for a comparative analysis. SNPs due to the cytoplasmic differences between AD1 and D8 were identified in the mitochondrial genes atp1, atp4 and atp8, and cox1, cox2, and cox3 (Table 3). Furthermore, SNPs among AD1, D8 and D2 were discovered in atp1 and cox1 (Table 3). No SNPs in the partially sequenced atp9 were detected between AD1 and D8 cytoplasms, although SNP may be present in the region where the sequencing of this gene was not performed in this study.
As indicated in Table 3, the 5′ UTR region of atp1 has two consecutive nucleotide polymorphisms between the CMS/restorer (CMS-D8 cytoplasm) lines and the maintainer line (AD1 cytoplasm). The CMS/restorer lines possess two adenines (AA) and the maintainer line has a cytosine and a guanine (CG) in these positions. Additionally, atp1 has one nucleotide polymorphism which is an adenine in the D8 (CMS/restorer lines) cytoplasm at the 1,365 nt position from the start codon ATG and a thymine in the AD1 (maintainer line) and D2 (restorer line) cytoplasm at the same position (Table 3, Supplementary Fig. 1).
The atp4 gene has a thymine in the CMS/restorer lines at the 181 nt position from the 5′ primer although the maintainer line possesses a cytosine at this position (Supplementary Fig. 1 and Table 3).
In atp6, no SNP between AD1 and D8, including full sequence of coding region and non-coding region, was detected. However, nine nucleotide insertion-deletion (InDel) sequences, i.e. “AATTGTTTT” at the 59–67 bp positions from the start codon of atp6 of the CMS and restorer lines with the D8 cytoplasm were inserted, while this sequence was not detected in the maintainer line with the AD1 cytoplasm (Supplementary Fig. 1 and Table 4).
In atp8, an adenine at the 134 nt position from the 5′ primer is present in the CMS/restorer lines, but a cytosine exists in the maintainer line at the identical position. This SNP causes amino acid alteration from arginine (aga) to isoleucine (atc) in the CMS/restorer lines (Supplementary Fig. 1 and Table 3).
In the partially sequenced atp9 with 170 bp, neither SNP nor InDel between AD1 and D8 cytoplasm was identified (Supplementary Fig. 1). The atp9 gene generally possesses shorter nucleotide sequences ranging from 200 to 300 bp in plants (Giegé and Brennicke 1999).
In cox1, the CMS/restorer lines contain an adenine at the 1,210 nt position from the 5′ primer, but the maintainer line possesses a cytosine (Supplementary Fig. 1 and Table 3). Furthermore, the D2 restorer line D2RB418 with the D2 cytoplasm possesses an adenine at the position of 742 nt from the 5′ primer based on the AD1 and D8 sequences, while the other three lines with AD1 or D8 cytoplasm contain a cytosine at the position (Table 3). It appears that the mtDNA from the D2 cytoplasm possessed more sequence variations from the AD1 and D8 cytoplasms (Supplementary Fig. 1).
The sequence of the cotton cox2 gene contains two exons, 700 bp of the upstream region and 83 bp of the downstream region, separated by a 1,506 bp intron region. This length of intron in cotton is relatively larger than that of many other plant species. The two exons do not show any sequence variation between AD1 and D8 cytoplasm, while in the intron region (1,506 bp), the CMS/restorer lines have two consecutive nucleotides, a cytosine and a thymine at the 1,427 and 1,428 positions from the start codon, but the maintainer line has two consecutive adenines in the same positions (Supplementary Fig. 1 and Table 3).
In cox3, the D8CMS/restorer lines have an adenine at the 113 nt position from the 5′ primer, but the maintainer line has a cytosine in the same position, resulting in amino acid alteration from isoleucine in the CMS/restorer lines to leucine in the maintainer line (Supplementary Fig. 1 and Table 3).
Development of single nucleotide polymorphic (SNP) and InDel markers
SNP genotyping was first conducted using primer extension method (Supplementary Fig. 3). Almost all the primer pairs yielded monomorphic fragments of the same sizes, indicating that this method in designing primers (only one or two consecutive nucleotide mismatch in the 3′ terminus of primers, see Supplementary Table 1) failed in efficiently detecting the SNPs.
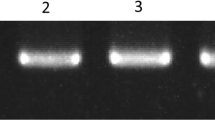
The AS-PCR was then used to develop PCR-based SNP markers (Table 1; Fig. 2a, c). This PCR-based SNP marker development was accomplished by modifying allele-specific and nonspecific primers. The artificial mismatched bases (T to C or C to A) different from genomic DNA sequences were incorporated into the third or fourth position from the 3′ end of the primers of both the specific and nonspecific primers. Eight out of eight SNPs (100 %) using eight primer pairs for the two cytoplasms, AD1 (8518) and D8 (CMS8518), or three cytoplasms, AD1 (8518), D8 (CMS8518/restorer8518) and D2 (restorer B418) cytoplasms were successfully discriminated between two different alleles by utilizing artificially mismatched primers on the agarose-based electrophoresis (Fig. 2a, c). One InDel identified in the downstream region of atp6 was also discriminated (Fig. 2b).
The SNP markers and an InDel marker detected in mitochondrial genes. a SNP markers based on the presence or absence of each PCR product was analyzed on 2 % agarose gels. b InDel marker discriminated based on the insertion/deletion of nine nucleotides in atp6. The InDel is also confirmed by 2 % of agarose gel. Five genotypes on ST474 or 8518 nuclear background composed of either AD1 or D8 cytoplasms were used to discriminate different alleles. c The SNP markers developed to discriminate three cytoplasms, i.e., AD1, D8 and D2
For atp1, the SNP observed in the 3′ region of atp1 was discriminated using allele 1 and 2 specific primers with three cytoplasms, composed of AD1 (maintainer line) and D2 (restorer line) cytoplasms with allele 1 (thymine) and D8 (CMS/restorer lines) cytoplasm with allele 2 (adenine) (Fig. 2c). This position is located at the 1,365th nucleotides downstream (455th amino acid) from the start codon (Table 3). The SNP was tested using four cotton genotypes. Products in allele 1 with AD1 and D2 cytoplasms and in allele 2 with D8 cytoplasm were successfully amplified and discriminated (Fig. 2c), as expected.
Additionally, The two consecutive nucleotide polymorphisms between AD1 and D8 cytoplasm in the 5′ UTR region of atp1 gene were located at the five and four nucleotide upstream from the start codon and are composed of two consecutive adenines (allele 2) in the CMS/restorer lines, while cytosine and guanine nucleotides (allele 1) are present in the maintainer line (Table 3). The maintainer 8518 with the AD1 cytoplasm had amplifications using only allele 1 specific primers; however, there is no amplification observed in allele 2 in this genotype. D8CMS8518 with the D8 cytoplasm had amplification in only allele 2, and no amplification was detected in allele 1 (Fig. 2a), as expected.
For atp4, the SNP between allele 1 (cytosine) in maintainer line and allele 2 (thymine) in CMS/restorer lines are detected at the position of 182 nt from the primer position at the 5′ end of sequenced length (Table 3). 8518 with AD1 cytoplasm has an intense band observed in allele 1, while there is no amplification detection in allele 2. D8CMS8518 had no bands in allele 1, and this banding pattern in allele 2 were observed (Fig. 2a), as expected.
The InDel phenomenon in atp6 was verified by agarose-based electrophoresis (Fig. 2b).
For atp8, the SNP containing allele 1 (cytosine) in maintainer line and allele 2 (adenine) in D8 CMS/restorer lines at the 134 nt (44th aa) downstream from the primer position at the 5′ end of sequenced length are identified (Table 3). This alteration of one nucleotide at the 44th aa position causes serine into arginine. Maintainer line with AD1 cytoplasm amplified only allele 1 specific cytoplasm; however, D8CMS8518 with the D8 cytoplasm had amplifications using allele 2 specific primers, but the amplification of allele 1 was not detected (Fig. 2a).
In cox1, the sequence variation was also utilized to develop a SNP marker using the two genotypes. The SNP is located at the 1,210 nt (403th aa) downstream from the primer position at the 5′ end of sequenced length in this gene, and composed of allele 1 (cytosine) in maintainer line and allele 2 (adenine) in CMS/restorer lines (Table 3). 8518 with AD1 cytoplasm was amplified by using allele 1 specific primer, while the amplification in allele 2 was not detected. D8CMS8518 was amplified using allele 2 specific primers, as expected. This result is identical to other genes, such as atp1 3′, atp1 5′, atp4 and atp8, and cox1, cox2 and cox3 (Fig. 2a). Furthermore, adenine at the 742 nt (247th aa) from the 5′ primer of the sequences in D2 cytoplasm is present, while cytosine in both AD1 and D8 is present at the position (Table 3). B418 with D2 cytoplasm was amplified in allele 1, while 8518 with AD1 cytoplasm and CMS/restorer with D8 cytoplasm were intensely amplified in allele 2, resulting in the allele discrimination of D2 from AD1 to D8 (Fig. 2c).
In cox2, SNP at the two consecutive positions, 1,427 and 1,428 nt from the start codon in an intron region of this gene consists of allele 1 (two adenines) in maintainer line and allele 2 (a cytosine and a thymine) in CMS/restorer lines (Table 3). 8518 with AD1 cytoplasm was amplified in allele 1 as the rest of the genes were also amplified in the same allele 1 (maintainer line). D8CMS8518 with D8 cytoplasm was also amplified in the corresponding allele 2 as other genes were amplified in allele 2 (Fig. 2a).
In cox3, the SNP, which is located at the 113 nt (38th aa) downstream from the primer position at the 5′ end of the sequenced length, was utilized for the gene amplification and can discriminate two different alleles between AD1 and D8 cytoplasms using two genotypes including 8518 with AD1 cytoplasm, and D8CMS8518 with D8 cytoplasm (Fig. 2a). Allele 1 is composed of a cytosine in the maintainer line and allele 2 consists of an adenine in the CMS/restorer lines (Table 3). In 8518 with AD1 cytoplasm, the amplification in allele 1 was detected, while there is no band in allele 2. In D8CMS8518 with D8 cytoplasm, the amplification in allele 2 was identified while the amplification in allele 1 was not observed.
Therefore, all eight SNPs and an InDel detected in mitochondrial genes were discriminated between AD1 and D8 cytoplasms. Two of them (T/A alleles for atp1_3′ and A/C alleles for cox1; Table 2) can be used together to distinguish among the three cytoplasms, i.e., AD1, D8 and D2.
Discussion
Primer extension method was first tested for its discrimination power between two alleles in mitochondrial genes between CMS-D8 and AD1 cytoplasms in this study. The result showed that the primer extension method using two different primers which are distinguishable based on only one 3′ nucleotide bases of the primer sequences could not discriminate two alleles due to the amplification of two alleles in most of the genes (Supplementary Fig. 3). This result indicates that only one nucleotide mismatch at the 3′ allele position of the primers is not sufficient to discriminate two alleles, as similar results were also reported in cotton by Pang et al. (2012). The one nucleotide mismatch could not inhibit the start of PCR, indicating that additional nucleotide mismatch is required to inhibit PCR from the 3′ end of the nonspecific primers. The modified AS-PCR, incorporating additional mismatch base in the last four bases from the 3′ end of a primer, obtained satisfactory results in this study.
Among many markers developed, the PCR-based SNP marker system has been prevalent as a rapid, simple, inexpensive, and reliable genotyping of SNPs (Newton et al. 1989). SNPs are known as the simplest and essential genetic variation and recognized as the most conventional tool for development of DNA markers (Westermeier et al. 2009). Small insertions–deletions (InDels) are also accessible for many plant species including important crops such as rice (Nasu et al. 2002; Feltus et al. 2004; Monna et al. 2006) and maize (Bhattramakki et al. 2002; Batley et al. 2003; Bi et al. 2006). The PCR-based SNP marker system is genotyped on the basis of the presence or absence of an amplified product, which demonstrates the presence of a specific allele or vice versa.
SNPs are an outstanding way to obtain high-quality markers useful for a quick identification of single base differences within the genome (Cooper et al. 1985; Ciarmiello et al. 2011). Therefore, SNPs are the most used molecular markers recently because of their highest abundance in the genome. The PCR-based marker method makes it possible for different cytoplasm types to be discriminated using a simple agarose electrophoresis. The PCR method, termed allele-specific PCR (AS-PCR), which is also recognized as allele-specific primer extension, has high specificity, repeatability and sensitivity for identifying cytoplasm type. AS-PCR is based on the formation of matched or mismatched primer-template formation. This PCR-based molecular marker system apparently abolishes the necessity to conduct phenotyping of the same breeding lines in multiple locations. Dominant markers selectively identify and amplify target SNP alleles from genomic DNA in a single step by including two allele-specific primers and one common primer in mitochondrial genes (Chen et al. 2010). Cotton breeders could now apply AS-PCR markers to select the desired traits with specific cytoplasm in their breeding lines based on cytoplasmic differences.
In cotton, SNP identification has been scarce compared to other crops such as soybean and maize, due to the lower efficiency for discriminating between genome-specific polymorphism and locus-specific polymorphism. Wu et al. (2011) utilized CMS cotton with D2 cytoplasm from G. harknessii in comparison with its isogenic maintainer line with AD1 cytoplasm to investigate sequence differences in the atp1 gene. They discovered two nucleotide differences at the 5′ untranslated region (5′UTR), −4 and −5 positions upstream of ATG codon, which is identical to our research results in terms of nucleotide differences between AD1 and D8 cytoplasm. This indicates that D2-2 and D8 cytoplasms are closer to one another than to the AD1 cytoplasm, which makes it more difficult to develop discriminating mtDNA markers between the two CMS (D2-2 and D8) cytoplasms.
In the present study, the practical goal was to develop more reliable and economical molecular markers to distinguish cytoplasm types of cotton to assist the hybrid cotton breeding process. SNPs between AD1 and D8 cytoplasms in mitochondrial genes, atp1, atp4, and atp8 and cox1 and cox2 and cox3, and among AD1, D8 and D2 cytoplasms in atp1 and cox1 were identified and PCR-based SNP markers have been successfully developed for the first time. We followed the PCR-based innovative SNP marker method of Hayashi et al. (2004), which proved to be simple, rapid and reliable. SNPs were detected simply by using allele-specific PCR primers, which have the corresponding 3′ terminal nucleotide for one allele and mismatch nucleotide for the other allele. As described by Hayashi et al. (2004), we incorporated artificial mismatched nucleotides into the third or fourth bases from the 3′ terminus in both the specific and nonspecific primers. Although several high-throughput genotyping of SNPs have been developed for genotyping of SNPs, they are still expensive and necessitate some specific instruments. SNP genotyping by simple PCR reaction employing allele-specific PCR requires only a simple electrophoresis-based assay, which is easily obtainable in any molecular biology lab and is also useful for many purposes which do not need genome-wide SNP markers. This is the first comprehensive effort to utilize mitochondrial gene-specific primers to perform PCR-based marker development for identifying cytoplasm types in mitochondria genes atp1, atp4, atp6, atp8, and cox1, cox2 and cox3. All allele-specific primers in mitochondrial genes successfully discriminated cytoplasm differences between AD1 and D8 cytoplasms, and among AD1, D8 and D2 cytoplasms. As the number of cytoplasm types increases, more PCR-based markers will be needed.
References
Batley J, Barker G, O’Sullivan H, Edwards KJ, Edwards D (2003) Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol 132:84–91
Bhattramakki D, Dolan M, Hanafey M, Wineland R, Vaske D, Register JC III, Tingey SV, Rafalski A (2002) Insertion-deletion polymorphisms in 30 regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol Biol 48:539–547
Bi IV, McMullen MD, Sanchez-Villeda H, Schroeder S, Gardiner J, Polacco M, Soderlund C, Wing R, Fang Z, Coe EH (2006) Single nucleotide polymorphisms and insertion-deletions for genetic markers and anchoring the maize fingerprint contig physical map. Crop Sci 46:12–21
Chen MH, Fjellstrom RG, Christensen EF, Bergman CJ (2010) Development of three allele-specific codominant rice Waxy gene PCR markers suitable for marker-assisted selection of amylose content and paste viscosity. Mol Breed 26:513–523
Ciarmiello LF, Piccirillo P, Pontecorvo G, De Luca A, Kafantaris I, Woodrow P (2011) A PCR based SNPs marker for specific characterization of English walnut (Juglans regia L.) cultivars. Mol Biol Rep 38:1237–1249
Cooper DN, Smith BA, Cooke H, Niemann S, Schmidtke J (1985) An estimate of unique sequence heterozygosity in the human genome. Hum Genet 69:201–205
Drenkard E, Richter BG, Rozen S, Stutius LM, Angell NA, Mindrinos M, Cho RJ, Oefner PJ, Davis RW, Ausubel FM (2000) A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol 124:1483–1492
Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res 14:1812–1819
Giegé P, Brennicke A (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci USA 96:15324–15329
Hayashi K, Hashimoto N, Daigen M, Ashikawa I (2004) Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor Appl Genet 108:1212–1220
Kwok S, Kellogg DE, McKinney N, Spasic D, Goda L, Levenson C, Sninsky JJ (1990) Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res 18:999–1005
Meyer VG (1975) Male sterility from Gossypium harknessii. J Hered 66:23–27
Monna L, Ohta R, Masuda H, Koike A, Minobe Y (2006) Genomewide searching of single-nucleotide polymorphisms among eight distantly and closely related rice cultivars (Oryza sativa L.) and a wild accession (Oryza rufipogon Griff.). DNA Res 13:43–51
Nasu S, Suzuki J, Ohta R, Hasegawa K, Yui R, Kitazawa N, Monna L, Minobe Y (2002) Search for and analysis of single nucleotide polymorphisms (SNPs) in rice (Oryza sativa, Oryza rufipogon) and establishment of SNP markers. DNA Res 9:163–171
Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 17:2503–2516
Pang MX, Percy RG, Stewart JM, Hughs SE, Zhang JF (2012) Comparative transcriptome analysis of Pima and Acala cotton during boll development using 454 pyrosequencing technology. Mol Breed 30:1143–1153
Sobrino B, Brión M, Carracedo A (2005) SNPs in forensic genetics: a review on SNP typing methodologies. Forensic Sci Int 154:181–194
Stewart JMcD (1992) A new cytoplasmic male sterility and restorer from cotton. In: Proceedings of beltwide cotton conferences, National Cotton Council, Memphis, p 610
Wang F (2008) Molecular analysis of cytoplasmic male sterility (CMS) and fertility restoration in cotton. PhD dissertation, New Mexico State University, Las Cruces
Wang F, Stewart JMcD, Zhang J (2007) Molecular markers linked to the Rf 2 fertility restorer gene in cotton. Genome 50:818–824
Wang F, Yue B, Hu J, Stewart JMcD, Zhang J (2009) A target region amplified polymorphism marker for fertility restorer gene Rf 1 and chromosomal localization of Rf 1 and Rf 2 in cotton. Crop Sci 49:1602–1608
Wang F, Feng C, O’Connell MA, Stewart JMcD, Zhang J (2010) RFLP analysis of mitochondrial DNA in two cytoplasmic male sterility systems (CMS-D2 and CMS-D8) of cotton. Euphytica 172:93–99
Weaver DB, Weaver JB (1977) Inheritance of pollen fertility restoration in cytoplasmic male-sterile upland cotton. Crop Sci 17:497–499
Westermeier P, Wenzel G, Mohler V (2009) Development and evaluation of single-nucleotide polymorphism markers in allotetraploid rapeseed (Brassica napus L.). Theor Appl Genet 119:1301–1311
Wu J, Gong Y, Cui M, Qi T, Guo L, Zhang J, Xing C (2011) Molecular characterization of cytoplasmic male sterility conditioned by Gossypium harknessii cytoplasm (CMS-D2) in upland cotton. Euphytica 181:17–29
Zhang JF, Stewart JMcD (2000) Economical and rapid method for extracting cotton genomic DNA. J Cotton Sci 4:193–201
Zhang JF, Stewart JMcD (2001a) Inheritance and genetic relationships of the D8 and D2–2 restorer genes for cotton cytoplasmic male sterility. Crop Sci 41:289–294
Zhang JF, Stewart JMcD (2001b) CMS-D8 restoration in cotton is conditioned by one dominant gene. Crop Sci 41:283–288
Zhang J, Stewart JMcD (2004) Identification of molecular markers linked to the fertility restorer genes for CMS-D8 in cotton. Crop Sci 44:1209–1217
Zhang X, Meng Z, Zhou T, Sun G, Shi J, Yu Y, Zhang R, Guo S (2012) Mitochondrial SCAR and SSR markers for distinguishing cytoplasmic male sterile lines from their isogenic maintainer lines in cotton. Plant Breed 131:563–570
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Schulman.
This work is dedicated to the memory of Dr. James McD. Stewart (Department of Crop, Soil and Environmental Sciences, University of Arkansas, Fayetteville, AR, USA) as the developer of the CMS-D8 system in cotton and for his numerous contributions to genetic and genomic studies of the system.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Suzuki, H., Yu, J., Wang, F. et al. Identification of mitochondrial DNA sequence variation and development of single nucleotide polymorphic markers for CMS-D8 in cotton. Theor Appl Genet 126, 1521–1529 (2013). https://doi.org/10.1007/s00122-013-2070-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2070-4