Abstract
Rapeseed (Brassica napus L.) is the leading European oilseed crop serving as source for edible oil and renewable energy. The objectives of our study were to (i) examine the population structure of a large and diverse set of B. napus inbred lines, (ii) investigate patterns of genetic diversity within and among different germplasm types, (iii) compare the two genomes of B. napus with regard to genetic diversity, and (iv) assess the extent of linkage disequilibrium (LD) between simple sequence repeat (SSR) markers. Our study was based on 509 B. napus inbred lines genotyped with 89 genome-specific SSR primer combinations. Both a principal coordinate analysis and software STRUCTURE revealed that winter types, spring types, and swedes were assigned to three major clusters. The genetic diversity of winter oilseed rape was lower than the diversity found in other germplasm types. Within winter oilseed rape types, a decay of genetic diversity with more recent release dates and reduced levels of erucic acid and glucosinolates was observed. The percentage of linked SSR loci pairs in significant (r 2 > Q 95 unlinked loci pairs) LD was 6.29% for the entire germplasm set. Furthermore, LD decayed rapidly with distance, which will allow a relatively high mapping resolution in genome-wide association studies using our germplasm set, but, on the other hand, will require a high number of markers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rapeseed (Brassica napus L.) is the leading European oilseed crop serving as source for edible oil and renewable energy. The species includes the subspecies B. napus ssp. napus, which comprises oilseed, fodder, and vegetable types and B. napus ssp. napobrassica, referred to as rutabaga or swede (Snowdon et al. 2007), which is utilized as root vegetable.
Crop breeding requires efficient germplasm organization, and this in turn is facilitated by knowledge of the population structure of the crop species under study. Population structure information can be gained not only from phenotypic or pedigree records, but also from variation at the DNA level (Varshney et al. 2005). Molecular markers such as restriction fragment length polymorphisms (RFLP), amplified fragment length polymorphisms (AFLP), random amplified polymorphic DNA (RAPD), and single sequence repeats (SSR) can be used for this purpose. Diers and Osborn (1994) fingerprinted 83 rapeseed inbreds, originating from Europe, Asia, America, and Australia, of winter, spring, as well as swede types using RFLP markers and observed distinct clusters for the former two. Chen et al. (2008) characterized patterns of allelic diversity in 72 B. napus genotypes from Australia and China, with controls from India, Europe and Canada, based on 55 SSRs. They identified distinct clusters for Chinese genotypes, Indian genotypes, as well as a set of genotypes with mixed origins, where the latter group shared many alleles with Australian genotypes. Hasan et al. (2008) examined the genetic structure among 94 B. napus gene bank accessions of winter and spring oilseed, fodder and vegetable types, originating from all continents except Africa. The most stable number of clusters was three, and each of them was dominated by (i) spring oilseed rape (OSR), (ii) fodder and vegetable, as well as (iii) winter OSR types. Nevertheless, no earlier study examined the population structure of a large, genetically diverse B. napus germplasm set representing species-wide as well as world-wide variation.
B. napus most likely originated from a few interspecific hybridizations between turnip rape (Brassica rapa L.) and cabbage (Brassica oleracea L.) that occurred spontaneously during medieval times (Iñiguez Luy and Federico 2011). This led to a relatively low genetic diversity in the species. The narrow genetic base together with the occurrence of two bottlenecks during breeding of modern oilseed rape, the selection for low erucic acid and low glucosinolate (GSL) content, led to a low genetic diversity in modern elite varieties (Becker et al. 1995). The drawback of a low genetic diversity is often a strong susceptibility to biotic and abiotic stresses (Kebede et al. 2010) as well as a reduced response to selection (cf. Falconer and Mackay 1996). To overcome such disadvantages, diverse germplasm has to be introgressed into elite germplasm (Haussmann et al. 2004). This, however, is facilitated by understanding the pattern of genetic diversity in the crop species under consideration. Hasan et al. (2006) found a high level of genetic diversity in vegetable types and low diversity in spring OSR and fodder types in a study of 96 genotypes with 30 SSR primer combinations.
As B. napus lacks wild relatives to its modern cultivars, its original parental diploid genomes, which hybridized spontaneously, are unknown (Iñiguez Luy and Federico 2011). It is therefore impossible to determine, based on the hybridization event between the founder genotypes, which parental species contributed more to the overall diversity or hints at more domestication events. However, diversity studies based on A and C genome-specific markers would provide information on which genome has a narrower diversity. Subsequently, genetic variation could be introduced into this specific genome using interspecific crosses (Chen and Heneen 1989).
The use of markers, which are linked to traits of interest, during the breeding process has the potential to increase the gain from selection (Collard et al. 2005). A promising method to identify such markers is association analysis (Thornsberry et al. 2001; Breseghello and Sorrells 2006; Stich et al. 2008). This, however, neccessitates the knowledge of extent and distribution of linkage disequilibrium (LD) of the germplasm set under consideration. Ecke et al. (2010) examined the extent and distribution of LD in a set of 85 winter rapeseed lines with 845 AFLP markers. This study found a low level of LD with a mean r 2 of 0.020 for unlinked and 0.122 for linked markers. LD decayed rapidly with distance between linked markers with the high levels of LD extending over about 2 cM. Apart from this study, little is known about the extent of LD in rapeseed and its causes, and no LD studies have been performed on a large and genetically diverse set of B. napus.
The objectives of our study were to (i) examine the population structure of a large and diverse set of B. napus inbred lines, (ii) investigate patterns of genetic diversity within and among different germplasm types, (iii) compare the two genomes of B. napus with regard to genetic diversity, and (iv) assess the extent of LD between SSR markers.
Materials and methods
Plant material and molecular markers
A set of 509 rapeseed inbred lines, assembled to maximize variation, was used in this study. Based on available information from gene banks, plant breeders, and our own observations the accessions were assigned to eight different germplasm types, namely winter oilseed rape (OSR) (183), winter fodder (22), swede (73), semi-winter OSR (7), spring OSR (204), spring fodder (4), vegetable (10), and so far unspecified rapeseed genotypes (6). From the whole set, 314 genotypes originated from Western Europe, 43 from Eastern Europe, 32 from Asia, 5 from Africa, 20 from Australia, 70 from North America, and 25 genotypes were of unknown origin (Supplementary Material S1).
A subset of 136 winter OSR genotypes was divided into three subgroups according to their seed quality with regard to levels of erucic acid and GSL, namely ++ (17), 0+/+0 (11), and 00 (108). Furthermore, 95 winter OSR genotypes were divided into the following four subgroups according to the time period of release: 1954–1979 (11), 1980–1989 (15), 1990–1999 (28), 2000–2007 (41) (Supplementary Material S1).
All 509 inbred lines were fingerprinted with SSR markers amplified by 89 primer combinations for loci evenly distributed across the B. napus genome, of which 31 SSRs were proprietary (primer combinations can be obtained via a material transfer agreement from Agriculture and Agri-Food Canada) and 58 SSRs were public (Supplementary Material S2). According to information from multiple segregating populations, 43 of the 89 primer combinations amplified specific polymorphic loci in the B. napus A genome and 44 amplified specific polymorphic loci in the B. napus C genome. Chromosome positions of two SSRs were unknown.
The examined germplasm was provided from seed banks as inbred material, however, one additional generation of selfing was carried out. Furthermore, to ensure homozygosity, phenotypes were checked for homogeneity. DNA was isolated from young leaves using a modified CTAB extraction protocol (Saghai-Maroof et al. 1984). SSR genotyping was performed using capillary electrophoresis of fluorescently labeled PCR products at the Max Planck Genome Centre Cologne following standard protocols. Only homozygous marker-genotype combinations were used in the statistical analysis described below.
Statistical analyses
Gene diversity D (Weir 1996), range and mean of the number of alleles per locus (Nb), and the number of germplasm type-specific alleles (Nbt) were calculated. An F ST analysis according to Wright (1965) was performed. Pairwise modified Roger’s distance (MRD) estimates between germplasm types were calculated according to Wright (1978). Standard errors of the MRD estimates were obtained through a bootstrap procedure with resampling across markers and genotypes. Associations among genotypes were revealed with principal coordinate analysis (PCoA) (Gower 1966) based on MRD estimates between pairs of inbred lines. All aforementioned analyses were performed for the entire germplasm set as well as for various subsets. In addition, calculation of D and PCoA analysis were performed for all markers as well as separately for the markers representing the A and C genomes, respectively.
A model-based cluster analysis MCLUST was carried out to assign genotypes to clusters. This was done based on the first 30 principal coordinates, using the best model with (a) the number of clusters as estimated by MCLUST, and (b) the number of clusters as chosen according to the results of the PCoA. For the genetic diversity measures, the following resampling procedure was performed to correct for the different sizes of the examined germplasm groups (Stich et al. 2005): a subset of N S genotypes was randomly selected from the original germplasm group and all genetic diversity measures were calculated. The procedure was repeated 1000 times and the results were averaged. Because of the small sample sizes of the germplasm types spring fodder (N = 4) and unspecified inbred lines (N = 6), the size of the subset of genotypes was chosen to be N S = 7, and the resampling procedure was not performed for the aforementioned two germplasm types. The same analyses were performed with N S = 73 for germplasm types and N S = 119 for MCLUST clusters.
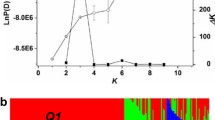
The model-based approach implemented in software package STRUCTURE 2.2 (Pritchard et al. 2000) was used to subdivide the rapeseed germplasm examined in our study into different subgroups. Because the estimated ‘log probability of data’ (LnP(D)) from STRUCTURE overestimates the number of subgroups when examining inbred lines (Pritchard and Wen 2004), we used the ad hoc measure \(\Updelta K\) (Evanno et al. 2005) to estimate the number of subgroups. In our study, the membership of each genotype was tested for the range of genetic clusters from K = 1 to K = 15 with the admixture model, without prior information on their origins. Each run consisted of a burn-in period of 100,000 steps followed by 100,000 Monte Carlo Markov Chain replicates as suggested by Pritchard and Wen (2004), assuming that allele frequencies are uncorrelated across clusters. The run of the estimated numbers of subgroups showing the maximum likelihood was used to assign inbreds to that subgroup for which they showed the highest membership probability.
The LD measure r 2 (Hill and Robertson 1968; square of the correlation of the allele frequencies) was calculated for linked and unlinked loci pairs, where linked loci were defined as loci being located on the same chromosome and unlinked loci were defined as loci being located on different chromosomes. The percentage of linked loci in significant LD was determined with two different significance thresholds, namely the 95% quantile of the r 2 value among unlinked loci pairs, and 0.1 as the minimal r 2 value to detect associations explaining 10% of the phenotypic variance in populations with at least 300 entries (Ersoz et al. 2007). The overall decay of LD was evaluated by nonlinear regression of r 2 according to Heuertz et al. (2006). In addition, the effective population size N e (Wright 1931) was calculated. The previously described resampling strategy was also used to obtain comparable estimates for LD, since the power to detect LD depends on the number of examined genotypes (Stich et al. 2005). The procedure was repeated 50 times and the results were averaged. All statistical analyses were performed with statistical software R (R Development Core Team 2011).
Results
The gene diversity D was 0.50 for the entire germplasm set (Table 1). D estimates ranged from 0.35 (semi-winter) to 0.51 (unspecified) for germplasm types and from 0.38 (MCLUST 1) to 0.50 (MCLUST 3) for MCLUST clusters of the three-cluster model. For the separate genomes, D was 0.58 for the A genome and 0.43 for the C genome. The mean Nb was 7.42. For the individual germplasm types and MCLUST clusters of the three-cluster model, the mean Nb ranged from 2.12 (spring fodder) to 5.76 (spring OSR) and from 5.07 (MCLUST 2) to 6.30 (MCLUST 3), respectively. Nbt varied from 1 (winter fodder, semi-winter OSR, and spring fodder) to 65 (spring OSR) for the germplasm types and from 36 (MCLUST 2) to 102 (MCLUST 3) for the MCLUST clusters of the three-cluster model. The overall F ST was 0.176 for the entire germplasm set when considering the germplasm types and ranged from 0.002 (spring fodder, unspecified) to 0.095 (spring OSR) for individual germplasm types. The overall F ST for for the entire germplasm set with regard to MCLUST clusters of the three-cluster model was 0.171 and ranged from 0.054 (MCLUST 3) to 0.109 (MCLUST 1) for individual MCLUST clusters (Table 1). The measures for genetic diversity calculated for the subgroups based on seed quality and release period resulted in a tendency for a higher genetic diversity with older genotypes (Table 2).
In the PCoA based on MRD estimates between all pairs of B. napus inbreds and 89 SSRs, the first two principal coordinates explained 15.5 and 8.5% of the molecular variance (Fig. 1). With respect to these two principal coordinates, winter types, spring types, and swedes were assigned to three major clusters. The average MRD estimates between pairs of inbreds from different germplasm types ranged from 0.16 (winter OSR vs. winter fodder) to 0.48 (winter OSR vs. semi-winter OSR) for the A genome (Table 3) and from 0.19 (winter fodder vs. winter OSR) to 0.39 (spring fodder vs. swede) for the C genome.
Principal coordinate analysis of 509 B. napus genotypes based on modified Roger's distance calculated from 89 simple sequence repeat marker loci. PC 1 and PC 2 are the first and second principal coordinates, respectively. The proportion of variance explained by the principal coordinates is given in parentheses. Genotypes were assigned to subgroups identified by three- (a) and eight-cluster (b) models of MCLUST. Colors represent the germplasm types and symbols represent MCLUST clusters
The first 30 principal coordinates, which were used for the model-based cluster analysis, explained 58.99% of the molecular variance. MCLUST assigned all genotypes to eight MCLUST clusters (Fig. 1). Although the number of estimated MCLUST clusters was identical to the number of germplasm types, the two classifications matched poorly (Table 4). The three-cluster model, however, resulted in a good accordance with the results of the PCoA (Fig. 1). The PCoA analyses based on the genome-specific markers revealed similar structures for the A and C genomes (Fig. 2). Different germplasm types clustered comparably to those detected in the PCoA with all 89 SSRs (Fig. 1). The correlation between pairwise MRD estimates of the A and C genomes was 0.59.
Principal coordinate analysis of 509 B. napus genotypes based on modified Roger’s distance calculated from 43 simple sequence repeat (SSR) marker loci mapping to the A genome (a) and 44 SSR marker loci mapping to the C genome (b) of B. napus. PC 1 and PC 2 are the first and second principal coordinates, respectively. The proportion of variance explained by the principal coordinates is given in parentheses. Colors represent the germplasm types
The percentage of linked SSR loci pairs in significant (r 2 > Q 95 unlinked loci pairs) LD was 6.29% for the entire germplasm set (Table 5), and ranged from 1.00 to 12.64% for the germplasm types which comprised at least seven genotypes. The percentage of linked loci pairs with r 2 > 0.1 was 5.14% for the entire set and covered a range from 1.18 to 81.62% for germplasm types. With regard to the entire set, the mean r 2 was 0.023 for linked loci and 0.019 for unlinked loci. For germplasm types consisting of seven or more genotypes, the mean r 2 ranged from 0.014 to 0.141 for linked loci pairs and from 0.009 to 0.154 for unlinked loci pairs. In the entire germplasm set, r 2 decayed with genetic distance (Fig. 3).
Plot of the squared allele frequency correlations (r 2, dots) versus genetic map distance (cM) between linked simple sequence repeat marker loci in the entire germplasm set. The solid line represents the nonlinear regression trend line of r 2 versus genetic map distance, whereas the dashed line shows the Q 95 unlinked loci pairs threshold. The inset gives an enhanced view of the r 2 decay over small distances
Discussion
Population structure of the rapeseed germplasm set
The overall fixation index F ST, which serves as an overall measure of population differentiation, was 0.176, indicating a moderate differentiation between germplasm types of our study. Cruz et al. (2007) found a \(\Upphi_{pt}, \) which is analogous to F ST (Rudh et al. 2007), of 0.11 in a set of 50 winter and spring rapeseed accessions from North America, Europe, Asia, and New Zealand. The stronger differentiation we found in our set might be explained by the inclusion of swede types in our study. However, Lombard et al. (2000) found with \(\Upphi_{pt} = 0.329\) a much greater differentiation in a set of 83 mostly European rapeseed cultivars, of which 68 were winter and 15 were spring cultivars. This observation could be due to the relatively small number of spring genotypes in their study, which were possibly strongly differentiated from the winter types.
Comparable studies on maize elite breeding material resulted in a similar (0.16) overall F ST value among heterotic pools (Van Inghelandt et al. 2010). However, rapeseed breeding mainly uses material of only one germplasm type. Therefore, our results suggest that rapeseed breeding material shows a considerably lower differentiation than that of maize.
Population structure in the germplasm set was examined with PCoA (Fig. 1) and software STRUCTURE 2.2 (Pritchard et al. 2000) (data not shown). Both methods indicated that the inbreds of our rapeseed germplasm set can be assigned to three clusters. Furthermore, the results of these two methods as well as the MCLUST analysis with three clusters indicated that one of the clusters was mainly made up of winter types, whereas the other two were made up of spring types as well as swede types, respectively (Fig. 1; Table 4). Our finding of distinct clustering of winter and spring types has been described previously by Diers and Osborn (1994) as well as by Hasan et al. (2006). The high genetic distance between spring and winter types can be explained not only by the breeding history but also by the adaptation of breeding material to distinct environments. Spring types have a low winter hardiness and do not require vernalisation to flower. Therefore, they are generally grown in regions with a short summer growing season, like Canada or Scandinavia, or where the winter is not sufficiently cold to induce flowering (e.g. Australia). On the other hand, winter and semi-winter oilseed rape cultivars are adapted to the temperate climate in Western Europe and Asia, respectively, where they are preferred to spring types due to their considerably higher yield potential (Kimber and McGregor 1995; Friedt and Snowdon 2009).
So far, the structuration of a large number of swede types in a germplasm set has not been studied. We observed a clear separation of swede types from the other two identified clusters (Fig. 1). This indicates that swedes most likely derived from different interspecific hybridisation events than those which led to today’s winter OSR and spring OSR.
Three of the four spring fodder genotypes examined in our study clustered together with the spring OSR inbreds (Fig. 1). However, the spring fodder inbred ‘Tira’ clustered with the winter fodder types. The same observation was made by Hasan et al. (2006), who classified this genotype as a winter form even though it is listed as spring fodder type. Furthermore, the average MRD estimate was particularly small between winter OSR and winter fodder types (Table 3). These findings can be explained by the fact that OSR as well as fodder types were derived from the corresponding winter or spring gene pool and differ only with respect to their growth type but not with respect to their ancestry. Similarly, we observed a low MRD between vegetables and winter fodder types, suggesting that these forms also share a common ancestry.
The MRD estimates between semi-winter OSR from China and the other germplasm types were especially high for the markers from the A genome. This is probably due to the breeding history of the inbreds of this germplasm type. Chinese semi-winter OSR genotypes were created by introgressing Chinese B. rapa into B. napus germplasm, which has been brought to China in the 1930–1940s. Since then, it has genetically diversified from OSR originating from regions other than Asia (Qian et al. 2006).
Within winter OSR, we did not observe further distinct subgroups. This finding is in accordance with the results of Ecke et al. (2010). Our observation could be explained by the fact that until recently, rapeseed has been bred by line-breeding and population breeding. In such breeding programs, population structure tends to be disregarded when choosing the parents of a cross. However, over the past few years hybrid breeding has received considerable attention in winter, spring, and semi-winter types alike, so that differentiated subpopulations will be developed in the various gene pools through recurrent, reciprocal selection, as has been the case for maize (Duvick et al. 2004) and sugar beet (Li et al. 2010).
Genetic diversity of the rapeseed germplasm set
In addition to population structure, information about the distribution of genetic diversity is important for practical breeding as well as genetic resource management. Across the 509 rapeseed inbreds, we observed a total gene diversity D of 0.50, a range of Nb of 2–18 and a mean Nb of 7.42 (Table 1). Hasan et al. (2006) found a smaller Nb for a set of 96 rapeseed inbreds of diverse germplasm types and origins. This finding is partly due to the lower number of inbreds examined by Hasan et al. (2006) compared to our study. However, when adjusting the size of our germplasm set to that of Hasan et al. (2006) by resampling simulations (data not shown), we observed with a mean Nb = 5.42 still more alleles per locus than Hasan et al. (2006). This finding can be explained by the composition of our germplasm set which is of more diverse origins and the fact that more germplasm types were covered in our study when compared with the material of Hasan et al. (2006). This observation, in turn, suggests that our germplasm set might be a valuable resource for pre-breeding and association mapping.
We observed a lower genetic diversity for winter OSR inbreds of more recent release than for inbreds which were released some decades ago (Table 2). Furthermore, we observed a lower genetic diversity for 00 seed quality winter OSR varieties than for the other seed quality subgroups (Table 2). These findings illustrate the effect of genetic bottlenecks on the genetic variability found in modern winter OSR inbred lines. The absence of erucic acid and low levels of GSL were introduced to modern OSR varieties from the spring cultivars ‘Liho’ and ‘Bronowski’, respectively (Friedt and Snowdon 2009). Spring OSR experienced similar bottlenecks as winter OSR. Therefore, it is an important goal of today’s rapeseed breeding to increase the genetic variability of modern winter and spring OSR germplasm.
With D = 0.44, we found a higher gene diversity for spring OSR compared to D = 0.38 for winter OSR (Table 1). This observation is in accordance with our finding of a larger effective population size for the former than the latter (Table 5). These results suggest that winter OSR types experienced a stronger bottleneck than spring OSR. Our observation was unexpected and in contrast to the results of Hasan et al. (2006), who found a higher allelic diversity for winter OSR than for spring fodder/OSR types. This finding is most likely due to the fact that the spring OSR sample size in our study was with N = 204 much higher than that of Hasan et al. (2006) (N = 21). This in turn increases the power for detecting allelic diversity. This result suggests, together with the observation of a high Nbt value for spring OSR, that genetic diversity of winter OSR could be increased by introgressing spring OSR germplasm. However, this has to be accompanied by phenotypic selection on winter hardiness and vernalization requirements.
The high D value of 0.44 (Table 1) found for vegetable types is in accordance with the results of Hasan et al. (2006). We further observed that swede types showed a high gene diversity, which confirms that this type is not only phenologically extremely diverse (Hasan et al. 2006), but also on the genetic level. The high gene diversity of germplasm types which today are of relatively low economic importance, namely vegetable, fodder, and swede types (Table 1), might reflect a part of the domestication process, as these germplasm types did not fully undergo the major genetic bottlenecks which occurred during OSR breeding (Ramanatha Rao and Hodgkin 2002). These points suggest, together with the observation that the swede germplasm formed a distinct subgroup from spring OSR and winter OSR, the idea of utilizing swede germplasm as a resource for diversity in corresponding breeding programs. However, the risk of transmission of unsuitable agronomic characters of swedes must be taken into consideration. Repeated backcrossing in order to remove unwanted alleles will be required (Basunanda et al. 2007). On the other hand, swedes may also possess interesting variation for tolerance to abiotic stresses, particularly drought.
The availability of genome-specific primer combinations makes it possible to dissect the genetic diversity not only on a genome-wide scale, but also separately for the A and C genomes. This allows the detection of genome-specific patterns of diversity, which in turn would allow to make inferences on possible sources of genetic diversity for breeding purposes. The ratio of D A genome/D C genome observed was higher than 1 for the entire germplasm set as well as all germplasm types (Table 1). This observation could be explained by a higher contribution from B. rapa than from B. oleracea genotypes to the B. napus diversity. Our finding suggests that the introduction of known genetic variability from B. oleracea, as described by Rahman et al. (2011), might be a strategy to broaden the genetic diversity specifically of the C genome.
The high ratio of D between the A and C genomes found for semi-winter OSR (Table 1) might be explained by the aforementioned introgression of Chinese B. rapa in B. napus in order to improve the adaptation of the latter to the local environment (Qian et al. 2006). The same explanation might be true for the high ratio of D between the A and C genomes observed for swede and spring OSR. On the other hand, this latter finding might reflect major individual domestication events of the respective germplasm types on the C genome. These events might have ocurred during selection for swollen hypocotyls in the case of swedes (Ramsay et al. 2001) and for low erucic acid and low GSL content in spring OSR (Becker et al. 1995). However, elucidation of these observations requires further research.
Extent of linkage disequilibrium and prospects for association studies
The resolution of genome-wide association studies (GWAS) depends on the extent and structure of LD detected in the germplasm under consideration. With regard to germplasm types, the percentage of linked loci pairs with r 2 > 0.1 covered a very large range from 1.18 to 81.62% (Table 5). The high percentage of linked loci pairs in LD with r 2 > 0.1, which occurred for germplasm types with small sample sizes, is caused by an overestimation of r 2 values. This is in line with the findings of Zapata et al. (2001), who described the effect of sample size on D′. On the other hand, we observed a decrease in the percentage of linked loci pairs with r 2 > Q 95 unlinked loci pairs with decreasing sample size. This is because the inflation of r 2 values between linked loci pairs is leveled by relating it to the r 2 values between unlinked loci. In this case, a small sample size reduces the power to observe LD and leads to a reduced percentage of linked loci pairs with r 2 > Q 95 unlinked loci pairs. These findings illustrate the importance of performing resampling simulations when comparing LD in germplasm sets of differing sizes, independent of the chosen significance threshold.
The mean r 2 for winter OSR types resampled to N = 85 was 0.015 for unlinked loci pairs (data not shown) and therewith in adherence to the finding of Ecke et al. (2010) (0.020). Nevertheless, the mean r 2 value for linked loci pairs was with 0.021 only slightly higher than that between unlinked loci pairs and considerably lower than the value of 0.122 reported by Ecke et al. (2010). This finding can be explained by the average marker density, which was remarkably lower in our study compared to that of Ecke et al. (2010) and too low for GWAS.
The expected r 2 declined to the significance threshold within about 1 cM in the entire germplasm set (Fig. 3). This value is in good accordance with the distance determined by Ecke et al. (2010), who found high levels of LD extending over about 2 cM. The rapid decay of LD with distance will allow a relatively high mapping resolution in GWAS using our germplasm set. Assuming a genome size of at least 2,000 cM, and aiming at a coverage of at least 1 marker per cM, more than 2,000 evenly distributed markers will be required. However, for detecting markers which explain only a low proportion of the phenotypic variation, r 2 values of at least 0.8 are necessary (Ersoz et al. 2007). Consequently, considerably more than 2,000 markers will be required for GWAS.
Summary
A large B. napus genetic diversity set representing a large range of geographical and morphological diversity showed genetic differentiation into three distinct clusters broadly representing winter, spring, and swede types. The genetic diversity of winter OSR was lower than the diversity found in other germplasm types, and within winter OSR types, a decay of genetic diversity with more recent release dates and reduced levels of erucic acid and GSL was observed. The extent of LD found in our study suggests that GWAS using this germplasm set will have a high mapping resolution, but, on the other hand, will require a high number of markers.
References
Basunanda P, Spiller TH, Hasan M, Gehringer A, Schondelmaier J, Lühs W, Friedt W, Snowdon RJ (2007) Marker-assisted increase of genetic diversity in a double-low seed quality winter oilseed rape genetic background. Plant Breed 126:581–587
Becker HC, Engqvist GM, Karlsson B (1995) Comparison of rapeseed cultivars and resynthesized lines based on allozyme and RFLP markers. Theor Appl Genet 91:62–67
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177
Chen BY, Heneen WK (1989) Resynthesized Brassica napus L.: a review of its potential in breeding and genetic analysis. Hereditas 111:255–263
Chen S, Nelson MN, Ghamkhar K, Fu T, Cowling WA (2008) Divergent patterns of allelic diversity from similar origins: the case of oilseed rape (Brassica napus L.) in China and Australia. Genome 51:1–10
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 142:169–196
Cruz VMV, Luhman R, Marek LF, Rife CL, Shoemaker RC, Brummer EC, Gardner CAC (2007) Characterization of flowering time and SSR marker analysis of spring and winter type Brassica napus L. germplasm. Euphytica 153:43–57
Diers BW, Osborn TC (1994) Genetic diversity of oilseed Brassica napus germ plasm based on restriction fragment length polymorphisms. Theor Appl Genet 88:662–668
Duvick DN, Smith JSC, Cooper M (2004) Long-term selection in a commercial hybrid maize breeding program. Plant Breed Rev 24:109–151
Ecke W, Clemens R, Honsdorf N, Becker HC (2010) Extent and structure of linkage disequilibrium in canola quality winter rapeseed (Brassica napus L.). Theor Appl Genet 120:921–931
Ersoz ES, Yu J, Buckler ES (2007) Applications of linkage disequilibrium and association mapping in crop plants. In: Varshney RK, Tuberosa R (eds) Genomics-assisted crop improvement, vol 1: genomics approaches and platforms. Springer, Dordrecht, pp 97–119
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics, 4th edn. Longman Group Ltd, London
Friedt W, Snowdon RJ (2009) Oilseed rape. In: Vollmann J, Rajcan, I (eds) Handbook of plant breeding, vol 4: oil crops breeding. Springer, New York, pp 91–126
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Hasan M, Friedt W, Pons-Kühnemann J, Freitag NM, Link K, Snowdon RJ (2008) Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus). Theor Appl Genet 116:1035–1049
Hasan M, Seyis F, Badani AG, Pons-Kühnemann J, Friedt W, Lühs W, Snowdon RJ (2006) Analysis of genetic diversity in the Brassica napus L. gene pool using SSR markers. Genet Resour Crop Evol 53:793–802
Haussmann BIG, Parzies HK, Presterl T, Sušić Z, Miedaner T (2004) Plant genetic resources in crop improvement. Plant Genet Resour 2:3–21
Heuertz M, De Paoli E, Källman T, Larsson H, Jurman I, Morgante M, Lascoux M, Gyllenstrand N (2006) Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway spruce [Picea abies (L.) Karst]. Genetics 174:2095–2105
Hill WG, Robertson A (1968) Linkage disequilibrium in finite populations. Theor Appl Genet 38:226–231
Iñiguez Luy FL, Federico ML (2011) The genetics of Brassica napus L. In: Bancroft I and Schmidt R (eds) Genetics and genomics of the Brassicaceae. Springer, New York Dordrecht Heidelberg London, pp 291–322
Kebede B, Thiagarajah M, Zimmerli C, Rahman MH (2010) Improvement of open-pollinated spring rapeseed (Brassica napus L.) through introgression of genetic diversity from winter rapeseed. Crop Sci 50:1236–1243
Kimber D, McGregor DI (1995) Brassica oilseeds: production and utilization. CAB International, Wallingford
Li JQ, Schulz B, Stich B (2010) Population structure and genetic diversity in elite sugar beet germplasm investigated with SSR markers. Euphytica 175:35–42
Lombard V, Baril CP, Dubreuil P, Blouet F, Zhang D (2000) Genetic relationships and fingerprinting of rapeseed cultivars by AFLP: consequences for varietal registration. Crop Sci 40:1417–1425
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen W (2004) Documentation for structure software: Version 2. http://pritch.bsd.uchicago.edu/software/readme_2_1/readme.html
Qian W, Meng J, Li M, Frauen M, Sass O, Noack J, Jung C (2006) Introgression of genomic components from Chinese Brassica rapa contributes to widening the genetic diversity in rapeseed (B. napus L.), with emphasis on the evolution of Chinese rapeseed. Theor Appl Genet 113:49–54
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rahman MH, Bennett RA, Yang RC, Kebede B, Thiagarajah MR (2011) Exploitation of the late flowering species Brassica oleracea L. for the improvement of earliness in B. napus L.: an untraditional approach. Euphytica 177:365–374
Ramanatha Rao V, Hodgkin T (2002) Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tissue Organ Cult 68:1–19
Ramsay LD, Bradshaw JE, Griffiths DW, Kearsey MJ (2001) The inheritance of quantitative traits in Brassica napus ssp. rapifera (swedes): augmented triple test cross analyses of production characters. Euphytica 121:65–72
Rudh A, Rogell B, Höglund J (2007) Non-gradual variation in colour morphs of the strawberry poison frog Dendrobates pumilio: genetic and geographical isolation suggest a role for selection in maintaining polymorphism. Mol Ecol 16:4284–4294
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Snowdon RJ, Lühs W, Friedt W (2007) Oilseed rape. In: Kole C (ed) Genome mapping and molecular breeding in plants, vol 2: oilseeds. Springer, Berlin Heidelberg, pp 55–114
Stich B, Melchinger AE, Frisch M, Maurer HP, Heckenberger M, Reif JC (2005) Linkage disequilibrium in European elite maize germplasm investigated with SSRs. Theor Appl Genet 111:723–730
Stich B, Möhring J, Piepho HP, Heckenberger M, Buckler ES, Melchinger AE (2008) Comparison of mixed-model approaches for association mapping. Genetics 178:1745–1754
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289
Van Inghelandt D, Melchinger AE, Lebreton C, Stich B (2010) Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor Appl Genet 120:1289–1299
Varshney RK, Graner A, Sorrells ME (2005) Genomics-assisted breeding for crop improvement. Trends Plant Sci 10:621–30
Weir BS (1996) Genetic data analysis II. Sinauer Associates, Sunderland
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395–420
Wright S (1978) Evolution and the genetics of populations, vol IV. The University of Chicago Press, Chicago, p 91
Zapata C, Carollo C, Rodriguez S (2001) Sampling variance and distribution of the D’ measure of overall gametic disequilibrium between multiallelic loci. Ann Hum Genet 65:395–406
Acknowledgements
The authors thank Wolfgang Ecke (University of Göttingen, Germany), the Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben (Germany), Nordic Gene Bank, Alnarp (Sweden), The Centre for Genetic Resources (Netherlands), and Warwick Horticulture Research International Genetic Resources Unit (UK) for providing the seeds of the examined germplasm. Agriculture and Agri-Food Canada kindly shared the proprietary Brassica SSR markers and their map positions. We are grateful to Sarah Nele Kaul and Bent Müller for the technical assistance, as well as to the Max Planck Genome Centre Cologne for providing the genotyping facilities. The funding of this work is kindly provided by the Deutsche Forschungsgemeinschaft (DFG) and the Max Planck Society. This work was performed in the framework of the ERA-NET PG project “ASSYST”. Finally, we thank the associate editor C. Quiros and three anonymous reviewers for their valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Quiros.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Bus, A., Körber, N., Snowdon, R.J. et al. Patterns of molecular variation in a species-wide germplasm set of Brassica napus . Theor Appl Genet 123, 1413–1423 (2011). https://doi.org/10.1007/s00122-011-1676-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1676-7