Abstract
Allelic variability of the aroma gene, betaine aldehyde dehydrogenase 2 (BADH2) was studied in a random subset of indigenous aromatic rice germplasm along with a few exotic aromatic accessions. Use of functional markers of four badh2 alleles identified that the test panel possessed only two alleles, badh2-E7 and badh2-p-5′UTR. Two other alleles, badh2.2 and badh2-E4-5.2 were absent. Based on the alleles present, four functional polymorphisms (FP) were detected, namely FP1 to FP4. 188 genotypes possessed FP1 having both the badh2-p-5′UTR and badh2-E7 (71.8%) alleles. The badh2 allele with FP1 is named badh2-E7-p. 39 genotypes (14.9%) possessed only the badh2-p-5′UTR allele (FP3), while three genotypes were found to carry only the badh2-E7 allele (FP2). We also found that 32 genotypes (12.2%) did not have any of the target aroma alleles tested in this study (FP4). Interestingly, for badh2-p-5′UTR marker, the expected 198 bp amplicon for the non-aromatic allele could not be detected among any of the genotypes tested. Instead, an amplicon of 456 bp length appeared with 100% presence in the non-aromatic checks. Notwithstanding, the 456 bp allele also showed a 16% presence among the aromatic lines. This article forms the first report of this allele, named badh2-p1, among aromatic rice. Quantification of 2-Acetyl-1-Pyrroline (2AP) content and sensory evaluation among the test genotypes showed that those with FP1 are highly aromatic than the genotypes carrying other types of FPs. But, a few strongly aromatic lines showed lower 2AP content. The BADH2 characterization carried out in this study is suggestive of identifying the additional gene(s)/ allele(s) governing aroma among the Indian fragrant rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aromatic rice forms a special group of rice genotypes with exquisite grain and organoleptic properties. Owing to this, aromatic rice is gaining popularity in both domestic and international markets (Sakthivel et al. 2009). Confined to a small sub-group within the cultivated rice, aromatic rice includes several ecotypes, of which Basmati rice is the most popular. Mainly grown in South Asia, particularly in countries such as India, Pakistan, Thailand, Bangladesh, Vietnam, Indonesia, Afghanistan and Iran, aromatic rice is considered a premium commercial segment in rice. Basmati rice, in particular, is traded and consumed predominantly in the Gulf nations, the USA and Europe (Siddiq et al. 2012). Occupying around 20% of the total rice traded, aromatic rice fetches three times far more revenue than all the remaining rice put together (Singh et al. 2000). On the economic front, the export of aromatic rice helps several rice-growing nations to boost their international trade (Giraud 2013). In traditional rice growing countries, aromatic rice is grown in specific niches and used for preparing speciality food and ceremonial offerings (Sulochana and Singaravadivel 2015), assuming social and religious significance (Roy et al. 2015). For instance, Basmati rice is confined to the north-western part of Indo-Gangetic plains (Singh et al. 2018).
India is endowed with a huge diversity of short and medium grain aromatic rice (Malik et al. 1994). Although distributed throughout the country, the aromatic rice remains limited to those regions where climatic conditions are conducive for production and retention of the desirable aroma and grain quality (Prodhan and Qingyao 2020; Okpala et al. 2021). Since the aroma expression requires a milder temperature (Mahajan et al. 2018), in India, most of the aromatic rice cultivation is confined to areas where a cooler climate prevails especially during grain filling stages (Cruz et al. 1989), such as in the case of Indo-Gangetic plains and adjoining Himalayan hill districts, where Basmati cultivation is confined. According to a classification proposed by Glaszmann (1987), based on isozyme diversity, aromatic rice including Basmati cultivars are mostly placed in Group V. The Group V genotypes also show great variation in grain length ranging between short and long. Reported to be originated from the foothills of the Himalayas, aromatic rice belonging to Group V has its centre of diversity extending from northwest Indian plains to Myanmar as its eastern limit (Khush 2000).
Because of the low yielding nature of aromatic cultivars, the commercial availability of rice is greatly constrained making it an expensive commodity (Giraud 2013). Because of its growing consumer preference, more recently, genetic improvement of aromatic rice has been identified as a major breeding target (Peng et al. 2018). This has kindled the interest among the rice breeders to explore the underlying genetic basis of aroma and other grain characteristics. Although there were conflicting earlier reports on the genetics of aroma, such as monogenic inheritance (Kadam and Padankar 1938; Jodon 1944) or not (Tripathi and Rao 1979; Dhulappanavar 1976), Ahn et al. (1992) confirmed the role of a single recessive gene, fgr through RFLP analysis. Subsequently, fgr was identified to be an aberrant version of the functional gene for the enzyme, betaine aldehyde dehydrogenase 2 (BADH2) (Chen et al. 2008). Belonging to the aldehyde dehydrogenase family, BADH2 contains 503 amino acids. The corresponding 6154 bp long BADH2 gene which is located on the long arm of chromosome 8 and possesses 15 exons and 14 introns (Bradbury et al. 2005a; Kovach et al. 2009). Functional BADH2 inhibits the synthesis of 2-Acetyl-1-Pyrroline (2AP) rendering rice grains non-aromatic (Chen et al. 2008), by catalyzing the oxidation of γ-aminobutyraldehyde (GAB-ald), to γ-aminobutyric acid (GABA). GAB-ald is the substrate for the production of Δ1-pyrolline that gets converted into 2AP (Okpala et al. 2019). When a gene mutation renders the BADH2 enzyme non-functional, accumulation of GAB-ald occurs in plants driving its conversion to 2AP (Bradbury et al. 2008; Hinge et al. 2016). Although rice grain aroma is attributed to more than 114 volatile compounds (Tsugita 1985; Champagne 2008), 2AP stands out as the prominent molecule (Buttery et al. 1983, 1986, 1988). Specific studies on Basmati and Thai Jasmine rice recognized 2AP as the major compound responsible for the aroma (Lorieux et al. 1996). The contents of 2AP and GABA show an inverse relationship in aromatic rice having an elevated 2AP content, while non-aromatic rice shows high GABA content (Chen et al. 2008). These findings conclusively support that the badh2 gene is responsible for conferring fragrance in the rice. Another gene, OsBADH1, a homolog of OsBADH2, is located on the long arm of chromosome 4, but is known to be involved in germination stage salt tolerance (Hasthanasombut et al. 2010; He et al. 2015) and information on its involvement in rice aroma seems limited (Amarawathi et al. 2008).
The aroma causing allele of the OsBADH2 gene is recessive to non-aromatic allele. The major aromatic allele of the gene, fgr, is also known as badh2-E7 or badh2.1. These names are found used widely and interchangeably in literature, but the name badh2-E7 is preferred the most, indicating the presence of an 8 bp deletion in the seventh exon of the BADH2 gene. The deletion is discontinuous at two sites 5 bp and 3 bp, leading to a frameshift and a premature termination. The result is a non-functional truncated protein of 251 amino acids (Bradbury et al. 2005a; Bourgis et al. 2008). Although several recessive allelic variants have been subsequently reported in the BADH2 gene, badh2-E7 remains the most prominent mutation among Indian aromatic rice, including Basmati and Jasmine rice (Singh et al. 2011).
At least twenty badh2 aromatic alleles have been reported in rice by sequence-based analyses (Withana et al. 2020; Chan-In et al. 2020), that include mutations in the coding and non-coding regions. Most of them are exonic mutations, that include nine deletions (badh2.1, badh2.2, badh2.3, badh2-E2.1, badh2-E2.2, badh2-E4-5.1, badh2-E4-5.2, badh2.5, badh2-E12), five insertions (badh2-E8, badh2.4, badh2-E13.1, badh2.7, badh2.8) and five substitutions (badh2.6, badh2-E1.2, badh2-E10.4, badh2.10, badh2.9). Among these, ten allelic variants, designated serially as badh2.1 to badh2.10 were reported in diverse rice genotypes (Kovach et al. 2009). The most prevalent allele, badh2-E7 is named badh2.1 in this classification. Mutations in the non-coding region, include a promoter insertion and a deletion in the allele, badh2-p-5′UTR reported from the japonica cultivar, Nankai 138 (Shi et al. 2014). This allele has an 8 bp insertion in the promoter region upstream from the start codon between base positions 1314 and 1315 and a 3 bp deletion in the 5′ end of the untranslated region (UTR). Later, a variant of this allele (badh2-p) was reported from an aromatic short grain rice landrace, Seeragasamba (also known as Jeeragasamba), having only the 8 bp promoter insertion (Bindusree et al. 2017). Other minor alleles reported were, badh2-E10.4 with one single nucleotide polymorphism (SNP) in exon 10, badh2-E2.2 having a 75 bp deletion in exon 2 and badh2-E4-5.1 having an 806 bp deletion between exons 4 and 5 (Shao et al. 2013). Another novel allele, badh2-E12 with a 3 bp deletion in exon 12 has also been reported (He and Park 2015).
Several functional markers have been developed for the badh2 alleles to aid molecular differentiation of aromatic and non-aromatic rice, as well as to aid marker-assisted introgression of aroma trait. The markers for the badh2.E7 allele target the deletion in the seventh exon (Bradbury et al. 2005b; Amarawathi et al. 2008; Sakthivel et al. 2009; Xu et al. 2011). Among the markers developed for badh2.E7, ‘nksbad2’ developed by Amarawathi et al. (2008) has been demonstrated as the most efficient in differentiating aromatic and non-aromatic genotypes. The amplicon of nksbad2 is an 82 bp fragment among aromatic genotypes, while a 90 bp fragment is produced from non-aromatic genotypes. Two other functional markers targeting badh2-E7 were also reported by Shi et al. (2008). To target badh2-p-5′UTR, Shi et al. (2014) have developed a marker, badh2-p-5′UTR. This marker, however, could not distinguish both badh2-p and badh2-p-5′UTR, since the marker targeted only the 8 bp insertion. Hence, in this study, we use badh2-p-5′UTR to designate the 206 bp aromatic allele amplified by the marker, badh2-p-5′UTR. Markers were also developed reportedly targeting another allele, badh2.2 (badh2-E2) which possessed a 7 bp deletion in exon 2 (Shi et al. 2008). Shao et al. (2011) developed a functional marker, FMbadh2-E4-5, based on an 803 bp deletion between exon 4 and 5 (badh2-E4-5.2) while Myint et al. (2012) developed a functional marker based on a badh2-E13.1 allele that carried a 3 bp insertion in exon 13. Other badh2 allele-specific markers reported include Badh2.7CAPS, a cleaved amplified polymorphic sequence (CAPS) based marker, for the identification of genotypes with badh2.7 allele in the Sri Lankan rice germplasm (Dissanayaka et al. 2014).
Although a few of these markers were tested among the aromatic lines across the world, most of the reports from India remain largely limited to a few aromatic genotypes. Since India has a highly variable aromatic rice germplasm, the BADH2 allelic diversity among Indian aromatic rice needs to be explored as there is a greater possibility of finding novel allelic variants in the Indian rice gene pool. Recognising this gap as a potential shortfall, we in this study, examined a few known badh2 alleles among a panel of indigenous and exotic aromatic rice genotypes, using functional markers. The panel included aromatic genotypes comprising short grain, Basmati and exotic genotypes.
Materials and methods
Plant materials
A panel of 266 rice genotypes comprising 133 short grain aromatic genotypes (120 indigenous genotypes from India and 13 exotic genotypes originating from Thailand and the Philippines), 107 Basmati, 22 exotic long grain aromatic rice genotypes and four non-aromatic checks namely, Pusa 44, JGL1798, Pusa 677 and Sharbati (Supplementary Table S1) were used in the study. The lines were field grown during Kharif 2018, the season between June to November, following an augmented block design with checks replicated three times. For raising the trial, the genotypes were germinated in a flat-bed nursery and transplanted after 25 days. The genotypes were line-transplanted (2 lines/genotype) during the first week of June maintaining a spacing of 0.15 m between the plants and 0.20 m between the rows. The experiment was managed with recommended agronomic practices (kvk.icar.gov.in/API/Content/PPupload/k0306_1.pdf). At maturity, the grains were harvested from five plants individually and dried under shade to 12% moisture level. After drying, the single plant harvest was cleaned off all the debris and non-filled grains. 20 g of filled grains from each plant was then pooled to constitute the 100 g bulk seed sample per genotype and stored at 4 °C in the refrigerator. The seeds were kept packed in Ziploc® freezer bags to prevent the loss of aroma till further processing and analysis.
Genotyping
To detect functional polymorphism (FP) in the BADH2 gene, aromatic rice genotypes were characterized for four alleles, badh2-E7, badh2-p, badh2.2 and badh2-E4-5.2 using corresponding functional markers (Table 1). DNA was extracted from leaf tissue of 15 days old seedlings from all the genotypes using the standard protocol (Murray and Thompson 1980), and their quality and quantity were checked on 1% agarose gel electrophoresis using the uncut λ-DNA ladder as standard. All the markers shared a similar annealing temperature (55 °C) and hence were subjected to the same amplification profile in the polymerase chain reaction (Ellur et al. 2016) using a Veriti™ Dx 96-Well Thermal Cycler. Diluted DNA of 25 ng/µl was used for PCR amplification using a standard portfolio. The amplicons were resolved in gel electrophoresis using 4% agarose containing 0.1% ethidium bromide and visualized under UV transillumination in a gel documentation system, GelDoc™ XR (Bio-Rad Laboratories Inc., USA). Based on the allelic pattern among the genotypes, functional polymorphisms (FP) were determined.
Sensory assessment of aroma
The stored seeds were evaluated for aroma one month after harvesting. For the determination of aroma, grains were dehusked and milled using a palm husker and 1 g of milled rice kernels were kept in 10 ml of 1.7% potassium hydroxide (KOH) at room temperature in covered petri plates for 10 min (Sood and Siddiq 1978). Sensory evaluation and scoring of the aroma were done by a panel of three experts. Each accession was scored on a 0–2+ scale, where 0 stands for the absence of aroma, 1 for mild aroma, 1+ for mild to moderate aroma, 2 for moderate aroma and 2+ for strong aroma. Non-aromatic rice genotypes, Pusa 44, Sharbati, Pusa 677 and JGL1798 were used as negative controls.
Quantification of 2AP
Based on the FP variation, 28 genotypes approximately representing 10% of the panel, and all the allelic combinations detected were used for quantification of 2AP. The analytical measurement employing gas chromatography-mass spectrometry (GC–MS) was used for the estimation of 2AP in rice (Hinge et al. 2016). 100 g seeds in storage for 30 days from each genotype were hulled using a laboratory huller and milled. The milled rice samples were then ground to a fine powder and 5 g of the powder was taken in a tightly sealed 20 mL glass vial to prevent the escape of volatiles. The vials were heated to 80 °C for 20 min and the separation of volatiles was carried out on a TG-5MS capillary column (30 m, 0.25 mm) used in a DSQII GC–MS system (ThermoFisher Scientific) equipped with a headspace autosampler. The oven temperature was kept at 35 °C initially for 5 min, which was then increased to 240 °C @ 3 °C/min and was held at 240 °C for 5 min. The inlet port was maintained at 250 °C in splitless mode with a constant 1 ml/min flow of 99.9% Helium gas. The ion source and interface temperatures were kept at 250 °C and 270 °C, respectively. The electron impact energy was set to 70 eV, and a range from 35 to 350 m/z was applied for the scanning. The GC–MS analysis was performed in triplicates for all the samples. The retention time of 2AP was recorded at 7.9 min. Peak areas were determined by integrating the peaks corresponding to 2AP (at m/z 83) in selected ion monitoring (SIM) mode. The 2AP in the rice samples was quantified by comparing peaks areas with that of the 2AP standard.
Sequence analysis of promotor mutation in badh2
The amplicons obtained from badh2-p-5′UTR were compared for the promoter sequence variation among 14 genotypes falling in different FP classes. FP1 and FP4 classes included six genotypes each, while the FP3 class was represented by two genotypes. The amplicons were eluted from the gel using standard protocols and Sanger sequenced using ABI 3730xl DNA Analyser (ThermoFisher, ABI 3730 XL DNA Analyser). The sequences are compared using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), a new multiple sequence alignment program (Madeira et al. 2019).
Comparative analysis of the single nucleotide variation in BADH2
Additionally, whole genome SNP data of a total of 76 aromatic rice varieties were collected from the 3K rice genome project (The 3,000 rice genomes project 2014). From the database, the SNP variants present in the BADH2 gene spanning the region from 20,38,0233 to 20,38,6542 bp of Chromosome 8 in all the 76 aromatic rice varieties were analysed. The SNPs from the genotypes were further categorized as synonymous (causes no change in amino acid) and non-synonymous (causes a change in amino acid). They were further designated as transitions (C/T and G/A) and transversions (C/G, T/A, A/C and G/T) SNPs. There were six genotypes among this set, that were included in the test panel in this study. SNPs were also compared for these genotypes.
Statistical analyses
The data were subjected to standard statistical analyses, including analysis of variance (ANOVA), mean comparison and correlation studies. ANOVA has been carried out using the completely randomised model, while group-wise analysis was carried out by the single factor approach. Pearson correlations were used for striking the relations between different factors. All the analyses were carried out using the R statistical environment (Base package).
Results
Variation in badh2 alleles in the aromatic rice genotypes
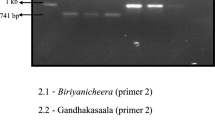
The genotyping results showed that, among the four functional markers used only two could reveal polymorphisms among the 262 aromatic genotypes (Supplementary Table S1). These markers were nksbad2 and badh2-p-5′UTR targeting the alleles badh2-E7 and badh2-p-5′UTR, respectively. Both badh2-E2 and badh2-E4-5 alleles were absent in the study panel (data not presented). The marker, nksbad2 produced two amplicons, 82 bp and 90 bp, respectively corresponding to the aromatic and non-aromatic alleles. Whereas, the marker badh2-p-5′UTR produced amplicons of size 206 bp and 456 bp (Fig. 1). The 206 bp fragment represented the aromatic allele. Although badh2-p-5′UTR was expected to produce a non-aromatic allele of 198 bp, no genotype in the present panel was found to amplify this fragment. Instead, a new allele of 456 bp was amplified with this marker, representing the non-aromatic allele. All four non-aromatic checks showed non-aromatic alleles for both the markers. Based on different combinations of alleles at two loci, four FPs were identified, FP1, FP2, FP3 and FP4 (Table 2). FP1 had a combination of two aromatic alleles, badh2-E7 and badh2-p-5′UTR, FP2 had badh2-E7 alone, FP3 had only badh2-p-5′UTR and FP4 had a combination of non-aromatic alleles from both the markers.
Representative gel picture for badh2-E7 and badh2-p-5′UTR allele in the aromatic genotypes resolved using markers nksbad2 and badh2-p-5′UTR, respectively. 82 bp and 90 bp are the aromatic and non-aromatic allele, respectively for the marker, nksbad2; 206 bp and 456 bp are the aromatic and novel non-aromatic allele, respectively for badh2-p-5′UTR. Two most common badh2 alleles present in the test panel were badh2-E7 and badh2-p-5′UTR. The allele possessing both the recessive mutations is renamed as badh2-E7-p. The 456 bp allele is named as badh2-p1. The genotypes are, 1. Dindli, 2. Sonachoor, 3. Kalanamak 1, 4. Pusa Basmati 1, 5. Gayasu, 6. Tulsiful, 7. Koliha, 8. Basmati Mehtrah, 9. Bastul, 10. Karnal Local, 11. Basmati 370, 12. Longku Labat, 13. Kalabhutia, 14. Haldi Chudi, 15. Popot, 16. Basmati 397, 17. Hansraj, 18. Anterved, 19. Hasan Serai, 20. Della, 21. Vasumati, 22. Dudhkhasa, 23. Basmati 376, 24. Kalimooch
Analysing the different classes of aromatic lines used, both badh2-E7 and badh2-p-5′UTR alleles were found in indica and japonica genotypes. In the panel, 175 genotypes (66.8%) exclusively possessed the badh2-E7 allele (82 bp), while a total of 71 genotypes were found to have the alternate allele (90 bp) (Table 2). Sixteen genotypes were heterozygous for this marker. The second marker, badh2-p-5′UTR, identified 220 genotypes (84.0%) possessing the badh2-p-5′UTR allele (206 bp), while 35 aromatic rice genotypes showed the alternate allele of 456 bp. Besides, seven heterozygotic genotypes amplifying both 206 bp and 456 bp bands were identified. The distribution of FPs among the test panel indicated that FP1 was present among 174 genotypes (66.4%), FP2 in one genotype, FP3 in 36 genotypes and FP4 in 32 genotypes. Within the FP1 group, there were 70 indigenous short-grain (ISG) aromatic lines, three exotic short-grain (ESG) lines, 82 Basmati (BAS) lines, and 19 exotic long-grain (ELG) lines. The single FP2 genotype was a Basmati cultivar, UPR 2828-7-2-1. The genotypes that carried FP3 included 18 ISG aromatic lines, five ESG genotypes, and 13 Basmati genotypes. The FP4 group included 32 aromatic genotypes comprising 18 ISG aromatic lines, four ESG lines, eight Basmati genotypes, and two ELG genotypes. Interestingly, the aroma pattern of the FP4 group displayed 26 mildly aromatic genotypes, five mild to moderately aromatic lines and one moderately aromatic genotype. Nineteen genotypes exhibited heterozygosity at either one or both the loci, resulting in combinations of FPs within an individual. There were ten genotypes with FP1/FP3 combination, three genotypes with FP3/FP4 combination, two genotypes (PKV Makarand and IR 75428-6-3) with FP2/FP3 and four genotypes having FP1/FP2/FP3/FP4. The last group had a heterozygous pattern at both the loci.
Fragrance variation among functional polymorphisms
The aroma assessment among the 28 genotypes representing four FPs, revealed distinct variations in aroma identified by sensory evaluation and grain 2AP content (Table 3). The 2AP content showed a highly significant correlation with the sensory score for aroma (r = 0.52). Among the markers, nksbad2 could resolve high aromatic and low/non-aromatic lines more efficiently than badh2-p-5′UTR. The most common FP, the FP1, had the highest average 2AP content of 32.0 ppb with a range of 15.7–90.5 ppb. Average scores based on sensory evaluation also indicated aroma ranging from mild to moderate (1 +) to strong (2 +) among the FP1 genotypes. The FP2 genotype (UPR 2828-7-2-1) had a 2AP content of 3.9 ppb with an aroma score of 1. The FP3 genotypes had the second-highest average 2AP content of 10.6 ppb with a range between 0.3 and 54.1 ppb. The aroma score of the FP3group ranged between 1 and 1+. The FP4 genotypes were predominantly mild aromatic with a sensory score ranging between zero and 1+. The average 2AP content among this group of genotypes was 1.3 ppb. Based on the presence of different alleles, the average 2AP content of genotypes carrying badh2-E7 (82 bp amplicon) was 28.5 ppb as against 34.8 ppb of those carrying the badh2-p-5′UTR alleles. The variation at an individual locus could be ambiguous, as their combination was found to produce better 2AP content and aroma, and different combinations of FPs were found to explain the variation better than the individual locus.
Among the genotypes, Nawab Bhog, an ISG genotype showed the highest 2AP content of 216.5 ppb and possessed all the FP combinations (Table 4). The second highest content of 2AP was recorded in Tulsiful, another ISG genotype with a 2+ sensory score, possessing a combination of FP1/FP3 (Fig. 2). The 2AP content in all the genotypes with FP1 was high, with two Basmati lines, IET 15835 and IET 13548 exhibiting the highest 2AP content of 90.5 and 44.0 ppb, respectively. Among the FP3 genotypes, interestingly, the ISG landrace, Kalanamak exhibited the highest 2AP content of 54.1 ppb as compared to the other genotypes with the same allelic profile. The remaining FP3 genotypes had relatively lower 2AP content that ranged from 0.3 ppb (RAU3055) to 5.1 (Basmati 370). All the non-aromatic checks used in the study showed an FP4 allelic profile.
Estimation of 2-Acetyl-1-Pyrroline (2AP) content using gas chromatography coupled mass spectrometry (GC–MS). The figure shows a representative GC–MS profile obtained from the aromatic genotype, Tulsiful. The peak retention time (7.90) for 2AP is highlighted in red. The remaining peaks indicate other volatile compounds present in the extract. 2AP is the compound responsible for aroma in rice grains
Sequence variation in the BADH2 promoter region
The sequences of the badh2-p-5′UTR amplicons confirmed the presence of the 456 bp allele among FP groups (Table 5). The FP1 and FP3 possessed an 8 bp insertion in the promoter region, but with a 252 bp deletion (Fig. 3). The FP 1, represented by the genotypes such as Pusa Basmati 1, Vallabh Basmati 22, Lal Basmati, KDML 105, Gangaballi and Jao Mali showed sequence similarity with the FP3 genotypes, Sonachoor and Dindli. All the genotypes belonging to FP1 and FP3 were aromatic types and showed significant promotor sequence variation from FP4 genotypes. Represented by six genotypes viz., Basmati Mahon 381, Basmati Surkh 161, Jalaka, AS-GPC-38, Sharbati and Pusa 44, the FP4 class, possessed a distinct 252 bp insertion in the region. It is interesting to note that, the first four genotypes in the FP4 are aromatic and the latter two (Sharbati and Pusa 44) were non-aromatic.
Sequence alignment of the genotypes for the amplicons of badh2-p-5′UTR marker. Functional polymorphisms (FP) show variation in alleles such as 252 bp insertion in FP4 (badh2-p1) highlighted in green and an 8 bp insertion (highlighted in yellow) in FP1 and FP3 (badh2-p-5′UTR). * denotes aligned sequences and – indicates deletion
SNPs in the BADH2 locus
A total of 15 SNPs was identified from the 3K SNP database among the six genotypes used in the present study with different badh2 alleles (Table 6; Supplementary Table S3). These genotypes included five aromatic genotypes (Basmati Surkh 161, Karnal Local, Hansraj, Basmati 385, Hasan Serai) and one non-aromatic genotype, Pusa 677. Among the 15 SNPs, 14 were synonymous and one was non-synonymous. The non-synonymous SNP identified at the position 20,38,2857 was a base transversion from T to A. Interestingly, this SNP showed distinct variation between aromatic and non-aromatic types with T in the former and A in the latter. Compared to the SNPs identified among the 76 aromatic genotypes in the 3K panel, however, this SNP did not show any conspicuous variation. Rather it was found that both the alleles at this SNP locus had a near equal distribution among the genotypes. Further, the 3K database did not provide the opportunity to study the promoter sequence variations in the badh2-p allele, because the sequencing depth was shallow (~ 7x) and the insertion length was large (253 bp).
Discussion
Despite being geographically limited within the subcontinent, Indian aromatic rice germplasm show extensive phenotypic diversity for various agronomic traits and adaptation (Ray et al. 2013). Several of these genotypes have had limited cross-breeding opportunities for being adapted to isolated geographic niches, accumulating specific alleles in the evolutionary process. However, information on the allelic diversity causing aroma variation remains limited among this group of rice cultivars. In aromatic rice, functional impairment of BADH2 gene due to mutations has been primarily implicated in the development of several allelic variants (Shao et al. 2011). This prompted us to look for the variation in the BADH2 locus, to understand the fragrance diversity among the Indian aromatic rice.
Origin of badh2 alleles
Recent investigations on the origin and evolution of rice aroma indicate a monophylogenetic origin for badh2 alleles (Bourgis et al. 2008; Kovach et al. 2009). The non-aromatic wild type allele, BADH2 has as many as twenty recessive allelic variants for aroma (Withana et al. 2020). Kovach et al. (2009) concluded that the japonica cultivar, Azucena share the same aroma haplotype with indica cultivars carrying the badh2-E7 allele, implying the origin from an ancient japonica source and its subsequent introgression to indica. Tracing the badh2 allele evolution, Shi et al. (2014) also emphasised that a single source phylogeny could explain the wide prevalence of badh2-E7 and badh2-p-5′UTR alleles. Furthermore, the presence of multiple alleles of the BADH2 gene also implies that mutations are key in evolving fragrance variation in rice. But, there was no information on how many of these variants are found in Indian rice.
Allelic variation for badh2 in aromatic rice
The badh2-E7 has been the most ubiquitous allele reported among the Indian aromatic genotypes. In a study using 24 rice genotypes, five fragrant genotypes namely Kalanamak 3119, Kasturi Basmati, Basmati LC 74-3, Tharunbhog and Jeeragasamba were reported not to possess the badh2-E7 allele (Rai et al. 2015). Similarly, out of 84 indica rice landraces validated for badh2-E7, 11 did not show the presence of this allele despite being aromatic (Chakraborty et al. 2016), suggesting the presence of other known or unknown mutations among Indian landraces. Further, the presence of badh2-p allele identified in Jeeragasamba could confirm that the 8 bp insertion is the key for aroma expression, even in the absence of badh2-E7 allele (Bindusree et al. 2017). They reported the presence of the 8 bp insertion prevalent among 30 out of the 76 aromatic landraces, further confirming it as the aromatic allele. In the current study too, we have identified 36 genotypes having only badh2-p-5′UTR but aromatic, including Basmati 370 and Kalanamak.
Being the most prevalent allele, the first step in our approach was to preclude the carrier genotypes of badh2-E7 from the test panel, so that other allelic variants could easily be identified. By this approach, we could identify that 72.9% of the panel possessed the badh2-E7 allele. However, when we screened the entire panel for the badh2-p-5′UTR allele in the next step, we not only could find that badh2-p-5′UTR was the most abundant allele having 86.6% prevalence, but also that 71.8% of the genotypes possessed both the alleles, badh2-p-5′UTR and badh2-E7 together enabling them to be classified under the FP1 group. Surprisingly, the proportion of genotypes that carried either of the alleles singly was low; 13.7% for badh2-p-5′UTR (FP3), and < 1% for badh2-E7 (FP2). To distinguish the FP1 allele having both the mutations together from those with single mutations (FP2 and FP3), we named the FP1 allele badh2-E7-p. Compared to the earlier report by Bindusree et al. (2017), the test panel in this study had a relatively higher proportion of both badh2-p-5′UTR and badh2-E7 alleles. Bindusree et al. (2017) reported that badh2-p had more prevalence (~ 40%) than badh2-E7 (~ 34%), either alone or in combination among 76 aromatic lines they have been studied. They have also reported the presence of badh2-p in both indica and japonica lines. However, this reported proportion could also be inclusive of badh2-p-5′UTR, because the BADH2 gene variant they studied included a -1326 bp upstream region. We could notice that badh2-E7 to badh2-p-5′UTR ratio in both the studies remained almost similar, with 0.87 in the earlier study and 0.84 in the current. Also, the proportion of genotypes with the badh2-E7-p allele was high in our study (71.8%) as against 22.4% reported by Bindusree et al. (2017). The reason for the reduced presence of badh2-E7 in Bindusree et al. (2017) could be attributed to the set of 76 aromatic genotypes that were selected from the 3000-genome panel (The 3,000 rice genomes project, 2014). This set, which included rice genotypes collected from 89 countries, represented 33.9% of Southeast Asia, 25.6% of South Asia and 17.6% of China. Whereas, our test panel was large (266 genotypes) and comprised prominently of Indian aromatic rice (86.6%), possibly leading to a high proportion of badh2-E7-p. This observation reinforces the earlier theory that the Group V genotypes to which most of the Indian aromatic rice belongs, remain highly conserved in their centre of origin (Glaszmann 1987; Khush 2000). We find that the two alleles, badh2-E7 and badh2-p-5′UTR, were present either singly or in combination among 87% of indigenous lines and 82.8% of the exotic lines. Among the indigenous set, 73.6% possessed the badh2-E7 allele, while 87.7% possessed the badh2-p-5′UTR allele. The proportion of indigenous genotypes that possessed the badh2-E7-p was 73.1%. However, there were 26 indigenous genotypes and six exotic types, that did not possess both the alleles (FP4), 18 of which belonged to the short-grain types, but with a mild aroma. These genotypes deserve further investigation for identifying the causal mutations. Among the exotic genotypes too, the allele status remained almost similar to that of indigenous genotypes, wherein 65.7% possessed badh2-E7, 80.0% carried badh2-p-5′UTR and 62.9% carried badh2-E7-p allele. Therefore, we conclude that the aroma variability in the current test panel was confined predominantly to the presence of two alleles, badh2-E7 and badh2-p-5′UTR and showed four FPs. In a recent report, Withana et al. (2020) identified badh2.7 as the next most prevalent allele in Asian rice after badh2-E7. However, they have not tested for the presence of the badh2-p allele. This allele, badh2.7 has an insertion of G in the 14th exon (Dissanayaka et al. 2014).
As observed in this study, not all the variations in the gene may cause aroma variation, such as the presence of a 456 bp amplicon among some of the aromatic and non-aromatic genotypes. Despite the use of a wide array of genotypes including tropical japonica cultivars, the expected non-aromatic amplicon for the marker, badh2-p-5′UTR, a 198 bp fragment, was not detected in this study. We consider this 456 bp allele novel because its presence among aromatic lines has never been reported before. We designate this allele as badh2-p1. There was only one instance of the report of badh2-p1 earlier, that too in a non-aromatic indica landrace, Velchi (Khandagale et al. 2017). The wildtype BADH2 does not possess the 8 bp insertion in the promoter region as found in the badh2-p-5′UTR allele (Shi et al. 2014). Remarkably, the allele reported by Khandagale et al. (2017) has an insertion of 258 bp in the promoter region leading to a 456 bp fragment but is devoid of the 8 bp insertion as that in badh2-p-5′UTR. However, the sequence analysis of the amplicons of the BADH2-p1 confirmed the presence of a consistent 252 bp insertion in the promoter region. This insertion was highly conserved among all the FP4 genotypes. Further, having found in all non-aromatic checks, as well as in some of the aromatic lines we conclude that BADH2-p1 was not particularly associated with the aroma. The role of badh2-p-5′UTR allele in imparting aroma among the FP1 and FP3 genotypes needs further investigation. We have found combinations of BADH2-p1 with both aromatic (82 bp) and non-aromatic (90 bp) fragments of badh2-E7. However, mild aroma noticed among the genotypes with non-aromatic allelic combinations may warrant additional investigations to elucidate underlying mechanisms.
The relationship between badh2 alleles and aroma
Among the genotypes with badh2-E7-p, the presence of the badh2-p-5′UTR allele was supplementary since the badh2-E7 leads to a truncated BADH2 protein. We could observe that the presence of both the alleles in FP1 imparted a strong aroma, with an average content of 32.0 ppb of 2AP. On the other hand, in the absence of the badh2-p-5′UTR allele, the badh2-E7 allele could produce only 3.9 ppb of 2AP with a mild to moderate aroma expression, indicating the prominence of having both the mutations together in aroma production. However, the badh2-p-5′UTR allele alone could produce an average 2AP content of 10.6 ppb, which is higher than that of badh2-E7, but also with mild to moderate aroma. Therefore, the potential role of promoter insertion in badh2-p-5′UTR in imparting aroma is undeniable but may be influenced greatly by other loci and/or external factors. This could be the reason for the higher variability of 2AP content in badh2-p-5′UTR carriers (FP3). A similar case was reported earlier in the japonica rice, Nankai 138, in which the badh2-p-5′UTR allele was found to yield elevated 2AP content (Wang et al. 2016). One of the major factors that influence gene expression under promoter mutation is the presence of cis-acting elements. Besides, the distance between the two cis-acting elements, their type, orientation and number can also play a crucial role (Hernandez-Gracia and Finer 2014). Hence, in Kalanamak and Basmati 370, it is a matter of investigation whether the presence of 8 bp promoter insertion is influenced by the presence of cis-acting elements leading to higher but varying levels of 2AP. Nevertheless, the possibilities of having additional gene(s)/ mutation(s) imparting aroma in Kalanamak have also been opined by Rai et al. (2015). Similarly, a significant role of promoter induced aroma gene expression imparting a wide variation cannot be ruled out among the FP1 genotypes and the genotypes having polymorphic combinations that included badh2-p-5′UTR. Remarkably, the presence of the 456 bp amplicon among 32 FP4 aromatic genotypes and four non-aromatic checks point towards the complexity of mutations in the promoter region in the BADH2 locus. It also implicates the existence of novel mutations hitherto not discovered in the BADH2 locus. Besides, there could also be other loci responsible for aroma variation in the FP4 genotypes. Since the marker badh2-p-5′UTR could detect the badh2-p1 allele, it can be concluded that this marker can distinguish multiple mutations in the promoter region. Therefore, this region warrants extensive investigations in the future for its role in aroma variability in the rice gene pool. Although little, yet, we have encountered 7.1% of genotypes with heterozygosity in either or both the FPs. This could be due to the natural heterogeneity observed in the indigenous landraces that were under community conservation, without undergoing genetic purification. These admixing can only be avoided by continuously controlled selfing for a few more generations. This calls for the purification of such genotypes under conservation breeding.
As described earlier as well as in this study, 2AP is unequivocally the major compound responsible for aroma in rice. Juxtaposing the sensory evaluation and 2AP content in the present aromatic panel, revealed a correlation of 0.52 between the two parameters. Strong correlations are seen in Basmati genotypes such as IET 13,548 and IET 15,835 which had a sensory scoring of two and 2AP values of 44 ppb and 90.5 ppb, respectively. Both these genotypes carried the badh2-E7-p allele. The values of 2AP obtained are comparable to the earlier reported values of 60 ppb (Buttery et al. 1983), and 61 ppb (Nadaf et al. 2006) in Basmati genotypes, however 2AP content as high as 588 ppb has also been reported (Tava and Bocchi 1999). Although found to be associated with non-aroma, the carriers of the allele BADH2-p1 also had lower levels of 2AP among the genotypes with a mild aroma.
SNP variation in badh2 gene
More than 85% similarity for the SNPs identified for the aromatic genotypes identified in the present study with the aromatic genotypes in the 3K database points toward the conservation of the SNPs in the aromatic genotypes. Coming to the occurrence of non-synonymous mutation (T to A) in the non-aromatic genotype Pusa 677 with the BADH2-p1 allele, its presence was detected in 50% of the aromatic genotypes of the 3K panel. Therefore, it cannot be considered conclusive evidence for providing aroma. However, it was absent in the three aromatic genotypes possessing the BADH2-p1 allele and two genotypes with badh2-E7 and badh2-p-5′UTR alleles used in the current study. The absence of this deletion in fragrant Hasan Serai could be a possible mutation conferring aroma and coincides with the earlier report of the non-aromatic nature of the BADH2-p1 allele (Khandagale et al. 2017). This indicates the importance of the genotypes with BADH2-p1 allele and its screening in the aromatic genotypes. Further studies on the mutation reported in this study could pave the way for revealing the aroma pattern in the genotypes possessing the BADH2-p1 allele.
Conclusion
The present study characterized the allelic variations at BADH2 locus amongst a diverse set of fragrant rice germplasm predominantly belonging to the Indian sub-continent along with a few exotic aromatic genotypes. A high frequency of occurrence of the badh2-p-5′UTR allele (86.6%) was observed in the aromatic rice landraces as compared to the badh2-E7 allele (72.9%). However, there was a significant portion of genotypes that possessed both the alleles together (badh2-E1-p). Genotypes with the badh2-E7 allele exhibited higher levels of 2AP correlating well with the sensory evaluation. The presence of BADH2-p1 allele of 456 bp with a 252 bp promoter insertion among the aromatic genotypes, was confirmed with the sequence analysis. The consistent presence of this allele among the non-aromatic genotypes points towards its non-involvement in conferring the aroma. Our observations implicate the complexity of allelic variation in BADH2 locus, which requires in depth studies on how they drive differential aroma levels in some of the aromatic genotypes. This leads to the necessity of further elucidation of badh2 diversity in aromatic rice.
Availability of data and materials
All the data used in this paper are provided in the supplementary information.
Code availability
Not applicable.
References
Ahn S, Bollich C, Tanksley S (1992) RFLP tagging of a gene for aroma in rice. Theor Appl Genet 84:825–828. https://doi.org/10.1007/BF00227391
Amarawathi Y, Singh R, Singh AK, Singh VP, Mohapatra T, Sharma TR, Singh NK (2008) Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol Breed 21:49–65. https://doi.org/10.1007/s11032-007-9108-8
Bindusree G, Natarajan P, Kalva S, Madasamy P (2017) Whole genome sequencing of Oryza sativa L. cv. Seeragasamba identifies a new fragrance allele in rice. PLoS ONE 12:e0188920. https://doi.org/10.1371/journal.pone.0188920
Bourgis F, Guyot R, Gherbi H, Tailliez E, Amabile I, Salse J, Lorieux M, Delseny M, Ghesquière A (2008) Characterization of the major fragrance gene from an aromatic japonica rice and analysis of its diversity in Asian cultivated rice. Theor Appl Genet 117:353–368. https://doi.org/10.1007/s00122-008-0780-9
Bradbury LMT, Fitzgerald TL, Henry RJ, Jin QS, Waters DLE (2005a) The gene for fragrance in rice. Plant Biotechnol J 3:363–370. https://doi.org/10.1111/j.1467-7652.2005.00131.x
Bradbury LMT, Henry RJ, Jin QS, Reinke RF, Waters DL (2005b) A perfect marker for fragrance genotyping in rice. Mol Breed 16:279–283. https://doi.org/10.1007/s11032-005-0776-y
Bradbury LM, Gillies SA, Brushett DJ, Waters DL, Henry RJ (2008) Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol Biol 68:439–449. https://doi.org/10.1007/s11103-008-9381-x
Buttery R, Ling LC, Juliano BO, Turnbaugh JG (1983) Cooked rice aroma and 2-acetyl-1-pyrroline. J Agric Food Chem 31:823–826. https://doi.org/10.1021/jf00118a036
Buttery RG, Ling LC, Mon TR (1986) Quantitative analysis of 2-acetyl-1-pyrroline in rice. J Agric Food Chem 34:112–114. https://doi.org/10.1021/jf00067a031
Buttery R, Turnbaugh J, Ling L (1988) Contribution of volatiles to rice aroma. J Agric Food Chem 36:1006–1009. https://doi.org/10.1021/jf00083a025
Chakraborty D, Deb D, Ray A (2016) An analysis of variation of the aroma gene in rice (Oryza sativa L. subsp. indica Kato) landraces. Genet Resour Crop Evol 63:953–959. https://doi.org/10.1007/s10722-016-0414-z
Champagne ET (2008) Rice aroma and flavour: a literature review. Cereal Chem 85:445–454. https://doi.org/10.1094/cchem-85-4-0445
Chan-in P, Jamjod S, Yimyam N, Rerkasem B, Pusadee T (2020) Grain quality and allelic variation of the Badh2 gene in thai fragrant rice landraces. Agronomy 10:779. https://doi.org/10.3390/agronomy10060779
Chen S, Yang Y, Shi W, Ji Q, He F, Zhang Z, Cheng Z, Liu X, Xua M (2008) Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell 20:1850–1861. https://doi.org/10.1105/tpc.108.058917
Cruz NDL, Kumar I, Kaushik RP, Khush GS (1989) Effect of temperature during grain development on stability of cooking quality of cooking quality components in rice. Japan J Breed 39:299–306. https://doi.org/10.1270/jsbbs1951.39.299
Dissanayaka S, Kottearachchi NS, Weerasena J, Peiris M (2014) Development of a CAPS marker for the badh2. 7 allele in Sri Lankan fragrant rice (Oryza sativa). Plant Breed 133:560–565. https://doi.org/10.1111/pbr.12201
Dhulappanavar CV (1976) Inheritance of scent in rice. Euphytica 25:659–662. https://doi.org/10.1007/BF00041603
Ellur RK, Khanna A, Gopala Krishnan S, Bhowmick PK, Vinod KK, Nagarajan M, Mondal KK, Singh NK, Singh K, Prabhu KV, Singh AK (2016) Marker-aided incorporation of Xa38, a novel bacterial blight resistance gene, in PB1121 and comparison of its resistance spectrum with xa13 + Xa21. Sci Rep 6:srep29188. https://doi.org/10.1038/srep29188
Giraud G (2013) The world market of fragrant rice, main issues and perspectives. Int Food Agribus Manag Rev 16:1–20. https://doi.org/10.2204/ag.econ.148577
Glaszmann JC (1987) Isozymes and classification of Asian rice varieties. Theor Appl Genet 74:21–30. https://doi.org/10.1007/bf00290078
Hasthanasombut S, Ntui V, Supaibulwatana K, Mii M, Nakamura I (2010) Expression of Indica rice OsBADH1 gene under salinity stress in transgenic tobacco. Plant Biotechnol Rep 4:75–83. https://doi.org/10.1007/s11816-009-0123-6
He Q, Park YJ (2015) Discovery of a novel fragrant allele and development of functional markers for fragrance in rice. Mol Breed. https://doi.org/10.1007/s11032-015-0412-4
He Q, Yu J, Kim TS, Cho YH, Lee YS, Park YJ (2015) Resequencing reveals different domestication rate for BADH1 and BADH2 in rice (Oryza sativa). PLoS ONE. https://doi.org/10.1371/journal.pone.0134801
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. https://doi.org/10.1016/j.plantsci.2013.12.007
Hinge VR, Patil HB, Nadaf AB (2016) Aroma volatile analyses and 2AP characterization at various developmental stages in Basmati and non-basmati scented rice (Oryza sativa L.) cultivars. Rice 9:38. https://doi.org/10.1186/s12284-016-0113-6
Jodon NE (1944) The inheritance of flower fragrance and other characters in rice. J Am Soc Agron 36:844–848. https://doi.org/10.2134/agronj1944.00021962003600100005x
Kadam BS, Patankar VK (1938) Inheritance of aroma in rice. Chron Bot 4:496–497
Khandagale KS, Zanan RL, Mathure SV, Nadaf AB (2017) Haplotype variation of badh2 gene, unearthing of a new fragrance allele and marker development of non-basmati fragrant rice ‘Velchi’ (Oryza sativa L.). Agri Gene 6:40–46. https://doi.org/10.1016/j.aggene.2017.09.003
Khush GS (2000) Taxonomy and origin of rice. In: Singh RK, Singh US, Khush GS (eds) Aromatic rices. Oxford and IBH Publishing Co. Pvt. Ltd, New Delhi, pp 5–13
Kovach MJ, Calingacion MN, Fitzgerald MA, McCouch SR (2009) The origin and evolution of fragrance in rice (Oryza sativa). Proc Natl Acad Sci USA 106:14444–14449. https://doi.org/10.1073/pnas.0904077106
Li J, Zhang FC, Wang WQ, Huang LY (2006) Advance in the study of higher plant promoter. Lett Biotechnol 17:658–661
Lorieux M, Petrov M, Huang N, Guiderdoni E, Ghesquiere A (1996) Aroma in rice: genetic analysis of quantitative trait. Theor Appl Genet 93:1145–1151. https://doi.org/10.1007/bf00230138
Mahajan G, Matloob A, Singh R, Singh VP, Chauhan BS (2018) Basmati rice in the Indian subcontinent: strategies to boost production and quality traits. Adv Agron 151:159–213. https://doi.org/10.1016/bs.agron.2018.0
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acid Res 47:W636–W641. https://doi.org/10.1093/nar/gkz268
Malik SS, Dikshit N, Das AB, Lodh SB (1994) Studies on morphological and physico- chemical properties of local scented rice. Oryza 31:106–110
Murray HG, Thompson WF (1980) Rapid isolation of high molecular weight DNA. Nucleic Acids Res 8:4321–4325. https://doi.org/10.1093/nar/8.19.4321
Myint KM, Arikit S, Wanchana S, Yoshihashi T, Choowongkomon K, Vanavichit A (2012) A PCR-based marker for a locus conferring the aroma in Myanmar rice (Oryza sativa L.). Theor Appl Genet 125:887–896. https://doi.org/10.1007/s00122-012-1880-0
Nadaf AB, Krishnan S, Wakte KV (2006) Histochemical and biochemical analysis of major aroma compound (2-acetyl-1-pyrroline) in Basmati and other scented rice (Oryza sativa L). Curr Sci 91:1533–1536
Okpala E, Mo Z, Duan M, Tang X (2019) The genetics and biosynthesis of 2-acetyl-1-pyrroline in fragrant rice. Plant Physiol Biochem 135:272–276. https://doi.org/10.1016/j.plaphy.2018.12.012
Okpala NE, Potcho MP, Imran M, An T, Bao G, He L, Li L, Tang X (2021) Starch Morphology and metabolomic analyses reveal that the effect of high temperature on cooked rice elongation and expansion varied in indica and japonica rice cultivars. Agronomy 11:2416. https://doi.org/10.3390/agronomy11122416
Peng B, Zuo YH, Hao YL, Peng J, Kong DY, Peng Y, Nassirou TY, He LL, Sun YF, Liu L, Pang RH (2018) Studies on aroma gene and its application in rice genetics and breeding. J Plant Stud 7:29–40. https://doi.org/10.5539/jps.v7n2p29
Prodhan ZH, Qingyao S (2020) Rice aroma: a natural gift comes with price and the way forward. Rice Sci 27:86–100. https://doi.org/10.1016/j.rsci.2020.01.001
Rai VP, Singh AK, Jaiswal HK, Singh SP, Singh RP, Waza SA (2015) Evaluation of molecular markers linked to fragrance and genetic diversity in Indian aromatic rice. Turk J Bot 39:209–217. https://doi.org/10.3906/bot-1405-117
Ray A, Deb D, Ray R, Chattopadhayay B (2013) Phenotypic characters of rice landraces reveal independent lineages of short-grain aromatic indica rice. AoB Plants. https://doi.org/10.1093/aobpla/plt032
Roy S, Banerjee A, Mawkhlieng B, Misra A, Pattanayak A, Harish G (2015) Genetic diversity and population structure in aromatic and quality rice (Oryza sativa L.) landraces from North-Eastern India. PLoS ONE 10:e0129607. https://doi.org/10.1371/journal.pone.0129607
Sakthivel K, Sundaram R, Shobha Rani N, Balachandran S, Neeraja C (2009) Genetic and molecular basis of fragrance in rice. Biotechnol Adv 27:468–473. https://doi.org/10.1016/j.biotechadv.2009.04.001
Shao GN, Tang A, Tang SQ, Luo J, Jiao GA, Wu JL, Hu PS (2011) A new deletion mutation of fragrant gene and the development of three molecular markers for fragrance in rice. Plant Breed 130:172–176. https://doi.org/10.1111/j.1439-0523.2009.01764.x
Shao G, Tang S, Chen M, Wei X, He J, Luo J, Jiao G, Hu Y, Xie L, Hu P (2013) Haplotype variation at Badh2, the gene determining fragrance in rice. Genomics 101:157–162. https://doi.org/10.1016/j.ygeno.2012.11.010
Shi WW, Yang Y, Chen SH, Xu ML (2008) Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol Breed 22:185–192. https://doi.org/10.1007/s11032-008-9165-7
Shi Y, Zhao G, Xu X, Li J (2014) Discovery of a new fragrance allele and development of functional markers for identifying diverse fragrant genotypes in rice. Mol Breed 33:701–708. https://doi.org/10.1007/s11032-013-9986-x
Siddiq EA, Vemireddy LR, Nagaraju J (2012) Basmati rices: genetics, breeding and trade. Agric Res 1:25–36. https://doi.org/10.1007/s40003-011-0011-5
Singh RK, Singh US, Khush GS, Rohilla R, Singh JP, Singh G, Shekhar KS (2000) Small and medium grained aromatic rices of India. In: Singh RK, Singh US, Khush GS (eds) Aromatic rices. Oxford and IBH Publishing Co. Pvt. Ltd, New Delhi, pp 155–177
Singh AK, Gopalakrishnan S, Singh VP, Prabhu KV, Mohapatra T, Singh NK, Sharma TR, Nagarajan M, Vinod KK, Devinder S (2011) Marker assisted selection: a paradigm shift in Basmati breeding. Indian J Genet Plant Breed 71:120–128
Singh V, Singh AK, Mohapatra T, Gopala Krishnan S, Ellur RK (2018) Pusa Basmati 1121—a rice variety with exceptional kernel elongation and volume expansion after cooking. Rice 11:19. https://doi.org/10.1186/s12284-018-0213-6
Sood BC, Siddiq EA (1978) A rapid technique for scent determination in rice. Indian J Genet Plant Breed 38:26
Sulochana S, Singaravadivel K (2015) A study on phytochemical evaluation of traditional rice variety of Tamil Nadu-‘Maappillai Samba’ by GC-MS. Int J Pharma Bio Sci 6:606–611
Tava A, Bocchi S (1999) Aroma of cooked rice (Oryza sativa): comparison between commercial Basmati and Italian line B5–3. Cereal Chem 76:526–529. https://doi.org/10.1094/cchem.1999.76.4.526
The 3,000 rice genomes project (2014) The 3,000 rice genomes project. GigaScience 3:7. https://doi.org/10.1186/2047-217X-3-7
Tripathi RS, Rao MJBK (1979) Inheritance and linkage relationship of scent in rice. Euphytica 28:319–323. https://doi.org/10.1007/BF00056589
Tsugita T (1985) Aroma of cooked rice. Food Rev Int 1:497–520
Wang J, Xu X, Luo Q, Li J (2016) Mutation in the non-coding sequence of badh2 gene reduces its transcription and translation in fragrant rice, ‘Nankai 138.’ Crop Sci 56:1–6. https://doi.org/10.1235/cropsci2014.03.0243
Withana W, Kularathna R, Kottearachchi N, Kekulandara D, Weerasena J, Steele K (2020) In silico analysis of the fragrance gene (badh2) in Asian rice (Oryza sativa L) germplasm and validation of allele specific markers. Plant Genet Resour Charact Util. https://doi.org/10.1017/S1479262120000015
Xu XL, Zhao GC, Li JY (2011) Development of molecular markers used to identify two types of fragrant rice and analysis of mutation sites of Badh2 gene in 24 varieties of fragrant rice. Plant Divers Resour 33:667–673
Acknowledgements
Author’s wish to acknowledge the support rendered by the staff of Division of Genetics, IARI for conduct of experiments, and ICAR-Indian Institute of Rice Research, Hyderabad for the supply of seed materials for the germplasm panel used. This work is a part of the doctoral research of the senior author.
Funding
The experiments are funded by the Department of Biotechnology through the research project entitled “Metabolic and molecular profiling of aromatic rice germplasm of India for gaining insights about aroma” (BT/PR13888/AGR/02/723/2010).
Author information
Authors and Affiliations
Contributions
Conceptualization of research (GKS, AKS), designing of the experiments (GKS, AKS), contribution of experimental materials (GKS, SMM, KS), execution of field/ lab experiments and data collection (GDS, GKS, HB, MN), analysis of data and interpretation (GDS, GKS, KKV, RKE, AK) and preparation of manuscript (GDS, GKS, KKV). All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Consent to participate
All the authors have given consent to participate in the research being published.
Consent for publication
All the authors have seen the manuscript and have given consent to publish.
Ethics approval
No animal or human subject are used in the research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, G., Gopala Krishnan, S., Kumar, A. et al. Molecular profiling of BADH2 locus reveals distinct functional allelic polymorphism associated with fragrance variation in Indian aromatic rice germplasm. Physiol Mol Biol Plants 28, 1013–1027 (2022). https://doi.org/10.1007/s12298-022-01181-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-022-01181-9