Abstract
PK23-2, a line of six-rowed barley (Hordeum vulgare L.) originating from Pakistan, has resistance to Japanese strains I and III of the barley yellow mosaic virus (BaYMV). To identify the source of resistance in this line, reciprocal crosses were made between the susceptible cultivar Daisen-gold and PK23-2. Genetic analyses in the F1 generation, F2 generation, and a doubled haploid population (DH45) derived from the F1 revealed that PK23-2 harbors one dominant and one recessive resistance genes. A linkage map was constructed using 61 lines of DH45 and 127 DNA markers; this map covered 1268.8 cM in 10 linkage groups. One QTL having a LOD score of 4.07 and explaining 26.8% of the phenotypic variance explained (PVE) for resistance to BaYMV was detected at DNA marker ABG070 on chromosome 3H. Another QTL having a LOD score of 3.53 and PVE of 27.2% was located at marker Bmag0490 on chromosome 4H. The resistance gene on chromosome 3H, here named Rym17, showed dominant inheritance, whereas the gene on chromosome 4H, here named rym18, showed recessive inheritance in F1 populations derived from crosses between several resistant lines of DH45 and Daisen-gold. The BaYMV recessive resistance genes rym1, rym3, and rym5, found in Japanese barley germplasm, were not allelic to rym18. These results revealed that PK23-2 harbors two previously unidentified resistance genes, Rym17 on 3H and rym18 on 4H; Rym17 is the first dominant BaYMV resistance gene to be identified in primary gene pool. These new genes, particularly dominant Rym17, represent a potentially valuable genetic resource against BaYMV disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Barley yellow mosaic virus (BaYMV) causes severe disease in barley (Hordeum vulgare L.) in Asia and Europe. BaYMV is transmitted to barley roots through infection by the soil-borne fungus Polymyxa graminis during the winter, and it causes mosaic symptoms on growing leaves the following spring (Toyama and Kusaba 1970). To overcome this disease in fields contaminated with BaYMV, introduction of resistance into barley cultivars is the most efficient and economical procedure. At present, 14 resistance genes derived from H. vulgare and two dominant genes from Hordeum bulbosum have been reported. These resistance genes have been localized on chromosomes 1H (rym7), 2H (Rym16 Hb), 3H (rym4, 5, 6, and 10), 4H (rym1, 8, 9, 11, 12, and 13), 5H (rym3), 6H (rym15 and Rym14 Hb), and 7H (rym2) by genetic and molecular analyses (Ukai and Yamashita 1980; Graner and Bauer 1993; Ordon and Friedt 1993; Graner et al. 1995, 1999; Bauer et al. 1997; Konishi et al. 1997; Iida et al. 1999; Ruge et al. 2003; Werner et al. 2003; Le Gouis et al. 2004; Ruge-Wehling et al. 2006).
In Europe, BaYMV, BaYMV-2, and barley mild mosaic virus (BaMMV) cause disease in susceptible winter barley cultivars. Resistance in European winter barley cultivars is conferred by the resistance genes rym4 and rym5. In Japan, BaYMV has differentiated into distinct strains, designated Japanese strains I to V (Kashiwazaki et al. 1989; Sotome et al. 2010). Modern Japanese resistant cultivars have been bred to contain recessive resistance genes rym5, effective against Japanese strains I, II, IV and V, and rym3, effective against strains I, II and III. Cultivars harboring both rym3 and rym5 are resistant to all strains of BaYMV currently identified in Japan. However, in general, viruses such as BaYMV undergo frequent genome rearrangement, enabling them to defeat resistance genes in the host (Sotome et al. 2010). To address this threat, it is necessary to identify and introduce new resistance genes or to pyramid three or more of the existing resistance genes in breeding programs.
In the past, we collected wild barley (Hordeum vulgare L.) originating from Pakistan, and one of them was established as a line, PK23-2, by pure-line selection. We investigated the resistance of PK23-2 and revealed that PK23-2 harbored novel BaYMV resistance genes. Furthermore, PK23-2 appeared to contain at least one dominant resistance gene (Furusho et al. 2002). Here, we evaluated the resistance of PK23-2 and several types of progeny populations, including doubled haploid lines, and we investigated the inheritance of resistance by a molecular approach to identify genetic resources for use in barley breeding programs.
Materials and methods
Plant material
PK23-2, a six-rowed accession of H. vulgare from Pakistan, was established by pure-line selection (Furusho et al. 1997). Daisen-gold is a two-rowed cultivar of H. vulgare that is susceptible to all known Japanese strains (I–V) of BaYMV. To perform segregation analysis, F1 and F2 populations were bred from reciprocal crosses between Daisen-gold and PK23-2, and a set of doubled haploid lines (DH45) was produced from the F1 by the Hordeum bulbosum method (Furusho et al. 1990). To test for dominance of the resistance genes in PK23-2, we chose seven resistant DH45 lines (DH45-1, -10, -63, -67, -68, -69, and -72) based on heading stage for cross with Daisen-gold. F1 populations were produced by making crosses between each resistant DH45 line and Daisen-gold. Moreover, to classify the recessive resistance gene discovered in PK23-2 (see Results), a genetic complementation test was performed using crosses between DH lines containing only the recessive resistance gene and two-rowed barley materials containing known resistance genes to Japanese strains I, II and III of BaYMV: Chikukei ym1-1 (rym1, 4H), Harushizuku (rym3, 5H).
Evaluation of resistance to BaYMV
The parental lines PK23-2 and Daisen-gold were sown each year from 2006 to 2010. The F1 and F2 populations were sown in 2006, and the corresponding F3 population was sown in 2007. The DH45 lines were sown each growing season from 2008 to 2010. F1 populations from each resistant DH45 line (DH45-1, -10, -63, -67, -68, -69, and -72) and Daisen-gold were sown to clarify the dominance in the resistance genes in 2010. Among four DH lines (DH45-10, -63, -67 and -69) carried only the recessive resistance gene, each DH45-63 and -69 and two barley materials containing known resistance genes were crossed and the obtained F1 progenies were sown to test for allelism to known recessive resistance genes in 2010. These plant materials were sown in early November of each year and evaluated the following year. For PK23-2, Daisen-gold, each F1 and 61 lines of DH45, ten seedlings were scored in each year of evaluation. For F2 population, 106 individuals of the F2 were scored in 2007 of evaluation. The test field was contaminated with BaYMV Japanese strains I and III. Individual plants were scored as resistant or susceptible to BaYMV based on mosaic symptoms on the leaves; scoring was performed every 9–10 days (three times) during the reproductive stage. Plants showing mosaic symptoms at any of the three scoring times were scored as susceptible. Evaluations of resistance to BaYMV in 61 lines of DH45 were performed for 3 years. The presence of the virus was confirmed using the DAS-ELISA method (Usugi et al. 1984) when the symptoms were not unequivocal.
DNA extraction and QTL analysis for BaYMV resistance genes
The linkage map construction and QTL analysis were performed by using the 61 lines of the DH45 population. For dominance analysis of the resistance genes in PK23-2, we investigated graphical genotypes of seven resistant DH45 lines (DH45-1, -10, -63, -67, -68, -69, and -72). Genomic DNA was extracted from young leaves by using a DNeasy Plant Mini Kit (QIAGEN, Japan) according to the manufacturer’s protocol. Simple sequence repeat (SSR) markers (Ramsay et al. 2000) and sequence tagged site (STS) markers (Blake et al. 1996; Mano et al. 1999) were selected and designed to construct the linkage map and detect QTLs for resistance to BaYMV. When polymorphisms could not be detected using STS markers, the PCR products were digested with 11 endonucleases (AccII, AluI, AvaII, Csp6I, HaeIII, HinfI, Hinf6I, MboI, MnlI, MspI and TruI) to generate cleaved amplified polymorphic sequence (CAPS) markers. PCR amplifications were done in a final volume of 15 μL, each containing 0.5 units of Taq polymerase (TaKaRa, Japan); 0.3 μM of each primer, 250 μM of dNTPs, 1 × buffer supplied with the polymerase, and 50 ng genomic DNA. PCR amplification conditions for SSR markers were 5 min at 95°C; followed by 35 cycles of 1 min at 95°C, 1 min at 55°C, and 1 min at 72°C; followed by 7 min incubation at 72°C. For STS or CAPS markers, the annealing time and elongation time were 2 min and 3 min, respectively; the rest of the conditions were as described for the SSR markers. PCR was done with a PC-818 thermal cycler (ASTEC, Japan). Amplified fragments with SSR markers were separated in 3.0% agarose gel electrophoresis, and those with STS or CAPS markers were separated in 1.0% agarose gel electrophoresis.
Initially, 480 DNA markers were screened for polymorphism between Daisen-gold and PK23-2. The 127 DNA markers that detected polymorphisms between the parents were selected for use in constructing the linkage map. The segregation ratios of all markers were checked in the DH45 population by the χ 2 test using the MAPL software (Ukai et al. 1995). For construction of genetic map, recombination values were calculated based on the model of doubled haploid and converted into genetic map distances (cM) by Kosambi function. QTL analysis of resistance to BaYMV was done using MAPL with a log-likelihood (LOD) of 2.0 for the doubled haploid model.
Results
Segregation analyses for BaYMV resistance in the F1, F2, and DH45 populations
The field was effective for evaluation of the plant materials: Daisen-gold was severely infected by BaYMV, whereas no symptoms were detected on the leaves of PK23-2 and the F1 populations grown in the same field (Table 1). The reaction of the F1 populations against BaYMV revealed that PK23-2 had at least one dominant resistance gene. Of the 106 F2 individuals evaluated for resistance in the infected field, 90 were uninfected and the remaining 16 showed mosaic symptoms. This observed segregation was not different from the expected segregation ratio (13 resistant: 3 susceptible) for an F2 generation segregating for two unlinked resistance genes, one dominant and one recessive. Moreover, in the DH45 population, 47 lines were resistant and 14 lines were susceptible in the infected field. This observed segregation was not significantly different from a ratio of 3 resistant: 1 susceptible, as expected if PK23-2 harbored two unlinked resistance genes.
Map construction and QTL analysis
To construct a linkage map and perform QTL analysis for resistance to BaYMV, 61 lines of DH45 were screened with the 127 DNA markers found to be polymorphic between the parents. A total of 121 markers in ten linkage groups were assigned to the seven chromosomes of barley (Fig. 1). The linkage map covered 1268.8 cM. Two QTLs for resistance to BaYMV were detected: one on chromosome 3H and one on 4H. On chromosome 3H, the QTL at CAPS marker ABG070 with MboI had an LOD score of 4.07 and explained 26.8% of the phenotypic variance explained (PVE). On chromosome 4H, the QTL at SSR marker Bmag0490 had a LOD score of 3.53 and PVE of 27.2%.
Test for dominance of newly identified resistance genes
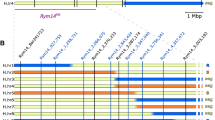
To test for dominance of the two resistance genes detected by QTL analysis, F1 populations derived from crosses between each resistant DH45 line (DH45-1, -10, -63, -67, -68, -69, and -72) and the susceptible Daisen-gold parent were examined for response to Japanese BaYMV strains I and III (Fig. 2). DNA marker analysis was used to identify the source of the DNA segments containing CAPS markers ABG070 with MboI and SSR marker EBmac0705 near the QTL on chromosome 3H, and CAPS marker MWG948 with TruI, SSR marker Bmag0490, and SSR marker Bmac0310 near the QTL on chromosome 4H. DH45-1, -68, and -72 contained segments from PK23-2 in both of these QTL regions associated with BaYMV resistance. In contrast, DH45-10, -63, -67, and -69 had segments from PK23-2 only in the QTL region on chromosome 4H; the region around the QTL on chromosome 3H was derived from Daisen-gold. F1 populations derived from DH45-1, -68, and -72 showed no mosaic symptoms, whereas F1 populations derived from DH45-10, -63, -67, and -69 were severely infected by BaYMV. These results revealed that the QTL on 3H behaved as a dominant gene and the QTL on 4H behaved as a recessive gene. Lines DH45-1, -68 and -72 had at least the dominant resistance gene, and lines DH45-10, -63, -67, and -69 had only the recessive resistance gene. To test for allelism to known sources of resistance, each DH45-63 and -69 were crossed to two barley lines known to carry rym1 or rym3 (Table 2). All of the F1 populations were susceptible in the contaminated field. These data showed that the recessive resistance gene in PK23-2 was different from rym1 and rym3.
Genotypes and phenotypes to BaYMV in resistant DH lines. Graphical genotypes for five DNA markers linked to BaYMV resistance QTLs on 3H and 4H in seven resistant doubled haploid (DH) lines. “F1” refers to the F1 generation derived from a cross between each resistant DH line and the susceptible parent Daisen-gold. White and gray rectangles indicate chromosome regions from Daisen-gold and PK23-2, respectively
Discussion
BaYMV disease is caused by infection with soil-borne BaYMV and results in severe yield loss in barley production. The only current procedure to prevent the disease is the introduction of resistance genes into cultivars. Prior to the present study, 14 recessive genes derived from H. vulgare and two dominant genes from H. bulbosum had been mapped to the seven chromosomes of barley. To overcome the threat of further differentiation of strains in BaYMV, it is necessary to introduce novel resistance genes derived from previously untapped genetic resources into modern cultivars. Until now, no completely dominant BaYMV resistance gene derived from H. vulgare had been reported. Dominant resistance genes have several advantages as preferable genetic resources in backcross breeding programs or production of F1 hybrid because of convenient introduction of resistance into objective cultivar. In this study, we focused on the unidentified sources of resistance in PK23-2, a line originating from Pakistan. PK23-2 showed no mosaic symptoms on its leaves in any year from 2007 to 2011 in the infected field contaminated with Japanese BaYMV strains I and III, whereas Daisen-gold consistently showed infection symptoms when grown at the same time and under the same conditions. Heterozygous F1 populations derived from reciprocal crosses between Daisen-gold and PK23-2 were completely resistant to BaYMV. The segregation of response to BaYMV in the F2 and DH populations was consistent with a model of one dominant resistance gene and one recessive resistance gene being present in PK23-2. Furthermore, QTL analysis and dominance testing revealed that the dominant gene, here named Rym17, and the recessive gene, here named rym18, are located adjacent to DNA marker ABG070 on chromosome 3H and Bmag0490 on chromosome 4H, respectively. The total PVE at nearest DNA markers located adjacent each QTL was 54.0%. We consider that the lower total PVE is due to distances between resistance genes and nearest DNA markers and also due to fewer numbers of DH lines and polymorphic DNA markers for high resolution QTL analysis. For this resolution, it needs further genetic analysis to reveal the details of two QTLs using large scale segregating population containing solely resistance gene. To date, there has been no report of the identification of a resistance gene at the distal part on 3H in H. vulgare. The reaction of the F1 populations from Daisen-gold and PK23-2 against Japanese BaYMV strains I and III revealed that Rym17 in PK23-2 conferred resistance to both strains of BaYMV. Moreover, the response of DH45-10, -63, -67, and -69, which carried only the recessive resistance gene revealed that rym18 was also resistant to the strains. In the previous studies, Rym14 Hb and Rym16 Hb were identified as dominant BaYMV resistance genes derived from H. bulbosum (Ruge et al. 2003; Ruge-Wehling et al. 2006), but use of these genes to improve commercial cultivars of H. vulgare carries the risk of linkage drag. In addition, it would be difficult to investigate or isolate these genes because of biased recombination in interspecific crosses. We consider that PK23-2 is a more useful genetic resource to overcome the threat of BaYMV disease because it can be used directly to investigate the genetics of resistance or to isolate the dominant resistance gene. In barley, the mechanism of recessive resistance conferred by rym4 and rym5 has been identified as eukaryotic translation factor 4E (Hv-elF4E) (Stein et al. 2005). In Arabidopsis, RTM1, RTM2, and RTM3, which are dominant resistance genes against tobacco etch virus (TEV), encode RTM proteins. These RTM proteins, multiprotein complex, are considered to confer the resistance mechanism by inhibition of the long-distance movement of TEV (Chisholm et al. 2000; Whitham et al. 2000; Cosson et al. 2010). Thereby, we consider the resistance mechanisms of Rym14 Hb, Rym16 Hb, and Rym17 differ from that of Hv-elF4E. It is interesting to clarify the dominant resistance mechanisms to overcome future threat of BaYMV. No DH line harboring only the PK23-2 allele of Rym17 was identified by the dominance test among seven resistant DH lines in this study, although we obtained several resistant DH lines containing this region from PK23-2 (Fig. 2). We will investigate the inheritance pattern of those resistant DH lines in an F2 segregation test, and from this material we hope to eventually isolate Rym17. The recessive resistance gene rym18 on chromosome 4H in PK23-2 was not allelic to rym1 or rym3, which resistance genes to Japanese strains I and III of BaYMV. In the future, we will test whether rym18 is allelic to resistance genes on chromosome 4H (Ordon et al. 2004). Moreover, identified Rym17 and rym18 will be investigated to resistance to other strains of BaYMV using resistant DH lines because of the future threat of further differentiation of strains in BaYMV. In conclusion, PK23-2 harbors one dominant BaYMV resistance gene (Rym17) and one recessive resistance gene (rym18), and it represents a potential resource for addressing the threat posed by future differentiation of BaYMV.
References
Bauer E, Weyen J, Schiemann A, Graner A, Ordon F (1997) Molecular mapping of novel resistance genes against barley mild mosaic virus (BaMMV). Theor Appl Genet 95:1263–1269
Blake TK, Kadyrzhanova D, Shepherd KW, Islam AKMR, Langridge PL, McDonald CL, Erpelding J, Larson S, Blake NK, Talbert LE (1996) STS-PCR markers appropriate for wheat–barley introgression. Theor Appl Genet 93:826–832
Chisholm ST, Mahajan SK, Whitham SA, Yamamoto ML, Carrington JC (2000) Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proc Natl Acad Sci USA 97:489–494
Cosson P, Sofer L, Le QH, Léger V, Schurdi-Levraud V, Whitham SA, Yamamoto ML, Gopalan S, Gall OL, Candresse T, Carrington JC, Revers F (2010) RTM3, which controls long-distance movement of potyviruses, is a member of a new plant gene family encoding a meprin and TRAF homology domain-containing protein. Plant Physiol 154:222–232
Furusho M, Hamachi Y, Yoshida T (1990) Variety difference in crossability between Japanese two-rowed barley and Hordeum bulbosum L. Japan J Breed 40:411–417
Furusho M, Tanifuji K, Gamo T, Ahmad Z, Shahid Masood M (1997) Collaborative exploration of wheat, barley and their wild relatives in Pakistan, 1996. Annual Report on Exploration and Introduction of PGR 13:59-86
Furusho M, Kai H, Tsukazaki M (2002) The dominant resistance gene to barley yellow mosaic virus from barley germplasms collected in Pakistan. Breed Res 4(2):296
Graner A, Bauer E (1993) RFLP mapping of the ym4 virus resistance gene in barley. Theor Appl Genet 86:689–693
Graner A, Bauer E, Kellermann A, Proeseler G, Wenzel G, Ordon F (1995) RFLP analysis of resistance to the barley yellow mosaic virus complex. Agronomie 15:475–479
Graner A, Streng S, Kellermann A, Proeseler G, Schiemann A, Peterka H, Ordon F (1999) Molecular mapping of genes conferring resistance to soil-borne viruses in barley. An approach to promote understanding of host–pathogen interactions. J Plant Dis Protec 106:405–410
Iida Y, Ban T, Konishi T (1999) Linkage analysis of the rym6 resistance gene to Japanese strain II of barley yellow mosaic virus (BaYMV-II) in barley. Barley Genet Newsl 29:31–32
Kashiwazaki S, Ogawa K, Usugi T, Omura T, Tsuchizaki T (1989) Characterization of several strains of barley yellow mosaic virus. Ann Phytopath Soc Jpn 55:16–25
Konishi T, Ban T, Iida Y, Yoshimi R (1997) Genetic analysis of disease resistance to all strains of BaYMV in a Chinese barley landrace, Mokusekko 3. Theor Appl Genet 94:871–877
Le Gouis J, Devaux P, Werner K, Hariri D, Bahrman N, Béghin D, Ordon F (2004) rym15 from the Japanese cultivar Chikurin Ibaraki 1 is a new barley mild mosaic virus (BaMMV) resistance gene mapped on chromosome 6H. Theor Appl Genet 108:1521–1525
Mano Y, Sayed-Tabatabaei B, Graner A, Blake T, Takaiwa F, Oka S, Komatsuda T (1999) Map construction of sequence-tagged sites (STSs) in barley (Hordeum vulgare L.). Theor Appl Genet 98:937–946
Ordon F, Friedt W (1993) Mode of inheritance and genetic diversity of BaMMV resistance of exotic barley germplasms carrying genes different from ‘ym4’. Theor Appl Genet 86:229–233
Ordon F, Friedt W, Scheurer K, Pellio B, Werner K, Neuhaus G, Huth W, Habekuss A, Graner A (2004) Molecular markers in breeding for virus resistance in barley. J Appl Genet 45:145–159
Ramsay L, Macaulay M, Ivanissevich S, MacLean K, Cardle L, Fuller J, Edwards K, Tuvesson S, Morgante M, Massari A, Maestri E, Marmiroli N, Sjakste T, Ganal M, Powell W, Waugh R (2000) A simple sequence repeat-based linkage map of barley. Genetics 156:1997–2005
Ruge B, Linz A, Pickering R, Proeseler G, Greif P, Wehling P (2003) Mapping of Rym14 HB, a gene introgressed from Hordeum bulbosum and conferring resistance to BaYMV in barley. Theor Appl Genet 107:965–971
Ruge-Wehling B, Linz A, Habekuß A, Wehling P (2006) Mapping of Rym16 HB, the second soil-borne virus-resistance gene introgressed from Hordeum bulbosum. Theor Appl Genet 113:867–873
Sotome T, Kawada N, Kato T, Sekiwa T, Nishigawa H, Natsuaki T, Kimura K, Meoka Y, Nagamine T, Kobayashi S, Wada Y, Yoshida T (2010) The current and new strains of barley yellow mosaic virus (BaYMV) in Tochigi prefecture. Jpn J Crop Sci 79:29–36
Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J 42:912–922
Toyama A, Kusaba T (1970) Transmission of soil-borne barley yellow mosaic virus. 2. Polymyxa graminis Led. as vector. Ann Phytopath Soc Jpn 36:223–229
Ukai Y, Yamashita A (1980) Induced mutation for resistance to barley yellow mosaic virus. Jpn J Breed 30:125–130
Ukai Y, Ohsawa R, Saito A, Hayashi T (1995) MAPL: a package of computer programs for construction of DNA polymorphism linkage map and analysis of QTL. Breed Sci 45:139–142
Usugi T, Kuwabara T, Tsuchizaki T (1984) Serological detection of barley yellow mosaic virus, wheat yellow mosaic virus and soil-borne wheat mosaic virus by ELISA. Ann Phytopath Soc Jpn 50:63–68
Werner K, Rönicke S, Gouis J, Friedt W, Ordon F (2003) Mapping of a new BaMMV-resistance gene derived from the variety ‘Taihoku A’. J Plant Dis Protec 110:304–311
Whitham SA, Anderberg RJ, Chisholm ST, Carrington JC (2000) Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 12:569–582
Acknowledgments
This work was conducted by the malting barley breeding program in Fukuoka Agricultural Research Center, and supported by a Grant-in-Aid from the Ministry of Agriculture, Forestry and Fisheries of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Graner.
Rights and permissions
About this article
Cite this article
Kai, H., Takata, K., Tsukazaki, M. et al. Molecular mapping of Rym17, a dominant and rym18 a recessive barley yellow mosaic virus (BaYMV) resistance genes derived from Hordeum vulgare L.. Theor Appl Genet 124, 577–583 (2012). https://doi.org/10.1007/s00122-011-1730-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1730-5