Abstract
The extent of genome redundancy exhibited by Brassica species provides a model to study the evolutionary fate of multi-copy genes and the effects of polyploidy in economically important crops. Phytoene synthase (PSY) catalyzes the first committed reaction of the carotenoid biosynthetic pathway, which has been shown to be rate-limiting in Brassica napus seeds. In Arabidopsis thaliana, a single PSY gene (AtPSY) regulates phytoene synthesis in all tissues. Considering that diploid Brassica genomes contain three Arabidopsis-like subgenomes, the objectives of the present work were to determine whether PSY gene families exist in B. napus (AACC) and its diploid progenitor species, Brassica rapa (AA) and Brassica oleracea (CC); to establish the level of retention of Brassica PSY genes; to map PSY gene family members in the A and C genomes and to compare Brassica PSY gene expression patterns. A total of 12 PSY homologues were identified, 6 in B. napus (BnaX.PSY.a-f) and 3 in B. rapa (BraA.PSY.a-c) and B. oleracea (BolC.PSY.a-c). Indeed, with six members, B. napus has the largest PSY gene family described to date. Sequence comparison between AtPSY and Brassica PSY genes revealed a highly conserved gene structure and identity percentages above 85% at the coding sequence (CDS) level. Altogether, our data indicate that PSY gene family expansion preceded the speciation of B. rapa and B. oleracea, dating back to the paralogous subgenome triplication event. In these three Brassica species, all PSY homologues are expressed, exhibiting overlapping redundancy and signs of subfunctionalization among photosynthetic and non-photosynthetic tissues. This evidence supports the hypothesis that functional divergence of PSY gene expression facilitates the accumulation of high levels of carotenoids in chromoplast-rich tissues. Thus, functional retention of triplicated Brassica PSY genes could be at least partially explained by the selective advantage provided by increased levels of gene product in floral organs. A better understanding of carotenogenesis in Brassica will aid in the future development of transgenic and conventional cultivars with carotenoid-enriched oil.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Brassiceae is one of the most phenotypically diverse tribes within the Brassicaceae family and comprises approximately 240 species, including important Brassica crops. Brassica napus L. (AACC; n = 19), the second largest oilseed crop worldwide (FAO 2011) is believed to have originated from the interspecific hybridization of two base “diploid” genomes, Brassica rapa L. (AA; n = 10) and Brassica oleracea L. (CC; n = 9) (U 1935; Iniguez-Luy and Federico 2011). These diploid progenitors are considered to be ancient polyploids and still exhibit highly replicated genomes, each containing three paralogous subgenomes (Lysak et al. 2005; Parkin et al. 2005; Xiong et al. 2011). These Brassica subgenomes are closely related to that of Arabidopsis thaliana (Lysak et al. 2005; Parkin et al. 2005) with high nucleotide sequence similarity found at the coding sequence (CDS) level; similarity that has provided an opportunity to use comparative genomics with A. thaliana to study the effects of polyploidy in the phenotypic divergence of the Brassiceae.
Most comparative studies have confirmed the extensive triplication at the genome level in diploid Brassicas, but gene contents were found to be variable, with paralogous regions exhibiting interspersed gene losses and insertions (O’Neill and Bancroft 2000; Rana et al. 2004; Park et al. 2005; Town et al. 2006; Yang et al. 2006). Interestingly, less than 10% of CDS of predicted gene models from A. thaliana were found to be retained as syntenic orthologues in each of the triplicated subgenomes in the recently sequenced B. rapa genome (Brassica rapa Genome Sequencing Project Consortium 2011). Thus, it is important to study on case-by-case basis those gene families that have been retained in spite of diploidization, a process by which a genome tends to return to its original gene complement, as previously reported (Yang et al. 2006).
Carotenoids are isoprenoid compounds synthesized by all photosynthetic organisms. In plants, carotenoids are synthesized and accumulated in plastids. They are found in chloroplasts of green tissues where they play vital roles in light harvesting and energy transfer preventing photo-oxidation during photosynthesis (Demmig-Adams and Adams 2002). Large amounts of carotenoids also accumulate in chromoplasts of mature fruits and flowers where they serve as pigments and precursors to a range of scents that attract pollinators and secure seed dispersal (Demmig-Adams et al. 1996). The role that carotenoids play in seed and root tissues is related to their function as precursors of the plant hormone abscisic acid (ABA) (Maluf et al. 1997; Hirschberg 2001). Several studies have shown that root carotenogenesis regulates the stress-induced production of ABA in response to drought and salt stress (Li et al. 2008b; Welsch et al. 2008; Arango et al. 2010).
Phytoene synthase (PSY) catalyzes the first committed reaction of the carotenoid biosynthetic pathway, the head to head condensation of two geranylgeranyl diphosphate (GGPP) molecules. Since GGPP also serves as a precursor of tocopherols, chlorophylls, plastoquinones and gibberellins, PSY regulation is highly controlled (von Lintig et al. 1997; Welsch et al. 2000, 2003; Rodríguez-Villalón et al. 2009; Cazzonelli and Pogson 2010; Toledo-Ortiz et al. 2010). PSY is encoded by a single copy gene in A. thaliana, therefore the flexibility and response capabilities of the carotenoid biosynthetic pathway are limited to regulating this rate controlling enzyme (Cazzonelli and Pogson 2010). Most plant species, however, seem to have a PSY gene family composed of two or three homologous genes (Bartley et al. 1992; Bartley and Scolnik 1993; Busch et al. 2002; Gallagher et al. 2004; Li et al. 2008a; Arango et al. 2010). In these plant species, functional diversification of PSY homologues provided a mechanism that allowed for the accumulation of carotenoids in non-photosynthetic tissues, mainly fruits, seeds and flowers, and also to respond to environmental stress (Li et al. 2008b; Welsch et al. 2008).
The objectives of the present work were to determine whether PSY gene families exist in B. napus and its diploid progenitor species, B. rapa and B. oleracea; to establish the level of retention of Brassica PSY genes; to map PSY gene family members in the A and C genomes and to compare Brassica PSY gene expression patterns. Undoubtedly, expression studies of retained multicopy genes in Brassica species should provide insight into the functional and evolutionary effects that gene duplication and polyploidy had on Brassica crop evolution. In addition, a better understanding of carotenogenesis will aid in the future development of transgenic and conventional B. napus cultivars with carotenoid-enriched oil.
Materials and methods
Plant materials and nucleic acid extractions
Brassica rapa cv. R500, a highly inbred annual yellow sarson, B. oleracea TO1000DH3, a double haploid rapid cycling line (Iniguez-Luy et al. 2009), B. napus cv. Westar and A. thaliana Col-0 were grown in a controlled greenhouse under 16-h-day/8-h-night cycle. For genomic DNA (gDNA) extraction, flower buds from each species were harvested and lyophilized. DNA isolation was conducted following the CTAB procedure described by Kidwell and Osborn (1992). For RNA extractions, the following vegetative tissues were collected: cotyledons, seedlings with 3–4 true leaves, young leaves from mature plants, roots from 10-day seedlings, developing seeds from 4 stages (S1: early stage, day 7 after petals fall; S2: elongating pod, day 14; S3: pods reach final size, day 21 and S4: ripening begins, day 28). Floral tissues included anthers and petals from three developmental stages (S1: green bud stage, S2: yellow bud stage and S3: fully expanded petals). Total RNA from all tissues was extracted using RNA-Solv Reagent (Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s instructions.
Cloning of PSY genes from Brassica
The Arabidopsis PSY gene (At5g17230) sequence was used to query the Brassica EST database in GenBank (National Center for Biotechnology Information) using the megablast program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify putative Brassica PSY homologues. Based on these sequences, specific oligonucleotide primers (Supplemental Table S1) were designed in the web-based program GeneFisher (http://bibiserv.techfak.uni-bielefeld.de/genefisher2/) to amplify Brassica gDNA sequences using high fidelity enzymes, Phusion® High-Fidelity DNA Polymerase (Finnzymes, Espoo, Finland) or KOD Hot Start DNA Polymerase (Novagen, Madison, WI, USA) according to the manufacturer’s instructions. Specific PCR products were cloned using the StrataClone Blunt PCR Cloning Kit (Agilent Technologies, Santa Clara, CA, USA) or pGEM-T Easy Vector System (Promega, Madison, WI, USA) and at least 10 colonies per clone were sequenced.
Brassica PSY gene sequences described in this paper have been submitted to GenBank under the following accession numbers: BraA.PSY.a-c: JF920031-JF920033, BolC.PSY.a-c: JF920034-JF920036 and BnaX.PSY.a-f: JF920037-JF920042.
Single-strand conformation polymorphism (SSCP) analysis
A primer pair (BnPSY SSCP1F and BnPSY SSCP1R; Supplemental Table S1) was designed to amplify a conserved Brassica PSY region comprising part of exon 1. Amplicon sizes varied among different Brassica PSY genes but were approximately 250 bp in length. PCR reactions (50 μl) were carried out using GoTaq Flexi DNA Polymerase (Promega, Madison, WI, USA). Amplification started with a 95°C denaturation step (5 min), followed by 30 cycles of 30 s at 95°C, 30 s at 45°C, and 30 s at 72°C, with a final 72°C extension of 5 min. The resulting PCR fragments were gel-purified (E.Z.N.A. Gel Extraction Kit, Omega Bio-Tek, Norcross, GA, USA) and eluted in 15 μl for subsequent SSCP analysis. For purified PCR products derived from gDNAs, 5 μl was added to 15 μl of SSCP loading buffer [95% formamide, 10 mm NaOH, 0.25% (w/v) xylene cyanol, 0.25% (w/v) bromophenol blue], whereas for PCR products derived from PSY clones (plasmids), 0.8 μl was mixed with 15 μl of loading buffer. Purified PCR products were then heated for 10 min at 96°C and immediately cooled on ice. A total of 9 μl of each sample was loaded onto a 0.7× mutation detection enhancement gel (MDE; Lonza, Rockland, ME, USA) for B. rapa and B. napus and 0.5× for B. oleracea. Samples were electrophoresed at 7 W constant power for 20 h at room temperature in a Fisher scientific sequencing apparatus. Following electrophoresis, gels were silver-stained according to Bassam et al. (1991) with some modifications. Briefly, gels were fixed in acetic acid 10% (30 min), washed in distilled water (20 min), soaked 30 min in silver nitrate (1 g/l), formaldehyde (0,06%), rinsed quickly in distilled water, and developed in a chilled solution of sodium carbonate (30 g/l), formaldehyde (0.06%) and sodium thiosulfate (1 mg/l). Development was stopped in 10% acetic acid solution.
Southern blot analysis
Southern blot analysis was conducted as described in Iniguez-Luy et al. (2009) with minor modifications. Briefly, 12 μg of gDNA from each species was digested to completion with EcoRI and EcoRV in separate reactions using 8 units enzyme/μg of DNA. Digests were electrophoresed in 0.8% agarose gels (1× TAE), run for 16 h at 40 V and then transferred onto Amersham Hybond-XL membranes (GE Healthcare, UK) using an alkaline transfer method. The DNA was fixed to the membrane by UV crosslinking followed by 2 h in a 90°C vacuum oven. One probe (BnaPSY73, 611 bp) was amplified by PCR using BnPSY15F and BnPSY18R primers (Supplemental Table S1). Probe labeling was conducted using the RediPrime II labeling kit (GE Healthcare, UK) and 25 ng of PCR product. Hybridization was carried out using modified Church and Gilbert buffer (0.5 M phosphate buffer, pH 7.2, 7% SDS, 10 mM EDTA) at 65°C overnight. The blot was washed in 65°C 2× SSC/0.1% SDS for 10 min at room temperature.
RT-PCR and cDNA-SSCP analysis
Total RNA (2 μg) from all tissues were treated with RQ1 DNAse (Promega, Madison, WI, USA) for 1 h at 37°C. First strand cDNA synthesis was carried out with oligo(dT) primer and M-MuLV reverse transcriptase in the presence of RNase inhibitor (New England BioLabs, Beverly, MA, USA), according to the manufacturer’s instructions. Contamination of cDNA samples with gDNA was tested by PCR for ACTIN (AF111812) with a primer pair (BnActinF and BnActinR; Supplemental Table S1) that generates a predicted 725-bp fragment for the cDNA and a 900-bp fragment for gDNA due to the presence of an intron. PCR reactions (100 μl) and SSCP analysis for every tissue were carried out as described above, with a single modification. For purified PCR products derived from cDNAs, 6 μl was added to 12 μl of SSCP loading buffer.
Sequence analysis
To estimate gene copy number in each Brassica species, sequencing reads from cloned PSY genes were assembled with the Phred/Phrap/Consed software package (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998). Nucleotide and protein sequence alignments were performed using ClustalW2 (Chenna et al. 2003). Gene structures were predicted based on alignments of the Arabidopsis PSY gene (At5g17230) to genomic and coding sequences and each Brassica PSY assembled gene (BraA.PSY.a-c, BolC.PSY.a-c and BnaX.PSY.a-f). Alternatively, gene structures were predicted in FGENESH (http://mendel.cs.rhul.ac.uk/mendel.php?topic=fgen) using the dicot trained matrix. The presence of chloroplast transit peptides was predicted using the ChloroP 1.1 server (Emanuelsson et al. 1999). Repetitive sequences were identified in RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) using the A. thaliana library. Amino acid variability was calculated in the Protein Variability Server (PVS) (Garcia-Boronat et al. 2008) using the Wu-Kabat coefficient (Kabat et al. 1977). A phylogenetic tree was obtained by analyzing the nucleotide sequence divergence of Brassica PSY genes using the neighbor-joining method (Saitou and Nei 1987) implemented in MEGA4 software (Tamura et al. 2007). Nucleotide replacement (K a) and synonymous (K s) substitutions were estimated using K-estimator 6.1 (Comeron 1999).
Genetic mapping of the B. rapa and B. oleracea PSY paralogues
A subset of 50 lines from two previously described mapping populations, BraIRRi and BolTBDH (Iniguez-Luy et al. 2009), were genotyped using five sets of informative primer combinations (Supplemental Table S1) that yielded specific fragments for each of the six Brassica PSY paralogues. PCR amplification and gel electrophoresis were carried out as described in previous sections. Linkage analysis and map construction were conducted separately for each population using JoinMap® v4.0 (Van Ooijen 2006). Briefly, linked loci were grouped using a LOD threshold of 5 and a maximum recombination fraction of 0.4. Grouped RFLP, SSR and specific PSY gene marker loci were designated using the international linkage group/chromosomes nomenclature as described by Iniguez-Luy et al. (2009). Map distances in centiMorgans (cM) were calculated using the Kosambi mapping function.
Results
Cloning and copy number estimation of Brassica PSY genes
In A. thaliana, phytoene synthase (PSY) is encoded by a single copy gene (AtPSY, At5g17230) located in the top arm of AtChr5 (Scolnik and Bartley 1994). This chromosomal region containing AtPSY (At5G07410–At5G18280) has been found to be triplicated in diploid Brassica genomes (Osborn et al. 1997; Lysak et al. 2005, Parkin et al. 2005). Based on synteny information, we identified in silico three collinear regions of predicted PSY gene loci by extrapolating position from adjacent loci and markers in the A and C genomes. These regions were found at chromosomes A2, A3 and A10 of B. rapa; C2, C3 and C9 of B. oleracea and A2, A3, A10, C2, C3 and C9 of B. napus. However, deletions of at least one of the triplicated genes are common at the microsyntenic level (Town et al. 2006). Therefore, PSY copy number could only be determined empirically, since fully assembled A and C genome sequences were not available at the time of beginning our work. In order to estimate PSY gene copy numbers in B. napus and its diploid progenitors, we followed different approaches including gene cloning, DNA-SSCP and Southern blot analyses.
We queried the GenBank B. napus EST database and identified 44 ESTs with at least 80% sequence identity to AtPSY full length cDNA (data not shown). Primers were designed (Supplemental Table S1) using this sequence information and different primer combinations were assayed in PCR reactions. A total of 53 amplicons were cloned for B. rapa, 44 for B. oleracea and 134 for B. napus. Sequencing reads from these cloned Brassica PSY genomic sequences were assembled into contigs. As a result, a total of 12 PSY genes were identified, 6 in B. napus (BnaX.PSY.a-f) and 3 in each of its progenitor species, B. rapa (BraA.PSY.a-c) and B. oleracea (BolC.PSY.a-c). Brassica PSY genes were named according to the rules of systematic gene nomenclature proposed by Ostergaard and King (2008).
DNA-SSCP analyses were also performed to estimate PSY gene copy number in these three Brassica species. PCR amplicons spanning part of PSY exon 1 (~250 bp) were resolved on MDE gels. Banding patterns obtained from B. rapa, B. oleracea and B. napus genomic DNAs (gDNA) were compared to those obtained from cloned Brassica PSY genes (Fig. 1). Similarly, this second approach revealed the existence of at least 6 PSY homologous genes in B. napus (AACC) and 3 PSY paralogues in each of the two diploid species carrying the parental genomes, B. rapa (AA) and B. oleracea (CC).
DNA-SSCP analysis of Brassica PSY gene families. DNA-SSCP analysis revealed the existence of at least three PSY paralogues in B. rapa (a), three in B. oleracea (b) and six in B. napus (c). PCR was performed on gDNA from B. rapa (Bra), B. oleracea (Bol) and B. napus (Bna). The generated amplicon spans part of PSY exon 1 (~250 bp). Cloned Brassica PSY genes (BraA.PSY.a-c, BolC.PSY.a-c and BnaX.PSY.a-f) were used as template controls to determine banding patterns (lanes to the left of each gDNA). Two strands are shown for each gene
Southern blot analysis further verified that PSY belongs to a gene family in B. napus and its two diploid progenitor species. A 611 bp probe comprising part of exon 1, intron 1 and exon 2 of BnaA.PSY.b was used to hybridize gDNAs digested with EcoRI and EcoRV (Fig. 2). Hybridization patterns for EcoRI digests confirmed that PSY is a single copy gene in Arabidopsis also detecting two bands in B. rapa and B. oleracea and three in B. napus. Hybridization patterns for EcoRV digests correlated with the number of genes cloned in this study, detecting three bands in B. rapa and B. oleracea and six in B. napus.
Southern blot analysis of PSY genes in B. napus and its two progenitor species. The presence of PSY gene families was detected when a probe (611 bp) comprising part of exon 1, intron 1 and exon 2 of BnaA.PSY.b was used. Twelve micrograms of gDNA from A. thaliana (At), B. rapa (Bra), B. oleracea (Bol) and B. napus (Bna) were digested to completion with EcoRI and EcoRV
Brassica PSY gene sequence analysis
We have cloned and sequenced 12 Brassica PSY genomic sequences (gDNA) ranging from 1,272 to 2,503 bp (Table 1). These regions contained six partial and six complete coding sequences (CDS) that ranged from 733 to 1,275 bp (Table 1). Sequence analysis of AtPSY and these 12 Brassica PSY genes revealed a strong conservation of gene structure, all full length ORFs containing 6 exons and 5 introns (Table 1; Fig. 3a). Exons exhibited identical or similar size (bp) whereas intron lengths were found to be less conserved, as expected (Table 1). Noticeably, BraA.PSY.c intron 2 and BnaA.PSY.e intron 3 were strikingly larger than their counterparts (Table 1). Repetitive element searchers within the PSY sequences identified a 227-bp fragment with homology to the canonical telomeric repeat ATREP18 [5′-(TTTAGGG) n -3′] in BnaA.PSY.e intron 3 (Supplemental Fig. S1).
Brassica PSY gene families. a Schematic representation of Arabidopsis PSY gene (AtPSY, At5g17230) and the assembled contigs generated for B. rapa (BraA.PSY.a-c), B. oleracea (BolC.PSY.a-c) and B. napus (BnaX.PSY.a-f). Exons and introns are drawn to scale and represented by boxes (E1–E6) and lines, respectively. Gene structures were deduced from FGENESH predictions and alignment analysis. For details, lengths of the exons and introns are shown in Table 1. b Structure of Brassica PSY proteins. Chloroplast transit peptide (TP) and phytoene synthase signatures (PROSITE patterns PS01044 and PS01045) are depicted
High protein sequence similarity has been maintained in these Brassica PSY gene families. All PSY proteins possess an N-terminal transit peptide (TP) for plastid targeting and two characteristic PSY signature motifs (PS01044 and PS01045) (Fig. 3b). Deduced protein sequences from complete ORFs varied in length from 414 to 424 amino acids and TPs ranged from 65 to 75 amino acids (Table 1). Interestingly, a Wu-Kabat variability plot of Brassica PSY protein sequences revealed that most divergence is found at the N-terminal region, which coincides with the location of the plastid TP (Supplemental Fig. S2). The level of replacement and synonymous site nucleotide divergence ratio (K a/K s) indicates that all members are likely undergoing purifying selection (Supplemental Table S2). This strongly indicates that PSY proteins have evolved under functional constraint.
Genome origins of the six BnaX.PSY homologues
Genome origins of the six BnaX.PSY genes were established based on sequence identity and phylogenetic analysis. The percentage of sequence identity for each pair of Brassica PSY genes was determined and orthologies inferred (Supplemental Table S3). Phylogenetic analysis revealed that three homeologous gene pairs, BnaA.PSY.a/BnaC.PSY.b, BnaA.PSY.f/BnaC.PSY.d and BnaA.PSY.e/BnaC.PSY.c, exist in B. napus, with each BnaX.PSY gene clustering with its predicted B. rapa or B. oleracea orthologue (Fig. 4). Sequence similarities between orthologous (93–100%) and homoelogous gene pairs were higher (92–96%) than those observed between pairs of paralogous genes in both diploid species (88–91%) and pairs of homologous genes in B. napus (85–92%) (Supplemental Table S3).
Phylogenetic relationship among Brassica PSY genes. The evolutionary history was inferred using the neighbor-joining method. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances. There were a total of 938 positions in the final dataset covering exons 1–3. Analyses were conducted using MEGA4 (Tamura et al. 2007). Using the estimated divergence time of 15–20 million years ago (MYA) for the split between Arabidopsis and the tribe Brassiceae (Yang et al. 1999), the genome triplication date was estimated to be 11–15 MYA
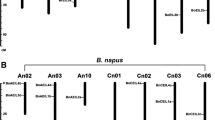
Segregation and linkage analyses using sets of specific molecular markers developed for each of the B. rapa and B. oleracea PSY genes (Supplemental Table S1) were conducted in order to locate each paralogue in the context of its genetic linkage position in the A or C genomes. PSY genes mapped to chromosomes A2, A3 and A10 of B. rapa and C2, C3 and C9 of B. oleracea (Fig. 5). In both diploid genomes, Brassica PSY map positions corresponded to the three collinear regions previously identified in silico by comparative mapping with Arabidopsis. In addition, evolutionary relationships of Brassica PSY genes could be further confirmed from genetic map positions, with pairs of orthologous genes (BraA.PSY.c and BolC.PSY.b, BraA.PSY.b and BolC.PSY.c, and BraA.PSY.a and BolC.PSY.a) mapping to syntenic regions in the A and C genomes (Fig. 5).
Retention of PSY syntenic orthologues in B. rapa and B. oleracea genomes. Map position of Brassica PSY genes on a B. rapa chromosomes A2, A3 and A10 and b B. oleracea chromosomes C2, C3 and C9. Marker locus names and distances (cM) are located to the right and left of each chromosome, respectively. Bold marker loci represent Brassica PSY genes. Map positions were calculated from two rapid cycling populations (BraIRRi and BolTBDH) described by Iniguez-Luy et al. (2009)
Brassica PSY gene expression analysis
In order to explore possible tissue-specific partitioning of PSY gene expression, we investigated the expression patterns of each of the 12 Brassica PSY homologues described in this work. Gene expression profiling was determined by RT-PCR followed by cDNA-SSCP analysis.
In B. rapa, all three PSY paralogues (BraA.PSY.a, BraA.PSY.b and BraA.PSY.c) were expressed in cotyledons, seedlings, mature leaves, roots and seeds (Fig. 6). Interestingly, expression of paralogues Bra.A.PSY.a and BraA.PSY.b was also detected in anthers and petals but no expression of BraA.PSY.c was detected in floral tissues (Fig. 6).
Analysis of PSY gene expression in B. rapa. a C cotyledon, Sd seedling, L leaf, R root, S developing seeds (stages 1–4); developing flowers, A anther (stages 1–2), P petal (stages 1–3). b BraA.PSY gene expression was determined by RT-PCR (upper panel). B. rapa ACTIN control (lower panel). c cDNA-SSCP analysis. RT-PCR products were examined by SSCP to elucidate the expression patterns of each B. rapa PSY paralogue. Bra, B. rapa gDNA control; BraA.PSY.a-c, cloned Brassica PSY gene controls; WC, water control
Similarly, in B. oleracea expression of all three PSY paralogues (BolC.PSY.a, BolC.PSY.b and BolC.PSY.c) was detected in cotyledons, seedlings, mature leaves, roots and seeds (Fig. 7). Paralogues BolC.PSY.a and BolC.PSY.c were found to be expressed in anthers and petals throughout flower development but paralogue BolC.PSY.b expression was only detected at the earliest stage (stage 1; Fig. 7).
Analysis of PSY gene expression in B. oleracea. a C cotyledon, Sd seedling, L leaf, R root, S developing seeds (stages 1–4); developing flowers, A anther (stages 1–2), P petal (stages 1–3). b BolC.PSY gene expression was determined by RT-PCR (upper panel). B. oleracea ACTIN control (lower panel). c cDNA-SSCP analysis. RT-PCR products were examined by SSCP to elucidate the expression patterns of each B. oleracea PSY paralogue. Bol, B. oleracea gDNA control; BolC.PSY.a-c, cloned Brassica PSY gene controls; WC, water control
In B. napus, expression of homeologues BnaC.PSY.a and BnaA.PSY.b was detected in all tissues (Fig. 8). Interestingly, homeologues BnaA.PSY.c and BnaC.PSY.e exhibited preferential expression in floral tissues (Fig. 8). Following an opposite trend, homoelogues BnaA.PSY.d and BnaC.PSY.f were preferentially expressed in cotyledons, seedlings, mature leaves, roots and seeds and barely detected in floral tissues (Fig. 8).
Analysis of PSY gene expression in B. napus. a C cotyledon, Sd seedling, L leaf, R root, S developing seeds (stages 1–4); developing flowers, A anther (stages 1–2), P petal (stages 1–3). b BnaX.PSY gene expression was determined by RT-PCR (upper panel). B. napus ACTIN control (lower panel). c cDNA-SSCP analysis. RT-PCR products were examined by SSCP to elucidate the expression patterns of each B. napus PSY gene. Non-informative gel space was cut out to reduce figure size. Bna, B. napus gDNA control; BnaX.PSY.a-f, cloned Brassica PSY gene controls; WC, water control
Discussion
PSY gene families in B. napus and its diploid progenitors
Phytoene synthase (PSY) catalyzes the first committed reaction of the carotenoid biosynthetic pathway and has been shown to be rate-limiting in B. napus seeds (Shewmaker et al. 1999). This enzyme is encoded by a single copy gene in Arabidopsis (Scolnik and Bartley 1994) but the existence of PSY gene families has been documented in several crop species including tomato (Bartley et al. 1992; Bartley and Scolnik 1993), tobacco (Busch et al. 2002), maize, rice, sorghum (Gallagher et al. 2004; Li et al. 2008a) and cassava (Arango et al. 2010). Taking into account that diploid Brassica genomes are highly redundant, each containing three Arabidopsis-like subgenomes, one of the objectives of this study was to determine the number of PSY genes present in B. napus (AACC) and its progenitor species, B. rapa (AA) and B. oleracea (CC).
Based on the high nucleotide sequence similarity found at the CDS level between Arabidopsis and Brassica species (Lysak et al. 2005; Parkin et al. 2005; Iniguez-Luy et al. 2009), we were able to clone a total of 12 Brassica PSY genes using an overlapping PCR strategy (Fig. 3). We identified three paralogous PSY genes in each of the diploid species, B. rapa (BraA.PSY.a-c) and B. oleracea (BolC.PSY.a-c) and three pairs of homoelogous PSY genes in B. napus (BnaX.PSY.a-f). With six members, B. napus has the largest PSY gene family described to date. Three independent methods (i.e. cloning, Southern blot and DNA-SSCP analyses) confirmed these findings (Figs. 1, 2, 3).
Sequence comparison between AtPSY and Brassica PSY genes revealed a highly conserved exon–intron structure exhibiting identity percentages above 85% at the CDS level (Table 1; Supplemental Table S3). All PSY proteins share two conserved sequence motifs (Fig. 3b) and possess a predicted N-terminal transit peptide (TP) for plastid targeting (Table 1). As seen in other PSY proteins (Busch et al. 2002; Gallagher et al. 2004; Welsch et al. 2008; Arango et al. 2010), this N-terminal TP region was found to exhibit the highest level of sequence divergence among Brassica PSY proteins (Supplemental Fig. S2). These TPs are sufficient and specific to target proteins to plastids, but the underlying molecular mechanisms are not fully understood, mainly because these peptides exhibit high sequence diversity and high heterogeneity (Li and Chiu 2010). Previous studies have shown that protein import is facilitated by multimeric protein complexes (translocons) in the outer (Toc) and inner (Tic) envelope membranes of plastids that recognize and bind TPs (Bauer et al. 2000). In Arabidopsis, AtToc159 has been proposed to be a receptor with specificity for photosynthetic proteins, whereas AtTOC132 and AtToc120 have specificity for non-photosynthetic proteins (Kubis et al. 2004). Functional characterization of TPs would help determine whether Brassica PSY proteins are preferentially targeted to distinct plastids (e.g. chloroplasts vs. chromoplasts).
The level of replacement and synonymous site nucleotide divergence ratio (K a/K s) revealed that all Brassica PSY genes are likely undergoing purifying selection. Moreover, K a/K s ratios between PSY family members were found to be <0.35 for all pairs tested, which strongly indicates high function constraint of protein evolution (Supplemental Table S2). This is not surprising; structural and functional analyses in several plant species have shown that PSY together with isopentenyl diphosphate isomerase (IPI) and geranylgeranyl diphosphate synthase (GGPS) form a plastid envelope membrane-associated multi-enzyme complex (Schledz et al. 1996; Fraser et al. 2000). Such complex has been proposed to be essential for optimizing biosynthesis in vivo, by enabling the channeling of hydrophilic precursors to phytoene, avoiding unfavorable equilibria and isolating intermediate metabolites from competing reactions (Fraser et al. 2000). This high functional constraint might be one of the reasons why overexpression of PSY transgenes from different sources exerted different metabolic effects in transgenic plants (Ducreux et al. 2005; Paine et al. 2005) since different PSY proteins could differ in their ability to form fully functional protein complexes leading to different balances of produced carotenoids (Lindgren et al. 2003; Osborn et al. 2003). In this context, the battery of Brassica PSY genes presented in this paper could be used in genetic engineering strategies aimed at enhancing carotenoid content in oilseed crops with the potential of reaching higher levels than previously reported (Shewmaker et al. 1999; Paine et al. 2005).
Brassica PSY gene family expansion dates to paralogous subgenome triplication event
In this study, a total of 12 Brassica PSY genes were identified (Fig. 3). Gene copy number, sequence identity, genetic map positions and phylogenetic relationships indicate that these PSY gene family members correspond to three paralogous copies in B. rapa (BraA.PSY.a-c) and B. oleracea (BolC.PSY.a-c) and three homeologous gene pairs (BnaA.PSY.a/BnaC.PSY.b, BnaA.PSY.f/BnaC.PSY.d and BnaA.PSY.e/BnaC.PSY.c) in B. napus (Figs. 3, 4, 5; Supplemental Table S3). Using the estimated divergence time of 15–20 million years ago (MYA) for the split between Arabidopsis and the tribe Brassiceae, the phylogenetic relationship among the studied PSY genes places the genome triplication in the Brassica ancestor 11–15 MYA. This estimation is consistent with the reported paralogous subgenome triplication of diploid Brassica species (Yang et al. 2006). In addition, the fact that the highest levels of sequence identities were found between pairs of orthologous genes (Supplemental Table S3) suggests that most divergence in this gene family occurred before the speciation of B. rapa and B. oleracea. Altogether, these data indicate that PSY gene family expansion preceded the speciation of B. rapa and B. oleracea and all studied Brassica PSY genes evolved from the same ancestral gene.
Brassica PSY gene family members exhibit overlapping redundancy and early signs of subfunctionalization
Due to the highly conserved nature of these 12 Brassica PSY genes, we characterized their individual tissue-specific and developmental patterns of expression using cDNA-SSCP analysis (Figs. 6, 7, 8). In photosynthetic tissues (chloroplast-rich) we followed PSY expression in cotyledons, seedlings and mature leaves. To study chromoplast-rich tissues, we dissected flower buds and flowers at three different developmental stages and followed PSY expression in anthers and petals. It is worth mentioning that chloroplast to chromoplast transition occurs early during petal development (Weston and Pyke 1999; Egea et al. 2010). Petals and anthers collected at stage 1 are typically green and petals and anthers at stages 2 and 3 should be considered to have transitioned and contain mainly chromoplasts (Figs. 6a, 7a, 8a).
In these three Brassica species, all PSY homologues are expressed, exhibiting overlapping redundancy and signs of subfunctionalization. In B. rapa and B. oleracea, expression of orthologous gene pairs BraA.PSY.a/BolC.PSY.a and BraA.PSY.b/BolC.PSY.c was detected in all tissues, whereas expression of orthologous genes BraA.PSY.c and BolC.PSY.b was not detected in chromoplast-rich stages of petal development (Figs. 6, 7). In B. napus, expression of homeologues BnaC.PSY.a and BnaA.PSY.b were detected in all tissues, but homeologous gene pairs BnaA.PSY.c/BnaC.PSY.e and BnaA.PSY.d/BnaC.PSY.f exhibited preferential expression in chromoplast and chloroplast-rich tissues, respectively (Fig. 8). Previous studies have used cDNA-SSCP analysis to follow paralogous gene expression in cotton (Adams et al. 2003) and barley (Federico et al. 2006), describing the occurrence of organ-specific reciprocal paralogue silencing. This reciprocal silencing of genes that encode nearly identical proteins could be functionally and selectively important for dosage effect reasons (Birchler et al. 2001, 2005; Osborn et al. 2003; Freeling and Thomas 2006; Edger and Pires 2009).
Retention of triplicated PSY genes in B. napus and its diploid progenitors
In Arabidopsis, the flexibility and response capabilities to control the flux of the carotenoid biosynthetic pathway are limited to regulating PSY, a single copy enzyme (Scolnik and Bartley 1994). In species that contain more than one PSY gene, such as the grasses, tomato and potato to name a few, gene duplication has resulted in the subfunctionalization of gene expression (Bartley et al. 1992; Bartley and Scolnik 1993; Li et al. 2008b). Since carotenoids and chlorophylls are required to accumulate in a defined stoichiometric ratio in chloroplasts and the synthesis of both pigments share GGPP as a common substrate (Maass et al. 2009; Cazzonelli and Pogson 2010), the subfuntionalization of PSY expression provided a mechanism that allowed for PSY overexpression in flowers, fruits, seeds or tubers, where PSY has been shown to be rate-limiting (Maass et al. 2009; Shewmaker et al. 1999) without the detrimental effects that excessive carotenoid accumulation throughout the plant would have caused on photosynthesis (Busch et al. 2002).
Carotenoid accumulation in chromoplasts of flowers and fruits, albeit not essential for plant metabolism, contributes to plant fitness (Ehrenreich and Purugganan 2006; Galpaz et al. 2006; Howitt and Pogson 2006). Thus, functional retention of Brassica PSY genes could be explained by the selective advantage provided by increased levels of gene product (Force et al. 1999; Gu et al. 2003; Osborn et al. 2003) in chromoplast-rich tissues and concomitant carotenoid accumulation in petals. Dosage effects have been observed for many genes, including key regulators of developmental processes, in both diploid and polyploid species (Birchler et al. 2001, 2005; Osborn et al. 2003; Freeling and Thomas 2006; Edger and Pires 2009). The existence of convergent PSY duplications among monocotyledonous and dicotyledonous species as well as the subfunctionalization of PSY expression among photosynthetic and non-photosynthetic organs is consistent with this idea. Clearly, the case of Arabidopsis represents an exception to the gene balance hypothesis (Birchler and Veitia 2010) since PSY exists as a single copy gene in spite of genome duplications (Blanc et al. 2003). Notably, Arabidopsis bears white flowers that do not contain chromoplasts (Pyke and Page 1998). Additionally, carotenoid synthesis under stress conditions could have been adaptive in many species (Gallagher et al. 2004; Arango et al. 2010). In this regard, it remains to be established whether stress-induced production of ABA in roots is mediated by particular PSY homologues in these Brassica species.
References
Adams KL, Cronn R, Percifield R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100:4649–4654
Arango J, Wüst F, Beyer P, Welsch R (2010) Characterization of phytoene synthases from cassava and their involvement in abiotic stress-mediated responses. Planta 232:1251–1262
Bartley GE, Scolnik PA (1993) cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J Biol Chem 268:25718–25721
Bartley GE, Viitanen PV, Bacot KO, Scolnik PA (1992) A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J Biol Chem 267:5036–5039
Bassam BJ, Caetano-Anollés G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403:203–207
Birchler JA, Veitia RA (2010) The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol 186:54–62
Birchler JA, Bhadra U, Bhadra MP, Auger DL (2001) Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol 234:275–288
Birchler JA, Riddle NC, Auger DL, Veitia RA (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21:219–226
Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13:137–144
Brassica rapa Genome Sequencing Project Consortium (2011) The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics. doi:10.1038/ng.919
Busch M, Seuter A, Hain R (2002) Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol 128:439–453
Cazzonelli CI, Pogson BJ (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15:266–274
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500
Comeron JM (1999) K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15:763–764
Demmig-Adams B, Adams WW 3rd (2002) Antioxidants in photosynthesis and human nutrition. Science 298:2149–2153
Demmig-Adams B, Gilmore AM, Adams WW 3rd (1996) Carotenoids 3: in vivo function of carotenoids in higher plants. FASEB J 10:403–412
Ducreux LJ, Morris WL, Hedley PE, Shepherd T, Davies HV, Millam S, Taylor MA (2005) Metabolic engineering of high carotenoid potato tubers containing enhanced levels of beta-carotene and lutein. J Exp Bot 56:81–89
Edger PP, Pires JC (2009) Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res 17:699–717
Egea I, Barsan C, Bian W, Purgatto E, Latché A, Chervin C, Bouzayen M, Pech JC (2010) Chromoplast differentiation: current status and perspectives. Plant Cell Physiol 51:1601–1611
Ehrenreich IM, Purugganan MD (2006) The molecular genetic basis of plant adaptation. Am J Bot 93:953–962
Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
FAO (2011) Oilcrops, oils and meals market assessment. http://www.fao.org/economic/est/commodity-markets-monitoring-and-outlook/oilcrops/en/ (accessed 25 July 2011)
Federico ML, Iniguez-Luy FL, Skadsen RW, Kaeppler HF (2006) Spatial and temporal divergence of expression in duplicated barley germin-like protein-encoding genes. Genetics 174:179–190
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545
Fraser PD, Schuch W, Bramley PM (2000) Phytoene synthase from tomato (Lycopersicon esculentum) chloroplasts—partial purification and biochemical properties. Planta 211:361–369
Freeling M, Thomas BC (2006) Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res 16:805–814
Gallagher CE, Matthews PD, Li F, Wurtzel ET (2004) Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol 135:1776–1783
Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J (2006) A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18:1947–1960
Garcia-Boronat M, Diez-Rivero CM, Reinherz EL, Reche PA (2008) PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res 36:W35–W41
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421:63–66
Hirschberg J (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4:210–218
Howitt CA, Pogson BJ (2006) Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ 29:435–445
Iniguez-Luy FL, Federico ML (2011) The genetics of Brassica napus L. In: Bancroft I, Schmidt R (eds) Genetics and genomics of the Brassicaceae. Springer, New York, pp 291–322
Iniguez-Luy FL, Lukens L, Farnham MW, Amasino RM, Osborn TC (2009) Development of public immortal mapping populations, molecular markers and linkage maps for rapid cycling Brassica rapa and B. oleracea. Theor Appl Genet 120:31–43
Kabat EA, Wu TT, Bilofsky H (1977) Unusual distributions of amino acids in complementarity-determining (hypervariable) segments of heavy and light chains of immunoglobulins and their possible roles in specificity of antibody-combining sites. J Biol Chem 252:6609–6616
Kidwell KK, Osborn TC (1992) Simple plant DNA isolation procedure. In: Beckman J, Osborn TC (eds) Plant genomes: methods for genetic and physical mapping. Kluwer, The Netherlands, pp 1–13
Kubis S, Patel R, Combe J, Bédard J, Kovacheva S, Lilley K, Biehl A, Leister D, Ríos G, Koncz C, Jarvis P (2004) Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16:2059–2077
Li HM, Chiu CC (2010) Protein transport into chloroplasts. Annu Rev Plant Biol 61:157–180
Li F, Vallabhaneni R, Wurtzel ET (2008a) PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol 146:1333–1345
Li F, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET (2008b) The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol 147:1334–1346
Lindgren LO, Stålberg KG, Höglund AS (2003) Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol 132:779–785
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15:516–525
Maass D, Arango J, Wüst F, Beyer P, Welsch R (2009) Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS One 4:e6373
Maluf MP, Saab IN, Wurtzel ET, Sachs MM (1997) The viviparous12 maize mutant is deficient in abscisic acid, carotenoids, and chlorophyll synthesis. J Exp Bot 48:1259–1268
O’Neill CM, Bancroft I (2000) Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J 23:233–243
Osborn TC, Kole C, Parkin IA, Sharpe AG, Kuiper M, Lydiate DJ, Trick M (1997) Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thaliana. Genetics 146:1123–1129
Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, Lee HS, Comai L, Madlung A, Doerge RW, Colot V, Martienssen RA (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet 19:141–147
Ostergaard L, King GJ (2008) Standardized gene nomenclature for the Brassica genus. Plant Methods 20:4–10
Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL, Drake R (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol 23:482–487
Park JY, Koo DH, Hong CP, Lee SJ, Jeon JW, Lee SH, Yun PY, Park BS, Kim HR, Bang JW, Plaha P, Bancroft I, Lim YP (2005) Physical mapping and microsynteny of Brassica rapa ssp. pekinensis genome corresponding to a 222 kbp gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol Genet Genomics 274:579–588
Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781
Pyke KA, Page AM (1998) Plastid ontogeny during petal development in Arabidopsis. Plant Physiol 116:797–803
Rana D, van den Boogaart T, O’Neill CM, Hynes L, Bent E, Macpherson L, Park JY, Lim YP, Bancroft I (2004) Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J 40:725–733
Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M (2009) Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J 60:424–435
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schledz M, al-Babili S, von Lintig J, Haubruck H, Rabbani S, Kleinig H, Beyer P (1996) Phytoene synthase from Narcissus pseudonarcissus: functional expression, galactolipid requirement, topological distribution in chromoplasts and induction during flowering. Plant J 10:781–792
Scolnik PA, Bartley GE (1994) Nucleotide sequence of an Arabidopsis cDNA for phytoene synthase. Plant Physiol 104:1471–1472
Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY (1999) Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J 20:401–412
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Toledo-Ortiz G, Huq E, Rodríguez-Concepción M (2010) Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci USA 107:11626–11631
Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, Vigouroux M, Trick M, Bancroft I (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18:1348–1359
U N (1935) Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Van Ooijen JW (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V. Wageningen, Netherlands
von Lintig J, Welsch R, Bonk M, Giuliano G, Batschauer A, Kleinig H (1997) Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings. Plant J 12:625–634
Welsch R, Beyer P, Hugueney P, Kleinig H, von Lintig J (2000) Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 211:846–854
Welsch R, Medina J, Giuliano G, Beyer P, Von Lintig J (2003) Structural and functional characterization of the phytoene synthase promoter from Arabidopsis thaliana. Planta 216:523–534
Welsch R, Wüst F, Bär C, Al-Babili S, Beyer P (2008) A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol 147:367–380
Weston EL, Pyke KA (1999) Developmental ultrastructure of cells and plastids in the petals of wallflower (Erysimum cheiri). Ann Bot 84:763–769
Xiong Z, Gaeta RT, Pires JC (2011) Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc Natl Acad Sci USA 108:7908–7913
Yang YW, Lai KN, Tai PY, Li WH (1999) Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J Mol Evol 48:597–604
Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi BS, Kim JA, Jin M, Park JY, Lim MH, Kim HI, Lim YP, Kang JJ, Hong JH, Kim CB, Bhak J, Bancroft I, Park BS (2006) Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18:1339–1347
Acknowledgments
We would like to thank Dr. Heiko Becker and an anonymous referee for their helpful and constructive comments which have improved the contents of this manuscript. The authors would also like to thank Cristell Navarro and Hector Urbina for bioinformatics support and Fernando Westermeyer and Daniela Quezada for their technical assistance. This research was funded by CONICYT through Fondecyt project 1090726 and Agriaquaculture Nutritional Genomic Center, CGNA, CONICYT-Regional, GORE La Araucanía (R10C1001). We acknowledge INIA for its support providing infrastructure. Bilateral collaboration was supported by CONICYT No. 018/DRI/216.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Becker.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cárdenas, P.D., Gajardo, H.A., Huebert, T. et al. Retention of triplicated phytoene synthase (PSY) genes in Brassica napus L. and its diploid progenitors during the evolution of the Brassiceae . Theor Appl Genet 124, 1215–1228 (2012). https://doi.org/10.1007/s00122-011-1781-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1781-7