Abstract
Aromatic rice is an important commodity for international trade, which has encouraged the interest of rice breeders to identify the genetic control of rice aroma. The recessive Os2AP gene, which is located on chromosome 8, has been reported to be associated with rice aroma. The 8-bp deletion in exon 7 is an aromatic allele that is present in most aromatic accessions, including the most popular aromatic rice varieties, Jasmine and Basmati. However, other mutations associated with aroma have been detected, but the other mutations are less frequent. In this study, we report an aromatic allele, a 3-bp insertion in exon 13 of Os2AP, as a major allele found in aromatic rice varieties from Myanmar. The insertion is in frame and causes an additional tyrosine (Y) in the amino acid sequence. However, the mutation does not affect the expression of the Os2AP gene. A functional marker for detecting this allele was developed and tested in an aroma-segregating F2 population. The aroma phenotypes and genotypes showed perfect co-segregation of this population. The marker was also used for screening a collection of aromatic rice varieties collected from different geographical sites of Myanmar. Twice as many aromatic Myanmar rice varieties containing the 3-bp insertion allele were found as the varieties containing the 8-bp deletion allele, which suggested that the 3-bp insertion allele originated in regions of Myanmar.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The flavor and palatability of aromatic rice are preferred by consumers all over the world. This type of rice is in high demand, and it obtains a premium price in both domestic and international markets. Although most of the trade is from India, Pakistan and Thailand, aromatic rice is cultivated and prized in many other countries of the world. Rice grain aroma results from the production of many biochemical compounds (Petrov et al. 1996), but the most important compound is 2-acetyl-1-pyrroline (2AP), which is considered to be a potent aroma constituent (Buttery et al. 1982). A single recessive gene located on chromosome 8 has been identified as the gene responsible for the aroma trait (Bradbury et al. 2005a; Vanavichit et al. 2008) because it has a key role in the synthesis of 2AP (Bradbury et al. 2005a). The identified gene has been named differently by different groups as BAD2 (Bradbury et al. 2005a, 2008), BADH2 (Niu et al. 2008) and Os2AP (Vanavichit et al. 2008); the latter name is used in the present study. The most important sequence variation of the gene was found in exon 7 as an 8-bp deletion. This sequence deletion generates a premature stop codon that presumably results in loss of function. The loss of Os2AP activity may cause 2AP accumulation. Although the unique genotype (8-bp deletion) determines the aromatic biosynthetic capability in most aromatic rice strains, the aroma content in different aromatic rice varieties does not always correlate with this deletion. The trait may be limited or regulated by other genes or other regions of the known aromatic gene. A previous study has shown that a number of aromatic varieties, primarily from South and Southeast Asia, do not contain the 8-bp deletion and that 2AP is identified in both raw and cooked rice of these varieties (Fitzgerald et al. 2008). Researchers have suggested that the 8-bp deletion in Os2AP is not the only sequence variation that causes aroma and that there are other mutations that drive the accumulation of 2AP. Studies on the diversity of the gene in a large collection of varieties have shown that the 8-bp deletion in exon 7 is present in most aromatic accessions but that other less frequent mutations associated with aroma are also detected (Bradbury et al. 2005a; Bourgis et al. 2008; Shi et al. 2008; Kovach et al. 2009).
The 2AP aromatic compound is present in various parts of the rice plant, such as the stems, leaves and grains, but not in the roots (Lorieux et al. 1996; Yoshihashi 2002). The following sensory methods have been applied to determine the aroma in rice: chewing several seeds or cooking a sample of seeds from individual plants (Ghose and Butany 1952; Dhulappanavar 1976), heating leaf tissue in water or eluting the aroma from leaf tissue with diluted potassium hydroxide (KOH) (Sood and Siddiq 1978; Hien et al. 2006) and heating several half-cut seeds in fresh water (Wanchana et al. 2005). However, these sensory evaluation methods are not consistent or reliable because the aroma is subjected to human preference. Alternatively, an objective gas chromatography/mass spectrometry (GC/MS) method that directly assays 2AP content has been introduced, but the assay is expensive and time consuming (Chen et al. 2006). Molecular markers, especially functional markers, linked to economically important traits have been extensively exploited to assist breeders in selecting individuals with favorite traits, and these markers are useful with respect to the aromatic trait in rice breeding programs. With these markers, the difficulty in evaluating the aroma phenotype is minimized, and the accuracy of selecting desired plants is increased. After the discovery of the aroma gene, the following functional molecular markers were developed: Aromarker (Vanavichit et al. 2008), allele-specific amplification (ASA) marker (Bradbury et al. 2005b), FMbadh2-E2A and FMbadh2-E2B (Shi et al. 2008). However, these markers cover only two types of aromatic alleles, including an 8-bp deletion in exon 7 (for the first two markers) and a 7-bp deletion in exon 2 (for the latter marker).
Nakagahra (1978) estimated that the center of diversity of Asian rice may be located in mountainous regions on the northern side of Southeast Asia, such as Myanmar, Thailand and Yunnan in China. Myanmar is rich in natural resources, and plant genetic diversity is one of the natural resources. Myanmar is well known as the primary center of plant genetic resource (PGR) diversity for rice and is the secondary center of PGR diversity for other crops. Rice is one of the most important food crops in Myanmar because it is an important part of the daily diet. Rice is cultivated all over the country in various agroecological conditions, such as in lowland, flooded, deep water, dry land and upland conditions. Rice is grown extensively, and it covers an area of 8.28 million hectares, which is approximately 60 % of the total cultivated area. The consumption of rice per capita in Myanmar is approximately 210 kg per person per year, and this amount is the highest consumption level in Asia. The rice preferred by Myanmar people varies depending on locations within the country. However, the majority of Myanmar people prefer intermediate amylose rice (20–25 %), which has a soft texture and is aromatic. Good-quality aromatic rice is grown in different regions of Myanmar to meet the high local demands. The price of these varieties in local markets is three times the price of non-aromatic rice varieties. In Myanmar, different places require a specific quality of rice for their cooking quality traits. Some of the preferred rice varieties include the popular aromatic varieties that have a soft texture, high aroma and high kernel elongation, and other rice varieties are locally adapted aromatic rice varieties, which are preferred by local consumers. Locally adapted varieties vary not only in terms of aroma degree, grain shape and grain size, but also with regard to morpho-agronomic characteristics. However, molecular information regarding the aromatic gene that is present in these aromatic rice varieties is still limited. Although an alternative mutated allele (a 3-bp insertion in exon 13) of the Os2AP gene has been identified in Myanmar aromatic rice accessions (Kovach et al. 2009), there are still a large number of aromatic rice accessions that remain uncharacterized. Screening the diverse aromatic rice varieties in Myanmar will allow for additional information regarding aromatic genes to be gathered. In this study, we chose aromatic rice varieties collected from different locations of Myanmar to identify the aroma gene and aromatic allele specific to Myanmar aromatic rice varieties and to develop the functional marker for breeding programs and genetic resource conservation programs.
Materials and methods
Plant materials
Two sets of rice samples were used in this study. The first set was a group of 52 Myanmar landrace rice varieties that belong to Isozyme group V (Khush et al. 2003); this group was kindly provided by the Plant Biotechnology Center at the Myanma Agriculture Service in Myanmar. The first set used the tracking system of the International Rice Germplasm Collection (IRGC) accession numbers. The second set consisted of 25 diverse aromatic rice varieties from unknown Isozyme groups that were collected from different geographical regions of Myanmar; this set of rice varieties was provided by the Seed Bank at the Department of Agricultural Research in Myanmar. Based on the variety names and passport data, all varieties in the second set were considered to be aromatic rice.
PCR genotyping for Os2AP gene mutations
The 52 Myanmar varieties and the two control varieties, KDML105 (aromatic, indica group; Kovach et al. 2009) and ISL-10 (non-aromatic), were used to characterize the Os2AP gene mutations. All plants were grown in an experimental field at Kasetsart University (Nakhon Pathom, Thailand) in July of 2008. Leaf samples from all of the plants were collected from 3-week-old seedlings. Genomic DNA was extracted from 1 g of leaf tissue according to the DNA Trap method developed by the DNA Technology Laboratory at Kasetsart University (Nakhon Pathom, Thailand). Two PCR markers, Aromarker (Vanavichit et al. 2008) and FMbadh2-E2B (Shi et al. 2008), were developed for the detection of the 8-bp deletion in exon 7 and the 7-bp deletion in exon 2 of Os2AP, respectively. These markers were used to amplify genomic DNA from the 52 rice varieties. The amplified PCR products were resolved by 4.5 % polyacrylamide gel electrophoresis (PAGE) and visualized by silver staining.
Aroma evaluation by sensory test and 2AP quantification
Two sensory test methods were performed for aroma evaluation. For the first method, the aroma was evaluated from 1-month-old leaves by incubating cut leaves in a diluted KOH solution and noting the aroma according to the method of Sood and Siddiq (1978). For the second method, the aroma was evaluated by chewing premature and mature seeds. Both sensory methods were performed by five trained panels. The 2AP content was analyzed from two-month-old callus samples cultured on Murashige and Skoog media (1962) with a previously described GC–MS method (Wongpornchai et al. 2004) and from grain samples (5 g) using an improved GC–MS method developed for detecting 2AP in a small-scale sample (as little as 0.5 g; Mahatheeranont et al. 2001).
Reverse transcription PCR (RT-PCR)
Total RNA was extracted from callus, leaf and root samples from aromatic and non-aromatic varieties with a High Pure RNA Isolation Kit (Roche Diagnostics GmbH, Roche Applied Science, Germany). Reverse transcription polymerase chain reaction (RT-PCR) was performed according to the manufacturer’s protocol for the Superscript™ III First Strand Synthesis System for RT-PCR (Invitrogen™ Life Technologies, UK). AromarkerBig (5′ TGCTCCTTTGTCATCACACC 3′ and 5′ CCATGCAACCATCCTTTCTT 3′) was used as the Os2AP gene-specific primer. Actin primers (5′ AGGGCTGTTTTCCCTAGTATCGTG 3′ and 5′ GATGGCATGAGGAGGGGCAT 3′) were used as a positive internal control. The RT-PCR products were analyzed on a 1 % agarose gel in 1× TBE buffer.
Sequencing of the Os2AP gene in selected aromatic rice varieties
The full-length Os2AP gene was sequenced in aromatic varieties from Pathein Nyunt, Yangon Saba and Kyet Paung. Primer pairs designed to amplify fragments covering the entire Os2AP gene were provided by the Rice Gene Discovery Unit at Kasetsart University in Thailand. Amplified PCR products were purified by a Gel/PCR DNA Fragment Extraction Kit (Geneaid, Taiwan) and cloned into a pGEM®-T Easy cloning vector (Promega, Madison, WI). Plasmids from positive colonies were purified with a High Speed Plasmid Minikit QIAprep Spin Miniprep Kit (QIAGEN, USA) and then sequenced using ABI PRISM® BigDye™ Terminators (Applied Biosystem/Perkin-Elmer, San Jose, CA, USA). The sequences of each variety were assembled based on the identical overlapping regions with the CAP3 Sequence Assembly Program (Huang X and Madan A. 1999). The genomic sequence of the non-aroma allele of the Os2AP gene on chromosome 8 (GenBank Accession No. AP005537) of japonica rice (Oryza sativa japonica cultivar Nipponbare) was retrieved from the NCBI website (http://www.ncbi.nlm.nih.gov). Multiple sequence alignment of the Os2AP alleles among Myanmar aromatic rice varieties and the non-aromatic rice variety, Nipponbare, was performed using the CLUSTALW program (Thompson et al. 1994).
Development of an aroma-specific functional marker for Myanmar aromatic rice
The genomic sequence of the aromatic variety, Pathein Nyunt, was used as a reference sequence for the aroma allele with the 3-bp insertion. The PCR marker, 3In2AP, was developed to detect this sequence variation. The forward (5′ GTCCTGTTCAATCTTGCAGC 3′) and reverse (5′ CTTGATGCAACCATGTCATA 3′) primers were designed with Primer 3 (version 0.4; http://frodo.wi.mit.edu/primer3) by applying the constraints of 20 nucleotides in length and a GC content of 40–60 % to avoid primer self-complementarity. The PCR marker was verified by amplifying the genomic DNA extracted from the leaf, seed and callus samples of different aromatic and non-aromatic rice varieties. PCR was performed in a total volume of 10 μl containing 2 μl of genomic DNA (50 ng/μl), 1 μl of 10× buffer, 1 μl of 25 mM MgCl2, 2 μl of 1 mM dNTPs, 0.5 μl of each primer (5 μM) and 0.1 μl of Taq DNA polymerase (Fermatas; Life Science, USA). The PCR analysis was initiated by denaturation at 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min. A final incubation at 72 °C for 7 min was allowed for the completion of primer extension. A portion (2 μl) of the PCR products was resolved on a 4.5 % polyacrylamide gel and visually examined after silver staining.
Application of functional markers for genotyping F2 populations for aroma
To evaluate the accuracy and reliability of the newly developed marker, 3In2AP, 160 F2 plants derived from a cross of Pathein Nyunt (aromatic variety; 3-bp insertion) and ISL-10 (non-aromatic variety; isogenic line derived from KDML105 × CT9993 crosses) were genotyped. The PCR amplicons were resolved on a 4.5 % polyacrylamide gel, and the segregation ratio according to the banding patterns was observed. Phenotypic evaluation was also performed by a sensory test with the leaves of 160 F2 plants according to the method of Sood and Siddiq (1978) and by chewing the F3 seeds of individual F2 lines.
Screening aromatic alleles in aromatic rice germplasms from Myanmar
To validate the utility of the 3In2AP marker and to screen the aromatic rice germplasms from Myanmar, 25 diverse aromatic rice varieties collected from different geographical regions of Myanmar were analyzed. Two markers for the different aroma alleles (Aromarker for the 8-bp deletion and 3In2AP for the 3-bp insertion) were used to amplify the target regions of the Os2AP gene. KDML105 (an aromatic variety with the 8-bp deletion), Pathein Nyunt (an aromatic variety with the 3-bp insertion) and ISL-10 (a non-aromatic variety) were used as positive and negative control varieties. PCR was performed in a total volume of 10 μl containing 2 μl of genomic DNA (50 ng/μl), 1 μl of 10× buffer, 0.4 μl of 25 mM MgCl2, 2 μl of 1 mM dNTPs, 0.4 μl of each primer (5 μM) and 0.1 μl of Taq DNA polymerase. The PCR analysis was initialized by denaturation at 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 2 min followed by a 7-min incubation at 72 °C. A portion (2 μl) of the PCR products was subjected to electrophoresis on a 4.5 % polyacrylamide gel, and the banding pattern was examined after silver staining.
Results
Aroma evaluation and Os2AP genotyping of Myanmar rice varieties
A collection of 52 Myanmar landrace rice varieties that belong to Isozyme group V, which contains many aromatic rice varieties (Khush et al. 2003), was used for the characterization of Os2AP gene alleles. The aroma characteristics of the varieties were evaluated by two sensory test methods as follows: the incubation of cut leaves in diluted KOH and the chewing of several seeds. Only 3 Myanmar rice varieties (Pathein Nyunt, Yangon Saba and Kyet Paung) out of the 52 varieties were detected and confirmed by both sensory methods to be aromatic rice. As analyzed by GC–MS, the 2AP content was present at an intermediate level in the grains of the two aromatic varieties (0.92 ppm for Pathein Nyunt and 0.86 ppm for Yangon Saba) when compared to Thai Jasmine rice (1.3 ppm for KDML105) and another Myanmar rice (1.46 ppm for Kyet Paung). All of the 52 varieties were subsequently genotyped for the two aromatic gene-specific markers to detect the 8-bp deletion in exon 7 and the 7-bp deletion in exon 2. The deletions were not present in all of the non-aromatic varieties. Interestingly, out of the three aromatic rice varieties, only the Kyet Paung variety contained the 8-bp deletion in exon 7. The other two aromatic rice varieties, Pathein Nyunt and Yangon Saba, did not contain the 8-bp deletion or the 7-bp deletion.
Expression of Os2AP in the two Myanmar aromatic rice varieties
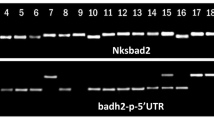
The expression of Os2AP was analyzed by RT-PCR in the two Myanmar aromatic rice varieties, Pathein Nyunt and Yangon Saba (these two varieties did not contain gene mutations in exon 2 or exon 7), to investigate whether this gene had a role in determining the aromatic trait in these two varieties. RT-PCR analysis demonstrated that the expression level of the Os2AP gene in the two varieties did not differ from that of the non-aromatic rice, ISL-10, but the gene expression level in KDML105 (the aromatic variety containing the 8-bp deletion) was highly reduced (Fig. 1).
RT-PCR analysis of Os2AP gene expression in callus, leaf and root samples. KDML 105 is the aromatic variety with the 8-bp deletion, and ISL-10 is a non-aromatic variety. Pathein Nyunt and Yangon Saba are aromatic varieties with the 3-bp insertion. The actin gene was used as a control because it was expressed at the same level in all tissues
Sequence variation in the Os2AP gene in the two aromatic Myanmar rice varieties
The Os2AP gene in the two aromatic rice varieties was sequenced further to investigate whether the other type of gene mutation was present in this gene, even though no effect on gene expression was observed. Gene sequencing revealed a 3-bp insertion (TAT) in exon 13 at the 1,257–1,259 position of the 1,512-bp cDNA sequence in both the Pathein Nyunt and Yangon Saba varieties. The Os2AP gene was also sequenced in the aromatic variety (Kyet Paung). The sequencing result confirmed the presence of an 8-bp deletion in this aromatic variety (Fig. 2a). The 3-bp insertion found in the two Myanmar aromatic rice varieties was an in-frame translation that caused an additional amino acid (tyrosine, Y) to be translated into the cDNA sequence of the Os2AP gene of the two aromatic varieties. The total amino acid sequence length encoded by the Os2AP gene in the two varieties was 504 amino acids (aa), and the normal length found in the non-aromatic varieties was 503 aa (Fig. 2b).
a The Os2AP gene structure. The functional gene consists of 15 exons and 14 introns with its start codon (ATG) in exon 1 and stop codon (TAA) in exon 15. The two aromatic alleles (the 8-bp deletion in exon 7 and the 3-bp insertion in exon 13) are shown in the nucleotide sequence alignments. Nipponbare is a non-aromatic rice variety. KDML105 is an aromatic rice variety containing the 8-bp deletion. Kyet Paung is a Myanmar aromatic rice variety containing the 8-bp deletion. Pathein Nyunt and Yangon Saba are Myanmar aromatic rice varieties containing the 3-bp insertion. b The multiple alignments of deduced protein sequences of Os2AP compared among aromatic and non-aromatic rice varieties. An additional amino acid, tyrosine (Y), in the Pathein Nyunt and Yangon Saba varieties is highlighted in red (color figure online)
Development of a functional marker specific to the 3-bp insertion in exon 13 of Os2AP
A new PCR marker, 3In2AP, was developed to detect the 3-bp insertion (TAT) in exon 13 of the Os2AP gene (Fig. 3a). The primer was validated by amplifying the genomic DNA of the two aromatic varieties, Pathein Nyunt and Yangon Saba, and KDML105 and ISL-10, which were used as controls. As a result, different banding patterns were observed between the two Myanmar aromatic varieties compared to KDML105 and ISL-10. The sizes of the PCR products amplified by the 3In2AP marker were 197 bp for aromatic varieties with the 3-bp insertion and 194 bp for KDML105 and ISL-10 (Fig. 3b).
a The schematic of the 3In2AP marker location in exon 13 of Os2AP. The sequence variation, 3-bp insertion (TAT) and expected sizes of the PCR products are shown. b The banding patterns of the 3In2AP marker. The following lanes are shown: lane 1 Pathein Nyunt (an aromatic rice with the 3-bp insertion), lane 2 Yangon Saba (an aromatic rice with the 3-bp insertion) and lane 3 KDML105 (an aromatic rice with the 8-bp deletion) and lane 4 ISL-10 (a non-aromatic rice). The PCR product sizes are indicated by arrows
Application of the 3In2AP marker for genotyping the F2 population
The aromatic variety, Pathein Nyunt, and non-aromatic variety, ISL-10, were used as parental lines to produce an F2 population that was segregated for aroma. The 3In2AP marker was used to genotype the parental lines and F1 plants. The polymorphic PCR bands were 197 bp in Pathein Nyunt and 194 bp in ISL-10, and both bands (197 and 194 bp) were observed in the F1 hybrid. The marker was then used to genotype 160 lines of the F2 population derived from a single F1 plant of the cross between Pathein Nyunt and ISL-10. As a result, 33 lines were identified as homozygous to the aromatic variety, Pathein Nyunt, and 80 lines were heterozygous. Moreover, 47 lines carried the allele of ISL-10 (Fig. 4). The segregation ratio of the three genotypes in the F2 population agreed with the expected ratio of 1:2:1 (χ 2 = 2.450; P = 0.2938), which corresponded to the Mendelian segregation of a single gene (Table 1). The phenotypic evaluation performed by a sensory test with the leaf samples of F2 plants showed that 33 lines were aromatic and 127 lines were non-aromatic. The numbers of the aromatic and non-aromatic F2 lines had a ratio of 1:3 (χ 2 = 1.633; P = 0.2013). The segregation of phenotypes also agreed with the Mendelian segregation ratio for a single recessive gene that controls a trait (Table 1). According to these results, the phenotypes and genotypes of all 160 F2 progenies were correlated. All 33 aromatic lines contained the same 3-bp insertion allele, which suggests that this single recessive allele is associated with the aroma phenotype (Fig. 4).
Application of the 3In2AP marker to screen aromatic rice germplasm from Myanmar
To screen the aromatic rice germplasm from Myanmar, Aromarker and 3In2AP were used to detect the two different aroma alleles in 25 aromatic rice varieties. The KDML105 and Pathein Nyunt aromatic varieties were used as positive control varieties, and ISL-10 was used as a negative control variety. The functional markers clearly showed that this set of Myanmar aromatic varieties carries either the 8-bp deletion or the 3-bp insertion aroma alleles; three varieties, which were not detected by the two functional markers, were exceptions. Sixteen aromatic varieties carried the 3-bp insertion aromatic allele, and six carried the 8-bp deletion aromatic allele (Table 2). The distribution of the aromatic rice varieties that contain the two different aromatic alleles in different geographical regions of Myanmar is shown in Fig. 5.
The geographical locations of the aromatic varieties collected in Myanmar. The number of varieties collected from each region is shown. (filled diamond) Collection site of aromatic rice varieties with the 3-bp insertion. ( filled circle ) Collection site of aromatic rice varieties with the 8-bp deletion
Discussion
A single recessive gene located on chromosome 8 controls the aroma trait in rice. After the gene was identified, many scientists have studied a large collection of aromatic rice varieties. The 8-bp deletion in exon 7, which leads to 2AP accumulation in aromatic rice, has been found to be the major allele in most aromatic accessions, and other types of mutations with less frequency have also been reported (Bradbury et al. 2005a; Bourgis et al. 2008; Shi et al. 2008; Kovach et al. 2009). Interestingly, the present study found that there were twice as many Myanmar aromatic rice varieties containing the 3-bp insertion allele as those containing the 8-bp deletion allele. It is likely that the aromatic rice varieties with the 3-bp insertion in the Os2AP gene, which results in a tyrosine insertion into the protein at position 420 of the 503 amino acid protein, contain less 2AP in the grains compared to the varieties that contain the 8-bp deletion. The Os2AP expression level in the aromatic varieties with the 3-bp insertion was not reduced compared to the non-aromatic variety, which suggests that the Os2AP protein could be produced in these aromatic lines. However, a recent study has demonstrated that the enzyme encoded by Os2AP from aromatic rice containing the 3-bp insertion exhibits less catalytic efficiency toward 4-aminobutanal (Wongpanya et al. 2011). The tyrosine (Y) insertion at position 420 could alter the conformation of a loop located near the NAD+ binding site and result in a slight reduction of the binding affinity for NAD+ and substrates (Wongpanya et al. 2011). The mutation may account for the accumulation of 1-pyrroline, the immediate substrate of 2AP and the generation of aroma in the Myanmar aromatic rice. It is possible that the Os2AP gene with the 3-bp insertion can partially function and that the Os2AP gene with the 8-bp deletion cannot function at all, which may explain the lower 2AP level in the varieties containing the 3-bp insertion compared with the variety containing the 8-bp deletion (KDML105).
The identification of multiple mutations for 2AP enables rice breeding programs to actively select for multiple genetic sources of 2AP in different aromatic rice varieties. The availability of a functional marker to detect the different mutations will facilitate marker-assisted selection aimed at developing the aromatic rice varieties. A functional marker to detect the 3-bp insertion was developed in this study. The validity of the newly developed marker, 3In2AP, was tested on F2 segregation lines of the Pathein Nyunt and ISL-10 cross. The 3In2AP marker predicted the phenotype of all the F2 plants segregated for aroma, and it clearly distinguished the homozygotes from heterozygous F2 plants. Due to its co-dominant nature, this marker can be potentially useful in testing large segregating materials of aromatic rice improvement programs through a marker-assisted breeding program. Moreover, the assay for genotyping rice aroma using the 3In2AP marker is a simple and robust method for screening rice populations for aroma. The PCR products can be easily and inexpensively analyzed by polyacrylamide gel electrophoresis (PAGE) with silver staining or by sophisticated high-throughput experiments.
The screening of aromatic varieties from Myanmar demonstrated that the majority of aromatic rice from seven different Myanmar regions had the 3-bp insertion in exon 13 and that several varieties from three other regions had the 8-bp deletion in exon 7. Most of the prominent aromatic varieties, such as Paw San Hmwe (MSB 2620), Paw San Yin (MSB 1128) and Paw San Bay Kyar (MSB 804), and landrace varieties, such as Pathein Nyunt (IRGC 33552) and Yangon Saba (IRGC 33858), contained the 3-bp insertion. The collection site of these varieties with the 3-bp insertion aromatic allele was the delta region of Myanmar. Moreover, the Paw San Bay Kyar (MSB 1863) and Taung Pyan Yin (MSB 1791) varieties collected from the same delta region carried the aroma allele with the 8-bp deletion. The locally adapted aromatic rice varieties (sticky aromatic rice) in the Eastern Plateau region, Kauk Hnyin Phyu (MSB 6097) and Kauk Hnyin Net (MSB 6801), contained the 3-bp insertion, and the Kauk Hnyin Hmwe (MSB 6701) variety contained the 8-bp deletion. In a recent study on the origin and evolution of aroma in rice, Kovach et al. (2009) reported that five Myanmar aromatic varieties, Paw San Hmwe (IRGC 97793), Paw San Hmwe (N/A), Yangon Saba (IRGC 33858), Emahta Longyu (IRGC 33064) and Balugyun IRRI (IRGC 32960), contain the 3-bp insertion and that three varieties, Bokehmwe (IRGC 32985), Ma Waine Ohn (IRGC 33357) and Paw San Hmwe (IRGC 33571), contain the 8-bp deletion in exon 7. The results of this study and previous studies demonstrate that the 3-bp insertion is the major aroma allele in Myanmar rice varieties and that most of the prominent aromatic rice varieties of Myanmar originated in the delta regions of Myanmar. In summary, the molecular information collected from Myanmar rice varieties can be used to study a large panel of aromatic rice varieties and their diversity at a whole genome level. Furthermore, the functional marker developed in this study can be used for aromatic rice breeding for improvement with respect to certain characteristics and also for the genetic resource conservation program.
References
Bourgis F, Guyot R, Gherbi H, Amabile I, Tailliez, Salse J, Lorieux M, Ghesquière A, Delseny (2008) Characterization of the major fragrance gene from an aromatic japonica rice and analysis of its diversity in Asian cultivated rice. Theor Appl Genet 117:353–368
Bradbury LMT, Fitzgerald TL, Henry RJ, Jin Q, Waters DLE (2005a) The gene for fragrance in rice. Plant Biotech J 3:363–370
Bradbury LMT, Henry RJ, Jin QS, Reinke RF, Waters DLE (2005b) A perfect marker for fragrance genotyping in rice. Mol Breed 16:279–283
Bradbury LMT, Gillies SA, Brushett DJ, Waters DLE, Henry RJ (2008) Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol Biol 68:439–449
Buttery RG, Ling LC, Juliano BO (1982) 2-Acetyl-1-pyrroline: an important aroma component of cooked rice. Chem Ind (Lond) 12:958–959
Chen SH, Wu J, Yang Y, Shi WW, Xu ML (2006) The fgr gene responsible for rice fragrance was restricted within 69 kb. Plant Sci 171:505–514
Dhulappanavar CV (1976) Inheritance of scent in rice. Euphytica 25:659–662
Fitzgerald MA, Sackville Hamilton NR, Calingacion MN, Verhoeven HA, Butardo VM (2008) Is there a second fragrance gene in rice? Plant Biotech J 6:416–423
Ghose RLM, Butany WT (1952) Studies on the inheritance of some characters in rice (Oryza sativa L.). Indian J Genet Plant Breed 12:26–30
Hien NL, Yoshihashi T, Sarhadi WA, Hirata Y (2006) Sensory test for aroma and quantitative analysis of 2-acetyl-1-pyrroline in Asian aromatic rice varieties. Plant Prod Sci 9:294–297
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Khush GS, Brar DS, Virk PS, Tang SX, Malik SS, Busto GA, Lee YT, McNally R, Trinh LN, Jiang Y, Shata MAM (2003) Classifying rice germplasm by isozyme polymorphism and origin of cultivated rice. IRRI Discussion Paper Series No.46. International Rice Research Institute, Los Banos, p 279
Kovach MJ, Calingacion MN, Fitzgerald MA, McCouch SR (2009) The origin and evolution of fragrance in rice (Oryza sativa L.). Proc Natl Acad Sci USA 106(34):14444–14449
Lorieux M, Petrov M, Hunag N, guiderdoni E, Ghesquière A (1996) Aroma in rice: genetic analysis of quantitative trait. Theor Appl Genet 93:1145–1151
Mahatheeranont S, Keawsa-Ard S, Dumri K (2001) Quantification of the rice aroma compound, 2-acetyl-1-pyrroline, in uncooked Khao Dawk Mali 105 brown rice. J Agric Food Chem 49:773–779
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nakagahra M (1978) The differentiation, classification and center of genetic diversity of cultivated rice by isozyme analyses. Trop Agr Res Ser 11:77–82
Niu X, Tang W, Huang W, Ren G, Wang Q, Luo D, Xiao Y, Yang S, Wang F, Lu BR, Gao F, Lu T, Liu Y (2008) RNAi-directed downregulation of OsBADH2 results in aroma (2-acetyl-1-pyrroline) production in rice (Oryza sativa L.) B. BMC Plant Biol 8:100
Petrov M, Danzart M, Giampaoli P, Fayre J, Richard H (1996) Rice aroma analysis: discrimination between a scented and non-scented rice. Sci Aliments 16:347–360
Shi W, Yang Y, Chen S, Xu M (2008) Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol Breed 22:185–192
Sood BC, Siddiq EA (1978) A rapid technique for scent determination in rice. Indian J Genet Plant Breed 38:268–271
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vanavichit A, Tragoonrung S, Toojinda T, Wanchana S, Kamolsukyunyong W (2008) Transgenic rice plants with reduced expression of Os2AP and elevated levels of 2-acetyl-1-pyrroline. US patent No. 7,319,181
Wanchana S, Kamolsukyunyong W, Ruengphayak S, Toojinda T, Tragoonrung S, Vanavichit A (2005) A rapid construction of a physical contig across a 4.5 cM region for rice grain aroma facilitates marker enrichment for positional cloning. Sci Asia 31:299–306
Wongpanya R, Boonyalai N, Thammachuchourat N, Horata N, Arikit S, Myint KM, Vanavichit A, Choowongkomon K (2011) Biochemical and enzymatic study of rice BADH wild-type and mutants: an insight into fragrance in rice. Protein J 30:529–538
Wongpornchai S, Dumri K, Jongkaewwattana S, Siri B (2004) Effects of drying methods and storage time on the aroma and milling quality of rice (Oryza sativa L.) cv. Khao Dawk Mali 105. Food Chem 87:407–414
Yoshihashi T (2002) Quantitative analysis on 2-acetyl-1-pyrroline of an aromatic rice by stable isotope dilution method and model studies on its formation during cooking. J Food Sci 67:619–622
Acknowledgments
We gratefully acknowledge the financial support of the Thai-French Trilateral Development Cooperation and Joint Fellowship Program for Doctoral Students under the Royal Golden Jubilee Program (Grant No. PHD/0123/2550), from the Thailand Research Fund (TRF) to K.M. Myint, and the Agricultural Research Development Agency (public organization), Thailand. The authors would also like to thank the Plant Biotechnology Center at the Myanma Agriculture Service (Yangon, Myanmar) and the Department of Agricultural Research at Yezin (Nay Pyi Taw, Myanmar) for kindly providing the rice materials used in this study. All experiments in this study complied with the current Biosafety Guidelines of Thailand.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Guiderdoni.
Rights and permissions
About this article
Cite this article
Myint, K.M., Arikit, S., Wanchana, S. et al. A PCR-based marker for a locus conferring the aroma in Myanmar rice (Oryza sativa L.). Theor Appl Genet 125, 887–896 (2012). https://doi.org/10.1007/s00122-012-1880-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1880-0