Abstract
Chromosome segment duplications are integral in genome evolution by providing a source for the origin of new genes. In the rice genome, besides an ancient polyploidy event known in the rice common ancestor, it had been identified that there was a special segmental duplication involving chromosomes 11 and 12, but the biological role of this duplication remains unknown. In this study, by using a set of chromosome segment substitution lines (CSSLs) and near isogenic lines (NILs) derived from the indica cultivar 9311 and japonica cultivar Nipponbare, a major QTL (qS12) resulting in hybrid male sterility was mapped within ~400 kb region adjacent to the special duplicated segment on the short arm of chromosome 12. Compared to the japonica cultivar Nipponbare, the two sides of the qS12 candidate region were inverted in the indica cultivar 9311. Among 47 of the 111 rice genotypes evaluated by molecular markers, the inverted sides were detected, and found completely homologous to indica cultivar 9311. These results suggested that the two inverted sides protect the sequence in the qS12 regions from recombination. On the short-arm of chromosome 12, two QTLs S-e and S25, in addition to qS12, were previously detected as a distinct segregation distortion and pollen semi-sterility loci. We propose these three hybrid sterility loci are the same locus, and the duplicated segment on chromosome 12 may play a prominent role in diversification, i.e., sub-speciation of cultivated rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Large chromosomal segment duplications are commonly detected during genome evolution in many species, such as Arabidopsis thaliana (The Arabidopsis Genome Initiative 2000), Sorghum bicolor (Paterson et al. 2009) and Oryza sativa (Yu et al. 2005). Benefit from the release of the whole genome sequence, extensive gene duplications had been discovered across the rice genome (Paterson et al. 2009; Wang et al. 2005a; Yu et al. 2005). There might be two independent duplications in rice genome, i.e., an ancient whole genome duplication (WGD) occurred ~70 million years ago (MYA) and a strange segmental duplication seemingly occurred recently between the distal regions of the short-arms of chromosomes 11 and 12.
The later duplication was estimated with a length of about 2.5–6.5 Mb by either genetic or physical mapping (Nagamura et al. 1995; Wu et al. 1998), and further confirmed by several studies (Jacquemin et al. 2009; Jiang et al. 2007; The Rice Chromosomes 11, 12 Sequencing Consortia 2005; Wang et al. 2005a, 2005b; Yu et al. 2005). Jiang et al. (2007) proposed that the appropriate length of the later duplicated DNA segments should be only ~2 Mb, based on the synonymous substitution rates (Ks) between conserved gene pairs of chromosomes 11 and 12. The relative chronology of this DNA duplication is unclear. Some studies implied that it might have happened recently, about 5–7 MYA based on the mean Ks value between the conserved gene pairs (Jacquemin et al. 2009; Jiang et al. 2007; Wang et al. 2005a, 2005b). But Paterson et al. (2009) and Wang et al. (2007, 2009, 2011) thought it should be an ancient duplication because it was also found in sorghum, wheat and other cereal genomes, and the high similarity between the two duplicated segments might be a consequence of gene conversion and illegitimate recombination.
Although the age of the unique duplication between the chromosomes 11 and 12 is controversial, there was no doubt about the rapid evolution and high frequency of sequence rearrangement within the pair of duplicated segments, especially in the duplicated segment on chromosome 12, which might facilitate the speciation in rice (Jiang et al. 2007; Wang et al. 2009, 2011). Therefore, several studies suggested that this special duplication might play a prominent role in rice genome evolution and diversification. But it remains unknown for the detail biological function as well as the evolutionary implications of these duplicated segments.
DNA segment duplications are considered very important for organism evolution, which could provide a hotbed for the evolution of many genes at once within the duplicated segment (McLysaht et al. 2002; Ohno et al. 1968). After gene duplication, one copy has the potential to evolve freely and rapidly, while the other copy maintains its original function (Lynch and Conery 2000; Lynch et al. 2001; Prince and Pickett 2002; Tocchini-Valentini et al. 2005). Although most new genes were reported without novel function, it had been suggested that duplicated genes still have a possibility in providing a source for the origin of reproductive barriers (Lynch and Conery 2000). And hybrid sterility serves as one of the major reproductive barriers (Mayr 1942).
In rice, partial hybrid sterility is a common phenomenon for hybrids between intraspecific crosses. Oka (1953) first described the partial sterility of the hybrid between indica and japonica. To date, several QTLs for intraspecific hybrid sterility has been mapped on rice genome. By using isogenic lines, Oka (1974) firstly identified sa1, sa2, sc1, sc2, etc., for hybrid sterility between distantly related varieties of cultivated rice. Then, many such QTLs for hybrid sterility had been identified and mapped (Ikehashi and Araki 1986; Wan et al. 1998; Sano et al. 1994; Sawamura and Sano 1996; Zhang and Lu 1996; Liu et al. 1997; Wang et al.1998; Yan et al.2000; Kubo and Yoshimura 2001; Zhuang et al. 2002; Ji et al. 2005; Qiu et al. 2005; Zhu et al. 2005; Zhao et al. 2006). Among them, two loci were successfully isolated (Chen et al. 2008; Ji et al. 2010; Long et al. 2008), and one locus causes male sterility and acts as a segregation distorter (Long et al. 2008). Besides, more than 30 QTLs for reproductive barriers were also identified from a genome-wide survey using different crosses in a F2 population between indica and japonica (Harushima et al. 2002). Among these identified QTLs, several were located in or near the region of the special duplicated segment on chromosome 12, which implied that the DNA duplication might lead to reproductive barriers (Win et al. 2009; Zhu et al. 2008). But up to now, none of the gene for hybrid sterility has been isolated or confirmed in this region.
In present study, the relationship between the hybrid sterility and this special DNA duplication on chromosome 12 was confirmed. By using a set of chromosome segment substitution lines (CSSLs) and their derived near-isogenic lines (NILs), a major QTL (qS12) responsible for partial hybrid sterility was mapped in an approximately 400 kb DNA region, which was located in the duplicated segment of rice chromosome 12. The results from a comparative genomics investigation indicated that the two sides of this candidate region in japonica cultivar Nipponbare were inverted from indica cultivar 9311.
Materials and methods
Plant materials
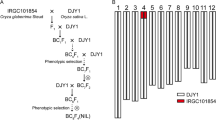
A total of 111 rice cultivars, including 58 indica and 53 japonica, were used in this study (Table S1). A set of 40 CSSLs, which were developed using japonica rice variety Nipponbare as the background parent, and indica rice variety 9311 as the donor parent, was used for the genome-wide survey of hybrid sterility loci. The genotype of each CSSL was firstly identified using molecular markers, and then confirmed with re-sequencing technology (Zhang et al. unpublished) according to the methods described by Huang et al. (2009) and Xu et al. (2010). Each CSSL was crossed with their parents, respectively, and the spikelet fertility was recorded as the phenotypic trait to survey the hybrid sterility loci. Subsequently, the CSSLs carrying the hybrid sterility locus were crossed with their background parent. After self-pollination of two or three generations, the heterozygous plants exhibiting partial sterility were selected and allowed to produce a set of NILs. The sterility trait was assayed every generation during NIL construction, which was used to further map the QTL qS12. Besides, a F2 population, derived from a cross between Nipponbare and 9311, was used to fine map and investigate the segregation ratio of qS12. All the rice materials were planted in the summer season at Mainland or in the winter season at Hainan of China, and the field management of rice plants was similar to that under normal rice production conditions.
Examination of spikelet and pollen fertility
In the summer of 2009, all CSSLs as well as their parents were planted in the Experiment Station at Yangzhou University, Yangzhou, Jiangsu Province, and their spikelet fertility was recorded. The ratio of fertile spikelets to total spikelets of the main panicles from three to five plants in each line was applied to calculate as the spikelet fertility score. The NILs were planted in the winter of 2009 at Sanya, Hainan province, and some NILs containing progenies generated from a cross between SY098 and SY025 were planted in the summer of 2010 in the Experiment Station of Wuhan University on Wuhan, Hubei province. The spikelet fertility score for NILs was the ratio of filled grains to total spikelets of three to four panicles. The pollen fertility examination was performed as described by Zhang and Lu (1996), and the average normal pollen ratio to total pollen of more than three spikelets was determined as the pollen fertility.
DNA extraction and molecular marker assay
Total genomic DNA was extracted from young fresh leave according to the method described by Murray and Thompson (1980). The molecular markers used in this study included SSR (Simple Sequence Repeat), CAPS (Cleaved Amplified Polymorphic Sequence) and STS (Sequence-Tagged Site) (Table S2), some of them were developed according to published polymorphism databases between the two sequenced rice varieties (McCouch et al. 2002; Shen et al. 2004; Wang et al. 2005b; Zhang et al. 2007). Molecular marker assays were performed as described previously (McCouch et al. 2002; Zhang et al. 2007).
Genome-wide survey of the hybrid sterility locus and data analysis
A genome-wide survey was performed by regression analysis with the CSSL package using QTL IciMapping software under the mathematical models described by Wang et al. (2006). Two mapping methods, Single Marker Analysis (SMA) and Inclusive Composite Interval Mapping (ICIM) of QTLs with additive (and dominance) effects (ICIM-ADD), were employed for power analysis. The parameter of threshold condition numbers for correlated marker variables was 1000, a LOD threshold of 2.0 (P = 0.05) for each method, and a 0.01 probability in ICIM-ADD step regression was used to reduce the Type-I error and identify suggestive QTLs. 1000 replicates were applied for the permutation tests, and tests of significance were performed using a t test or χ 2 test at a P < 0.01 level.
Fine mapping of the qS12 locus
To further map the qS12 candidate region, all polymorphic markers were physically mapped on chromosome 12 of the rice genome according to the Nipponbare whole genome sequence information (International Rice Genome Sequencing Project 2005). The physical distance of related linkage markers was used to determine the length of qS12 candidate region.
Results
Genome-wide survey of the hybrid sterility locus within CSSL populations
The phenotypes for the survey of the hybrid sterility locus were collected from two crosses between the CSSL and Nipponbare or 9311. The first cross was conducted to test the hybrid sterility of each CSSL directly, and the second one was served as a complementation test of the hybrid sterility of each CSSL after comparison with the F1 hybrid of the two initial parents. The homozygous (labeled as “0”) or heterozygous genotype (labeled as “2”) of the substituted chromosome segments in each CSSL was confirmed with 140 SSR or STS markers as well as the re-sequencing results. Based on the re-sequencing data of all introduced segments in CSSLs, the rice whole genome could be separated into fragments with 196 re-sequencing tags. These data were used to represent the genotype of each CSSL, and the average length of each substituted fragment was approximately 2 Mb.
According to both SMA and ICIM-ADD analyses for the first cross test (CSSL × Nipponbare), two QTLs for hybrid sterility, with the LOD scores of more than 2.0, were detected near marker M98 or M184, respectively (Table 1, Figure S1). The QTL near M98 was located in the region on chromosome 6 containing the cloned hybrid sterility gene S5 (Chen et al. 2008), which has been demonstrated as a major wide compatibility locus in rice (Ikehashi and Araki 1986; Ji et al. 2005; Qiu et al. 2005). The later QTL near M184, named as qS12, was detected in the region adjacent to the recently identified duplicated segment on chromosome 12. The data from SMA analysis gave a LOD score of 2.26 for the QTL qS12, while 7.53 from the ICIM-ADD analysis, which could explain the largest phenotypic variation effect (PVE) of 35.45 % for hybrid sterility (Table 1).
In the second cross test (CSSL×9311), the SMA analysis revealed two QTLs near the markers M46 and M183, respectively, for hybrid sterility with the LOD scores of 2.28 and 3.70 (Table 1, Figure S1). But only one QTL near M183 was identified after analysis by ICIM-ADD with a threshold LOD score of 7.25, which explained 40.60% of the phenotypic variance (Table 1). Interestingly, both markers M183 and M184 were located in the duplicated region on chromosome 12. Therefore, we proposed that the two QTLs around M184 and M183 detected in the two cross tests are the same locus, qS12. Taken together of the two analyses, in addition to the cloned hybrid sterility gene S5 on chromosome 6, qS12 seems as another major QTL for hybrid sterility in rice.
Further mapping of qS12 by using NILs
Of 40 CSSLs, nine contain the substituted chromosome segment covering the molecular marker M183. Based on the re-sequencing information of these nine CSSLs as well as their spikelet fertility phenotypes, the QTL qS12 was firstly determined to locate in a region between 0 and 4 Mb on the short distal arm of chromosome 12. Then, the qS12 region was confirmed and delimited by selected six recombinants from these CSSLs (Fig. 1).
We subsequently observed the spikelet fertility of rice plants in NILs around the qS12 locus derived from a cross between CSSL N21 with low spikelet fertility and its background parent Nipponbare. The spikelet fertility of a total of 339 offsprings from these NILs was recorded and the spikelet fertility of the background parent Nipponbare (73.4%) was set as a control. We further assayed the genotypes of 113 individual plants by using markers surrounding the qS12 locus. Among these 113 individuals, 53 were identified with low spikelet fertility (LF, <42%), and most (38 of 53) carried the heterozygous segment before marker MS102 (Figs. 2 and 3). Nearly all the other plants (n = 60) with high spikelet fertility (HF, >43%) exhibited a homozygous genotype following marker MS027 and before marker MS102 (Figs. 2 and 3). These results suggested that the qS12 locus might be located between the markers MS027 and MS102. Among the above 38 plants with low spikelet fertility, several individuals were identified heterozygous between markers MS027 and MS102. One recombinant SY025 with low spikelet fertility (22.65%) was detected to contain a heterozygous segment between markers MS062 and RM247 (Fig. 3, Figure S4). Thus, these results suggested that the plausible location of qS12 is between the markers MS062 and MS102 (Fig. 3).
a Physical maps of markers surrounding qS12, and mapping of qS12 with NILs grown in two different environments. SY rice plants grown in the city of Sanya, Hainan province; WH rice plants grown in the city of Wuhan, Hubei province; LF plants with low spikelet fertility, HF plants with high spikelet fertility. Black bars represent substituted segments from the donor parent, and gray bars represent chromosomal contributions from the background parent. MS designated markers were polymorphic between two initial parents; M designated markers were re-sequencing tags. b A hybrid cross with a heterozygous genotype resulting from a cross between SY025 and SY098 for most markers before RM247 exhibited high fertility (with the exception of the region between MS062 and MS102, which had a homozygous genotype); control plants with a heterozygous genotype before marker MS102 were partially sterile
Alternatively, qS12 may have an interaction with several genes to control the fertility phenotype, which might be located prior to marker MS062 or between markers MS102 and RM247. We therefore investigated these possibilities by crossing SY098 (containing a homozygous segment from 9311 before marker MS102) and SY025 (with a heterozygous genotype between marker MS062 and RM247). The genotypes and spikelet fertility of individuals derived from these two lines were assayed. All offsprings with a heterozygous genotype (n = 5) for most markers before RM247, with the exception of a homozygous region between MS062 and MS102, had a high fertility score (>90%). However, plants with a heterozygous genotype between markers MS062 and MS102 showed partial hybrid sterility. These results indicated that there was no locus responsible for partial sterility in the region prior to marker MS062 or the region from markers MS102 to RM247. Consequently, qS12 must be located in the region between markers MS062 and MS102 (Fig. 3 and Figure S5).
Physical mapping of qS12
Based on the genomic sequence information of the two parents Nipponbare and 9311, it was established that qS12 is located on a chromosomal duplicated segment with a length of approximately 400 kb in the Nipponbare rice genome (International Rice Genome Sequencing Project, 2005). In the genome of indica 9311, qS12 occupies an approximate length of 460 kb between 840 kb and 1,300 kb on chromosome 12 (Yu et al. 2005). Furthermore, we produced 450 plants derived from the NILs with heterozygous genotype in the qS12 candidate region, and 476 F2 plants derived from a cross between their two parents, Nipponbare and 9311. Among these populations, no recombinants were identified between markers MS075 and MS096 within the confirmed 400 kb region. Therefore, we could conclude that the qS12 candidate region is an approximately 400 kb DNA segment between the markers MS062 and MS102, and this target region is located on the duplicated segment of chromosome 12. The result suggested that the large duplicated segment on chromosome 12 might truly play an important role in rice genome evolution and diversification of rice subspecies.
Effects of the inverted sides of qS12 candidate region
As no recombinants between MS075 and MS096 were detected, we presumed that the recombination frequency in the qS12 candidate region would be very low. Interestingly, after a comparative genomics study between Nipponbare and 9311, it indicated that the two sides of the qS12 candidate region had just been inverted, and most of the center region remains parallel (Figure S3) (Ouyang et al. 2007). Consequently, we assumed that recombination in the qS12 candidate region is limited by the two inverted sides, which might maintain the qS12 sequence as a conserved element. Based on the genome sequence of the two rice cultivars Nipponbare and 9311, we designed a dominant marker XF28401 to identify one of the inverted sides of the qS12 region.
The marker XF28401 was applied to analyze 58 indica and 53 japonica cultivars (also including Nipponbare and 9311). The results showed that 29 (50%) indica and 18 (34%) japonica cultivars contain the inverted sides similar to 9311, while the other indica (29) and japonica (35) cultivars were found lacking the two inverted sides. These 111 cultivars were further assayed with another 10 co-dominant markers within the qs12 candidate region. All 47 cultivars with inverted sides showed complete identity to that of 9311 in the candidate region, but the cultivars lacking the two inverted sides exhibited several genotypic markers corresponding to Nipponbare. Among the 35 japonica cultivars lacking inverted sides, excluding six cultivars with an indica marker, 29 japonica cultivars were detected to have the same genotype in the candidate region of japonica variety Nipponbare by using 10 markers (Table S1). And an average homologous 67% hit to indica variety 9311 was generated for all 29 indica cultivars lacking the two inverted sides (Fig. 4). These results further supported that the two inverted sides in cultivars liking 9311 might protect the sequence in the qS12 region as a conserved element. In addition, the two inverted sides might be a primary cause for the low recombination frequency in the target region, and an obstacle for isolating qS12, which was evidenced by the absence of recombinant in this conserved region.
The qS12 candidate region was a segregation distortion locus exhibiting male semi-sterility
By using a NIL population derived from the plants with heterozygous region around qS12, the pollen fertility was carefully investigated. The results showed that the plants heterozygous for qS12, such as plants 322-09, 322-10, 324-01, and 324-08, exhibited a lower pollen fertility score (<50%) than that of their recipient parent plants (Fig. 5). The effects of the japonica qS12 allele were evaluated with a 476 F2 population derived from a cross between Nipponbare and 9311. After analyzing using markers MS075 and MS096 located within the qS12 region, 54 plants were selected to have the Nipponbare homozygous genotype. Besides, among a NILs population with 497 individuals generated from the plants with heterozygous qS12 region, only seven plants were screened with the homozygous genotype of Nipponbare by using the same two markers, which was significantly (P < 0.01) fewer than that from the F2 population. These data obviously showed that the segregation ratio among the three types of zygotes, i.e. homozygous dominant and recessive, and heterozygous, was not consistent with the 1:2:1 genotypic ratio (P < 0.001) (Table 2). It indicated that male gametes carrying the japonica qS12 allele might be largely sterile, so few zygotes with a homozygous qS12 genotype derived from japonica cultivar Nipponbare were observed in the two separate populations. Further more, In the region adjacent to qs12, Zhu et al. (2008) and Win et al. (2009) had identified two QTLs affecting F1 pollen sterility, S-e and S25, using SSR and RFLP markers, respectively. Their results showed that the male gametes carrying japonica alleles were responsible for the sterility. Moreover, Win et al. (2009) revealed that sterile pollen grain abnormalities caused by S25 occurred mainly at the late bicellular stage after initiation of starch accumulation. These data suggested that the qS12, S-e, and S25 loci might represent the same locus for hybrid sterility in rice, and function as a segregation distortion locus with male semi-sterility.
a and b pollen fertility assays and pollen fertility scores of heterozygous qS12 genotypes from NILs and rice plants of the recipient parent NIP (Nipponbare). The additional four plants have a qS12 heterozygous genotype from the NILs population. c Sketched map of the genotype of a qS12 heterozygous plant
Discussion
Following entire sequencing of the rice genome, an ancient whole-genome duplication (WGD) event was revealed by the comparative genomic studies. It was suggested that the duplication occur prior to the divergence of grasses approximately 70 MYA (Paterson et al. 2003; Wang et al. 2005a, 2005b; Yu et al. 2005). Furthermore, a ~2 Mb unique segmental duplication between chromosomes 11 and 12 was firstly estimated to have occurred 5–7.7 MYA based on the mean Ks value between conserved gene pairs (Jacquemin et al. 2009; Jiang et al. 2007; Wang et al. 2005a, 2005b). However, after a detailed analysis of the chromosome 11 and 12, Wang et al. (2011) pointed out that the age of this duplication might be misleading estimated in previous studies, and they suggested that the two conserved duplicated segments should have experienced a singular evolutionary history. Besides the high similarity, extensive gene losses and high frequency of sequence rearrangement rate were also observed in the pair of duplicated segments in several studies (Jiang et al. 2007; Wang et al. 2007, 2009, 2011). In addition, even non-homologous chromosome pairs have been found between these two duplicated segments on chromosomes 11 and 12 during the haploid rice meiosis by fluorescence in situ hybridization (Gong et al. 2010). Therefore, some authors suggested this duplication event might serve a significant role in rice genome evolution (Jiang et al. 2007; Wang et al. 2011). However, its impact on the phenotypic evolution of rice is still unknown.
Reproductive isolation is a criterion for the biological species concept (Mayr 1942), and the origin of reproductive barriers is integral in speciation (Johnson 2010), therefore hybrid sterility or partial sterility is often found between related species. Hybrid incompatibility loci have been detected in or near the duplicated segment of chromosome 12 (Harushima et al. 2002; Win et al. 2009; Zhu et al. 2008), but the exact locations of the loci are unclear. Furthermore, it is uncertain if the loci are in fact located in the duplicated segment. Consequently, a hybrid of this recent chromosome 12 duplicated segments, derived from indica and japonica resulting in hybrid partial sterility remains unknown. In the present study, using a set of CSSLs and several NILs, a major QTL (qS12) for partial hybrid male sterility was confirmed to locate in a ~400 kb region on the duplicated segment of chromosome 12. Adjacent to the candidate region of qS12, two QTLs for partial pollen sterility had also been identified (Zhu et al. 2008; Win et al. 2009). These results suggested that this unique segmental duplication might truly play a very important role on divergence between indica and japonica. The results were also congruent with the assumption that segmental duplication provides a source for the origin of reproductive barriers (Lynch and Conery 2003).
From the comparative study of the two varieties’ genomes (Nipponbare and 9311) (Ouyang et al. 2007), it indicated that the two sides of the candidate region in japonica cultivar Nipponbare were inverted from indica cultivar 9311. In this study, several cultivated rice varieties, with which two inverted sides consistent with the candidate region of 9311, were also detected. All of these cultivars were tested using markers within the candidate region, and found completely homologous to 9311. These results supported our hypothesis that the inverted sides may protect the sequence in the candidate region, and few recombination events can occur in the target area. Genetic studies implied that the rapid evolution of speciation genes were the major cause for hybrid sterility (Orr et al. 2004), so there may be one or more speciation genes lied in the region of qS12. Unfortunately, in terms of the inverted sides, recombinants could not be generated to map the candidate qS12 gene. Therefore, other crosses derived from non-inverted cultivars representing the two subspecies must be developed to isolate qS12, and more studies are necessary to understand the molecular mechanisms underlying the hybrid sterility at this locus.
Eventually, the qS12 locus will be very important for the rice breeders. We determined that almost all F1 hybrids derived from the crosses between CSSLs carrying qS12 and 9311 have increased fertility, especially CSSL N3, which had three substituted segments from the donor parent (Figure S3). The hybrid F1 plants between N3 and 9311 exhibited the highest fertility score (76.62%), compared with the F1 hybrids derived from the two initial parents grown in the same field (38.17%) (Figure S3). These results indicated that there might be fewer loci responsible for hybrid sterility in the F1 population than in the F2 population between indica and japonica. Thus, in new future, it is promised for application of the indica–japonica hybrid rice varieties. Interestingly, a percentage (34%) of the japonica varieties tested in this study possessed a qS12 like 9311 indica allele (Table S1). Therefore, crosses between japonica varieties with qS12 indica and japonica alleles should also result in partial sterility. Therefore, qS12 might also be a major locus for hybrid japonica incompatibility.
Wild rice has been under cultivation for thousands of years, with the earliest archeological evidence dating to 8000 BC in central and eastern China (Jiang and Liu 2006). This suggests that selection for different phenotypes began thousands of years ago, and population level diversification may be an artifact of human influences artificially selecting for desirable traits. Furthermore, molecular data suggests that Oryza was domesticated at least twice, O. sativa indica in eastern India and adjacent areas, and O. sativa japonica in southern China (Londo et al. 2006). Therefore, diversification may be the result of different geographic domestications. But some other studies believed that indica was domesticated primarily from its wild relatives, whereas japonica derived from indica (Chang 1976; Oka 1988). A broad range of genetic variability and genetically structured populations have been documented in O. sativa, and reproductive isolation in the form of genetic incompatibilities among interspecific hybrids. These observations suggest that speciation processes are operating in the species, resulting in the origin of two subspecies, indica and japonica. In present study, we propose that the QTLs S-e and S25, and qS12 on a recent duplicated segment of chromosome 12, may represent one segregation distortion, and pollen semi-sterility locus, which contribute to our molecular understanding of evolutionary diversification in Oryza sativa.
Abbreviations
- CSSL:
-
Chromosome segment substitution line
- CAPS:
-
Cleaved amplified polymorphic sequence
- ICIM:
-
Inclusive composite interval mapping
- ICIM-ADD:
-
ICIM of QTLs with additive effects
- Mb:
-
Mega base pairs
- MYA:
-
Million years ago
- NIL:
-
Near isogenic line
- PVE:
-
Phenotypic variation effect
- SMA:
-
Single marker analysis
- SSR:
-
Simple sequence repeat
- STS:
-
Sequence-tagged site
- WGD:
-
Whole genome duplication
References
Chang TT (1976) The origin, evolution, cultivation, dissemination, and diversifiction of Asian and African rices. Euphytica 25:435–441
Chen J, Ding J, Ouyang Y, Du H, Yang J, Cheng K, Zhao J, Qiu S, Zhang X, Yao J, Liu K, Wang L, Xu C, Li X, Xue Y, Xia M, Ji Q, Lu J, Xu M, Zhang Q (2008) A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica–japonica hybrids in rice. Proc Natl Acad Sci USA 105:11436–11441
Gong Z, Liu X, Tang D, Yu H, Yi C, Cheng Z, Gu M (2010) Non-homologous chromosome pairing and crossover formation in haploid rice meiosis. Chromosoma 120:47–60
Harushima Y, Nakagahra M, Yano M, Sasaki T, Kurata N (2002) Diverse variation of reproductive barriers in three intraspecific rice crosses. Genetics 160:313–322
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T, Dong G, Sang T, Han B (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19:1068–1076
Ikehashi H, Araki H (1986) Genetics of F1 sterility in remote crosses of rice. In: Rice Genetics. IRRI, PO Box 933, Manila, Philippines: 119-130
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jacquemin J, Laudie′ M, Cooke R (2009) A recent duplication revisited: phylogenetic analysis reveals an ancestral duplication highly-conserved throughout the Oryza genus and beyond. BMC Plant Biol 9:146
Ji Q, Lu J, Chao Q, Gu M, Xu M (2005) Delimiting a rice wide-compatibility gene S 5 n to a 50 kb region. Theor Appl Genet 111:1495–1503
Ji Q, Lu J, Chao Q, Zhang Y, Zhang M, Gu M, Xu M (2010) Two sequence alterations, a 136 bp InDel and an A/C polymorphic site, in the S5 locus are associated with spikelet fertility of indica-japonica hybrid in rice. J Genet Genomics 37:57–68
Jiang L, Liu L (2006) New evidence for the origins of sedentism and rice domestication in the lower Yangzi River, China. Antiquity 80:355–361
Jiang H, Liu D, Gu Z, Wang W (2007) Rapid evolution in a pair of recent duplicate segments of rice. J Exp Zool (Mol Dev Evol) 308B:50–57
Johnson NA (2010) Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet 26:317–325
Kubo T, Yoshimura A (2001) Linkage analysis of an F1 sterility gene in Japonica/Indica cross of rice. Rice Genet Newsl 2001
Liu K, Wang J, Li H, Xu C, Liu A, Li X, Zhang Q (1997) A genome-wide analysis of wide compatibility in rice and the precise location of the S5 locus in the molecular map. Theor Appl Genet 95:809–814
Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA 103:9578–9583
Long Y, Zhao L, Niu B, Su J, Wu H, Chen Y, Zhang Q, Guo J, Zhuang C, Mei M, Xia J, Wang L, Wu H, Liu Y-G (2008) Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc Natl Acad Sci USA 105:18871–18876
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate. Science 290:1151–1155
Lynch M, Conery JS (2003) The evolutionary demography of duplicate genes. J Struct Func Genomics 3:35–44
Lynch M, O’Hely M, Walsh B, Force A (2001) The probability of preservation of a newly arisen gene duplicate. Genetics 159:1789–1804
Mayr E (1942) Systematics and the Origin of Species. Columbia University Press, New York
McCouch S, Teytelman L, Xu Y, Lobos K, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:257–279
McLysaht A, Hokamp K, Wolfe K (2002) Extensive genomic duplication during early chordate evolution. Nat Genet 31:200–204
Murray M, Thompson W (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nagamura Y, Inoue T, Antonio BA, Shimano T, Kajiya H, Shomura A, Lin SY, Kuboki Y, Harushima Y (1995) Conservation of duplicated segments between rice chromosomes 11 and 12. Breed Sci 45:373–376
Ohno S, Wolf U, Atkin NB (1968) Evolution from fish to mammals by gene duplication. Hereditas 59:169–187
Oka HI (1953) The mechanism of sterility in the intervarietal hybrid. (Phylogenetic differentiation of the cultivated rice plants VI). Jpn J Breed 2:217–224
Oka HI (1974) Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Genetics 77:521–534
Oka HI (1988) Origin of cultivated rice: development in crop species. Jpn Sci Soc Press, Tokyo
Orr HA, Masly JP, Presgraves DC (2004) Speciation genes. Curr Opin Genet Dev 14:675–679
Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR (2007) The TIGR Rice genome annotation resource: improvements and new features. Nucleic Acids Res 35:D883–D887
Paterson AH, Bowers JE, Peterson DG, Estill JC, Chapman BA (2003) Structure and evolution of cereal genomes. Curr Opin Genet Dev 13(6):644–665
Paterson AH, Bowers JE, Bruggmann Rm, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman, Ware D, Westhoff P, Mayer KFX, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Prince V, Pickett F (2002) Splitting pairs: The diverging fates of duplicated genes. Nat Rev Genet 3:827–837
Qiu SQ, Liu K, Jiang JX, Song X, Xu CG, Li XH, Zhang Q (2005) Delimitation of the rice wide compatibility gene S5 n to a 40-kb DNA fragment. Theor Appl Genet 111:1080–1086
Sano Y, Sano R, Eiguchi M, Hirano HY (1994) Gamete eliminator adjacent to the Wx Locus as revealed by pollen analysis in rice. J Hered 85:310–312
Sawamura N, Sano Y (1996) Chromosomal location of gamete eliminator, S11(t), found in an Indica-Japonica hybrid. Rice Genet Newsl 13:70–71
Shen Y, Jiang H, Jin J, Zhang Z, Xi B, He Y, Wang G, Wang C, Qian L, Li X, Yu Q, Liu H, Chen D, Gao J, Huang H, Shi T, Yang Z (2004) Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol 135:1198–1205
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
The Rice Chromosomes 11, 12 Sequencing Consortia (2005) The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol 3:20
Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP (2005) Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc Natl Acad Sci USA 102:8933–8938
Wan J, Ikehashi H, Sakai M, Horisue H, Imbe T (1998) Mapping of hybrid sterility genes S17 of rice (Oryza sativa L.) by isozyme and RFLP markers. Rice Genet Newsl 15:151–154
Wang J, Liu K, Xu C, Li X, Zhang Q (1998) The high level of wide compatibility of variety ‘Dular’ has a complex genetic basis. Theor Appl Genet 97:407–412
Wang X, Shi X, Hao B, Ge S, Luo J (2005a) Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol 165:937–946
Wang X, Zhao X, Zhu J, Wu W (2005b) Genome-wide investigation of intron length polymorphisms and their potential as molecular markers in rice (Oryza sativa L.). DNA Res 12:417–427
Wang J, Wan X, Crossa J, Crouch J, Weng J, Zhai H, Wan J (2006) QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet Res 88:93–104
Wang X, Tang H, Bowers J, Feltus FA, Paterson AH (2007) Extensive concerted evolution of rice paralogs and the road to regaining independence. Genetics 177:1753–1763
Wang X, Tang H, Bowers JE, Paterson AH (2009) Comparative inference of illegitimate recombination between rice and sorghum duplicated genes produced by polyploidization. Genome Res 19:1026–1032
Wang X, Tang H, Paterson AH (2011) Seventy million years of concerted evolution of a homoeologous chromosome pair, in parallel, in major Poaceae lineages. Plant Cell 23:27–37
Win KT, Kubo T, Miyazaki Y, Doi K, Yamagata Y, Yoshimura A (2009) Identification of two loci causing F1 pollen sterility in inter- and intraspecific crosses of rice. Breeding Sci 59:411–418
Wu J, Kurata N, Tanoue H, Shimokawa T, Umehara Y, Yano M, Sasaki T (1998) Physical mapping of duplicated genomic regions of two chromosome ends in rice. Genetics 150:1595–1603
Xu J, Zhao Q, Du P, Xu C, Wang B, Feng Q, Liu Q, Tang S, Gu M, Han B, Liang G (2010) Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genomics 11:656
Yan C, Liang G, Zhu L, Gu M (2000) RFLP analysis on wide compatibility genes in rice variety ‘Dular’ of ecotype Aus (in Chinese). Acta Genet Sin 27:409–417
Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, Zhang J, Zhang Y, Li R, Xu Z, Li S, Li X, Zheng H, Cong L, Lin L, Yin J, Geng J, Li G, Shi J, Liu J, Lv H, Li J, Wang J, Deng Y, Ran L, Shi X, Wang X, Wu Q, Li C, Ren X, Wang J, Wang X, Li D, Liu D, Zhang X, Ji Z, Zhao W, Sun Y, Zhang Z, Bao J, Han Y, Dong L, Ji J, Chen P, Wu S, Liu J, Xiao Y, Bu D, Tan J, Yang L, Ye C, Zhang J, Xu J, Zhou Y, Yu Y, Zhang B, Zhuang S, Wei H, Liu B, Lei M, Yu H, Li Y, Xu H, Wei S, He X, Fang L, Zhang Z, Zhang Y, Huang X, Su Z, Tong W, Li J, Tong Z, Li S, Ye J, Wang L, Fang L, Lei T, Chen C, Chen H, Xu Z, Li H, Huang H, Zhang F, Xu H, Li N, Zhao C, Li S, Dong L, Huang Y, Li L, Xi Y, Qi Q, Li W, Zhang B, Hu W, Zhang Y, Tian X, Jiao Y, Liang X, Jin J, Gao L, Zheng W, Hao B, Liu S, Wang W, Yuan L, Cao M, McDermott J, Samudrala R, Wang J, Wong GK-S, Yang H (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3:266–281
Zhang G, Lu Y (1996) Genetics of F1 pollen sterility in Oryza sativa. In: Khush GS (ed) Rice Genetics III. IRRI, Manila, Philippines, pp 418–422
Zhang Z, Deng Y, Tan J, Hu S, Yu J, Xue Q (2007) A genome-wide microsatellite polymorphism database for the Indica and Japonica Rice. DNA Res 14:37–45
Zhao ZG, Wang CM, Jiang L, Zhu SS, Ikehashi H, Wan JM (2006) Identification of a new hybrid sterility gene in rice (Oryza sativa L.). Euphytica 151:331–337
Zhu S, Wang C, Zheng T, Zhao Z, Ikehashi H, Wan J (2005) A new gene located on chromosome 2 causing hybrid sterility in a remote cross of rice. Plant Breed 124:440–445
Zhu W, Li W, Ding X, Zhang Z, Zeng R, Zhu H, Zhang G (2008) Preliminary Identification of Pollen Sterility Gene S-e in Oryza Sativa (in Chinese). J South Chin Agric University 29:1–5
Zhuang CX, Fu Y, Zhang GQ, Mei MT, Lu YG (2002) Molecular mapping of S-c, an F-1 pollen sterility gene in cultivated rice. Euphytica 127:133–138
Acknowledgments
This work was supported in part by the National Basic Research Program (2007CB109005 and 2011CB100202), the National Special Program for Transgenic Research (2009ZX08009-008B), the National Natural Science Foundation (30521004 and 30971741), the Priority Academic Program Development from Jiangsu Government, and the Ministry of Education (307018 and NCET-07-0736) of China.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by A. Paterson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, H., Zhang, CQ., Sun, ZZ. et al. A major locus qS12, located in a duplicated segment of chromosome 12, causes spikelet sterility in an indica-japonica rice hybrid. Theor Appl Genet 123, 1247–1256 (2011). https://doi.org/10.1007/s00122-011-1663-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1663-z