Abstract
Hybrid breakdown is an important form of post-zygotic reproductive barriers, often arising from hybrid progeny between two varietal groups of Asian cultivated rice (Oryza sativa L.), indica and japonica. However, the genetic mechanism underlying the hybrid breakdown remains unclear. In the present study, a chromosomal segment substitute line (CSSL) population together with a backcross inbred line population, derived from the same cross of Sasanishiki/Habataki, were employed for genetic analysis of rice hybrid breakdown (spikelet fertility as index). Quantitative trait locus mapping results showed that, in both populations, qSF-12 was stably detected across different locations and growing seasons, which was found to interact with qSF-8. Subsequently, a CSSL line SL438 with low spikelet fertility was used to cross with Sasanishiki to generate the secondary F2 population for the mapping of qSF-12. The results showed that qSF-12 was restricted to a 137-kb long region on BAC clone AL928774, containing 11 predicted ORFs in total. The DNA sequencing revealed that a 3-bp insertion/deletion exists between two parents in the coding region of LOC_Os12g38850. Together with the RNA-seq data, it is suggested that LOC_Os12g38850is the putative candidate of qSF-12, which encodes a DUF1336 domain containing protein. These results provide an important clue to further dissect the mechanism of hybrid breakdown in rice and the linked markers will be useful in rice cross breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cultivated rice grown worldwide comprises two species, Oryza sativa L. (mainly in Asian) and Oryza glaberrima Steud. (in Africa). Asian cultivated rice (O. sativa L.) has plenty of varieties, with great diversity in morphological and physiological characteristics. Kato (1930) clarified Asian cultivated rice into two major groups or subspecies, indica and japonica, based on the inter-varietal cross progeny sterility. Due to the distant genetic differentiation between indica and japonica subspecies, a strong hybrid vigor (heterosis) in their F1 is frequently observed. Unfortunately, the reproductive isolation often accompanies, which not only limits the utilization of the strong hybrid vigor between the two subspecies, but also hinders the genetic communication between them.
Reproductive isolation plays an important role in speciation and maintaining species identity and thus has remained to be a key research interest in evolutionary biology for many decades in a wide range of organisms (Dobzhansky 1937; Coyne and Orr 2004). According to the stage of the barriers, reproductive isolation can be divided into two general categories: pre-zygotic and post-zygotic reproductive isolation. Hybrid sterility, a major form of post-zygotic reproductive isolation, has been the subject of intensive investigation, especially in rice hybrids between indica and japonica subspecies. Previous genetic studies reveal that there are a large number of genes or quantitative trait loci (QTL) affecting hybrid fertility. So far 16 genes have been identified to be responsible for embryo sac sterility, 30 genes for the pollen sterility, and 11 genes for the spikelet sterility (see review by Ouyang et al. 2010). For example, S5, a major locus encoding an aspartic protease and conferring female gamete abortion in indica/japonica hybrid, has been cloned and functionally characterized (Chen et al. 2008; Ji et al. 2012). Another locus, Sa, controlling pollen fertility in indica/japonica hybrids, has also been cloned. Sa comprises two adjacent genes, SaM and SaF, encoding a small ubiquitin-like modifier E3 ligase-like protein and a F-box protein, respectively (Long et al. 2008). The cloning and characterization of these genes provides useful information for understanding the molecular mechanism underlying hybrid sterility, which is useful for overcoming the reproductive barrier in rice breeding program.
In addition to hybrid sterility, hybrid breakdown (weakness or sterility of F2 and later generations) is another important form of post-zygotic reproductive barriers in rice (Kitamura 1962; Oka 1978; Yokoo 1984; Fukuoka et al. 1998; Kubo and Yoshimura 2002, 2005). Stebbins (1950) proposed that hybrid breakdown is controlled by complementary genes, and many findings support the hypothesis (Kubo and Yoshimura 2002, 2005; Fukuoka et al. 1998; Garavito et al. 2010). Perhaps due to the complicated mode of inheritance, the location information of genes controlling hybrid breakdown has not been determined until the successful utilization of molecular markers in plant researches. By using RFLP markers, the two complementary genes controlling hybrid weakness in the cross between the Nepalese cultivar Siborunauli and Thai cultivar Col. NO.15, hwd1 and hwd2, were assigned to chromosome 10 and 7, respectively (Fukuoka et al. 1998). Moreover, hybrid breakdown showing poor growth habit and complete sterile in the progeny of the cross between Asominori and IR24 was also observed, and two complementary genes, hwe1 and hwe2, were identifiedand located on chromosome 12 and 1, respectively (Kubo and Yoshimura 2002). However, in the same cross, three complementary genes, hsa1 (chromosome 12), hsa2 (chromosome 8) and hsa3 (chromosome 9), were identified to be responsible for the female gamete development (Kubo and Yoshimura 2005). Recently, S1 has been identified and delimited in a 27.8 kb long region on chromosome 6, controlling the female sterility in the progeny of cross between O. sativa L. and O. glaberrima Steud., and the gene interacting with S1 was also detected (Garavito et al. 2010). Likewise, two recessive complementary genes, hbd2 and hbd3, were identified to be responsible for weakness phenotype in the progeny of Habataki/Koshihikari (Yamamoto et al. 2007), and further mapping results showed that hbd2 encodes a casein kinase I (CKI1), and hbd3 gene was mapped to the NBS-LRR gene cluster region (Yamamoto et al. 2010).
Recently, Chen et al. (2014) identified two incompatible dominant loci, Hwi1 and Hwi2, controlling the hybrid weakness in the interspecific hybrids of rice. These two loci are located on chromosome 11 and 1, respectively. Hwi1 comprises two leucine-rich repeat receptor-like kinase genes, 25L1 and 25L2, specific to wild rice (Oryza rufipogon), resulting in hybrid weakness. Hwi2, a rare allele, is predominantly distributed in indica rice (O. sativa), encoding a secreted putative subtilisin-like protease. Pyramiding of Hwi1 and Hwi2 activates the autoimmune response in the basal nodes of hybrids, interrupting root formation and then impairing shoot growth.
Obviously many genes are involved in controlling hybrid breakdown, like those in hybrid sterility (Ouyang et al. 2010), and the genes responsible for hybrid breakdown are different in specific crosses. Up to date, few genes controlling hybrid breakdown in indica/japonica progeny have been cloned yet, which hinders us from understanding the genetic mechanism at molecular level and overcoming the problem in hybrid breeding.
In the present study, a chromosomal segment substitution line (CSSL) population and a backcross inbred line (BIL) population, derived from a cross between an indica variety Habataki and a japonica variety Sasanishiki, were employed to clarify the genetic nature of hybrid breakdown, and a secondary F2 population generated by crossing the CSSL line SL438 with its recurrent parent Sasanishiki was employed to finely map the genes responsible for hybrid breakdown and conduct the candidate gene analysis.
Materials and methods
Plant materials
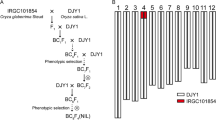
A total of 39 CSSLs and 85 BILs derived from the cross between a japonica variety Sasanishiki and an indica variety Habataki, were initially used as the population to detect the genes responsible for hybrid breakdown (spikelet fertility as index). The CSSL and BIL population, together with their genotype information, were introduced from the Rice Genome Project of the National Institute of Agrobiological Sciences, Japan (http://www.rgrc.dna.affrc.go.jp/stock.html).
A CSSL SL438 showing low spikelet fertility was selected as parent to cross with its recurrent parent Sasanishiki, and a secondary F2 population was developed and employed for gene mapping and candidate gene analysis.
In addition, six typical indica varieties (93-11, Minghui 63, Dular, Guichao 2, IR 24 and Nanjing 11) and six typical japonica varieties (Balilla, Nipponbare, Nongken 57, Zhonghua 11, Wuyujing 3 and Wuyunjing 8), were selected for DNA sequence analysis on candidate genes, together with Sasanishiki and Habataki.
The three population (CSSL, BIL and secondary F2), together with their parents were planted in the experimental farm of Yangzhou University in Yangzhou (32o24′N, 119o26′E) and Hainan (18o30′N, 110o1′E). The two parents, together with CSSL population were grown in four different environments, i.e., Hainan in 2007 and 2013, and Yangzhou in 2008 and 2012. The BIL population was planted in Yangzhou in 2009 and 2012,respectively. In addition, the F1 plants (SL438/Sasanishiki) was planted in Yangzhou summer season of 2008, and the F2 population was grown in the winter season in Hainan. The field plots in Hainan or Yangzhou are the same in those years. The dates of seed sowing in Hainan and Yangzhou were around November 15th and May 15th, respectively. The seedling plants were transplanted about a month later after seed sowing. Each l CSSL and BIL was transplanted in two rows and each row contained ten individuals at a spacing of 10 cm × 25 cm. A wide-row spacing of 30 cm was arranged between different lines. The fertilizing and controlling of diseases and insect pests were maintained with conventional field managements. Due to the consistency in genetic backgrounds, all individuals in each population headed uniformly, which ruled out the interference by uncontrolled environmental factors on the spikelet fertility to some extent.
Measurement of spikelet fertility
At maturity stage, panicles from the main stem of three healthy representative individuals in each CSSL, BIL and parents were collected for spikelet fertility evaluation. Spikelet fertility of plant was represented as the number of filled grains divided by the total number of filled and unfilled grains. The spikelet fertility of each line was represented by the mean of the three plants.
Histological experiments
Pre-flowering panicles of two parents and SL438 were collected for the pollen fertility examination. Pollen grains were stained by 1 % I2-KI solution, then examined and enumerated under the light microscope.
The embryo sac fertility measurements were conducted following the whole stain-clearing protocol described by Yang (1986). In brief, the pre-flowering panicles were collected and fixed with formalin acetic acid alcoholic solution at 4 °C. Before staining, the ovaries of samples were processed with an ethanol series (70, 50 and 30 %), and distilled water for 10 min each time. At room temperature, the samples were stained with Ehrlich’s hematoxylin for 40 min followed by steps of washing and dehydration with a series ethanol solution (30, 50, 70, 90 and 95 %), and finally stored in pure ethanol overnight. Then the samples were cleared with methyl salicylate for at least 1 day. The embryo sac fertility was examined under microscope. About twenty-five embryo sacs were examined for each plant.
DNA extraction and marker analysis
DNA was extracted from fresh–frozen leaves of each plant using CTAB according to Rogers and Bendch (1988). The simple sequence repeat (SSR) markers were mainly employed for the initial mapping. The primer sequences of SSR markers was downloaded from http://www.gramene.org and synthesized by Shanghai Sangon Inc. DNA amplification was performed by using a PCR machine (Biometra Corporation, USA), programmed for an initial 5 min denature at 94 °C, then followed by 33 cycles for 1 min at 94 °C, 1 min at 55 °C, 1.5 min at 72 °C, and a final 10 min at 72 °C. Each PCR reaction had 25 μL in volume, containing 10 mmol/L Tris–HCl (pH 8.3), 50 mmol/L KCl, 1.5 mmol/L MgCl2, 1 U Taq ploymerase, 4 nmol/L dNTP, 10 pmol/L primers, 20 ng DNA template. The PCR products were analyzed on 3 % agarose gels stained with ethidium bromide, and photographed with a GEL DOC 1000 system (BioRed Company). If there was no polymorphism detected on the agarose gel, the products were further analyzed on 6 % polyacrylamide gel stained with 0.1 % silver nitrate.
Newly sequence-tagged site marker development
According to the result from initial mapping of genes responsible for hybrid breakdown, the sequence-tagged site (STS) markers were developed basing on the diversity between the genomic DNA sequences of cv. Nipponbare and cv. 93-11 around the target region. The sequences of STS primers were designed by using Primer Premier 5.0 software. The PCR reaction and electrophoresis condition were the same as described above.
Statistical analysis and QTL mapping
The analysis of variance (ANOVA) procedure package in Matlab V7.0 was used for statistical analyses of the spikelet fertilities of all CSSLs and BILs in this study. The windows QTL IciMapping procedure was employed for identification of QTL responsible for spikelet fertility in CSSL and BIL populations (Li et al. 2007; Wang 2009; http://www.isbreeding.net/software/). The function CSL and MET were used for CSSL mapping and QTL analysis in BIL. Mapping function Kosambi and the 2.0 LOD score threshold were set in QTL identification. The linkage map information of both population are from the Rice Genome Project of the National Institute of Agrobiological Sciences, Japan (http://www.rgrc.dna.affrc.go.jp/stock.html). QTL nomenclature followed the recommendation by McCouch (2008).
For finely mapping the QTL on chromosome 12 controlling spikelet fertility, a secondary F2 population derived from a cross between SL438 and Sasanishiki was developed. Totally 2452 individuals with low spikelet fertility (<50 %) were selected for genotyping and partial linkage map construction.
Gene prediction and DNA sequence analysis
Gene prediction was performed using FGENESH (Salamov and Solovyev 2000). The genomic DNA fragments of candidate genes from two parents Sasanishiki and Habataki, as well as some typical indica/japonica varieties, were amplified and sequenced. Sequencing reaction was performed by Shanghai Sangon Inc.
Results
The spikelet fertilities of CSSL and BIL populations
The spikelet fertilities of the CSSL population and their parents were measured and showed in Fig. 1a. It was found that, the spikelet fertilities segregated dramatically in the CSSL population, ranging from 29.0 to 98.3 % in 2007 (Hainan), 32.0 to 95.2 % in 2008 (Yangzhou), 25.6 to 95.2 % in 2012 (Yangzhou) and 20.3 to 94.7 % in 2013 (Hainan), respectively. However, it was obvious that most CSSLs had normal spikelet fertilities when compared to recurrent parent Sasanishiki, only a hand of CSSLs showed low spikelet fertilities, such as SL438 and SL439 (~30 %). Similarly, in BIL population, the spikelet fertilities also segregated widely with more BILs skewed to the recurrent parent Sasanishki (Fig. 1b). Obviously, the hybrid breakdown occurred in the progeny of the cross of Sasanishiki/Habataki. It is notable that some CSSL lines, such as SL438 and SL439, exhibited partial sterile, but having normal plant height and the number of tillers, similar to its recurrent parent Sasanishiki (Fig. 2A, B).
The performance of spikelet fertilities in two populations. a The spikelet fertility performance of Sasanishiki/Habataki CSSL population in four locations.The spikelet fertilities of SL439 in 2012, Yangzhou and 2013, Hainan were missing. b Distributions of spikelet fertility of 85 Sasanishiki/Habataki BILs in Yangzhou 2009 and 2012. c Comparison of spikelet fertilities of four types in BIL population. SS, Sasanishiki homozygote at two loci; SH, homozygous for Sasanishiki at qSF-8 and homozygous for Habataki at qSF-12; HS, homozygous for Habataki at qSF-8 and homozygous Sasanishiki for at qSF-12; HH, Habataki homozygous at both loci
Phenotype comparison of Habataki (Ha),Sasanishiki (Sa) and SL438. A Gross morphology of Ha, Sa and SL438 at maturity stage. B Plant height comparison of Ha, Sa and SL438. C Panicle of Ha, Sa and SL438. D Comparison of spikelet fertilitiesof Ha, Sa and SL438.Values in (B) and (D) are means with SD (n = 10 plants). E Pollen grains of Ha, Sa and SL438 (stained by 1 % I2-KI solution). F Comparison of pollen fertilities of Ha, Sa and SL438. n = 3. G Fertile embryo sac with visible antipodal cell (a) and polar nucleus (P). Sterile embryo sac with degenerated cells (d). H Comparison of embryo sac fertilities of Ha, Sa and SL438. 25 embryo sacs were examined for each sample
Further, the ANOVAs of spikelet fertilities in both CSSL and BIL population were conducted, and the results clearly indicated that genotypes, year, and their interactions (genotype × location, genotype × year) significantly contributed to the total variances of spikelet fertilities with the exception of location treatment (Supplemental Table S1 and S2), suggesting that apart from genetic components, the environmental factors (such as temperature at booting stage) also have effects on the fertility performance.
The pollen and embryo sac fertilities in SL438
In order to analyze the reason causing the low spikelet fertility in SL438 (Fig. 2C, D), we further measured the pollen fertility and the embryo sac fertility of the two parents and SL438. The results showed that, the pollen fertility of SL438 was about 88.33 %, having no significant difference with those of Sasanishiki (93.11 %) and Habataki (92.32 %, Fig. 2E, F). However, the embryo sac fertility of SL438 (~25 %) was significantly lower than those of the two parents (95 and 90 %, respectively, Fig. 2G, H). The embryo sac fertility was approximately equivalent to the spikelet fertility in SL438, suggesting that the partial embryo sac abortion was the main reason for the low spikelet fertility in SL438.
QTL mapping for spikelet fertility in CSSL and BIL population
When a 2.0 LOD score threshold was applied, four QTL were detected in Hainan (2007) (Table 1), namely, qSF-1-1 (on chromosome 1), qSF-10-1 (on chromosome 10), qSF-12-1 and qSF-12-2 (on chromosome 12). However, only one QTL, qSF-12-1 (around RM6998) was detected on chromosome 12 in the two replicates of Yangzhou in 2008 and 2012. While in Hainan (2013), two QTL, qSF-5-1 (around the marker RM2744) and qSF-12-1 (around RM6998) were identified on chromosome 5 and 12, respectively. Obviously, of all the detected QTL for spikelet fertility in the CSSL population, only the QTL qSF-12 around the marker RM6998 on chromosome 12 could be repeatedly detected in the four seasonal environments, indicating that qSF-12 expressed stably across various seasonal environments.
For the BIL population, the result of QTL analysis showed that two QTL, qSF-8 (on chromosome 8) and qSF-12 (on chromosome 12), were identified in the two environments (Table 2). Basing on the linked marker information, qSF-12 seemed to be identical to that identified in CSSL population.
Furthermore, on the base of the spikelet fertilities of 85 BILs, we analyzed the relationship between two QTL, qSF-8 and qSF-12. The closely linked marker C1121 (qSF-8) and R1709 (qSF-12) were selected to group BILs into four types, SS (Sasanishiki type homozygous at both loci, designated as SS), HH (Habataki type homozygous at both loci), SH (homozygous for Sasanishiki at qSF-8 and homozygous for Habataki at qSF-12) and HS (homozygous for Habataki at qSF-8 and homozygous for Sasanishiki at qSF-12), and their average spikelet fertilities were compared. The spikelet fertilities of SH and HS type were 61.2, 54.6 % in 2009, 53.0 and 56.1 % in 2012, respectively, which were significantly lower than those of SS and HH type (Fig. 1c). The results clearly demonstrated that the epistatic effect played an important role in controlling rice hybrid breakdown.
Fine mapping of qSF-12
Based on the marker genotype information of SL437 (with high spikelet fertility, ~92.6 %), SL438 and SL439 (with low spikelet fertility, ~30 %) (Supplemental Fig. S1A), it was deduced that the introgression of the overlapped chromosomal segment from Habataki defined by molecular marker RM6998 and RM1300 to Sasanishiki background leads to the decrease of spikelet fertility. Hence, qSF-12 was considered to residue in this overlapped region governed by marker RM6998 and RM1300, which was consistent with the previous QTL mapping result.
A secondary F2 population containing nearly 10,000 individuals derived from a cross of SL438/Sasanishiki was generated for fine mapping of qSF-12 (Supplemental Fig. S1A). The spikelet fertility of F1 (SL438/Sasanishiki) was normal (92.5 %), while in F2 population, the spikelet fertilities segregated into two groups, normal spikelet fertilities (>70 %) and low ones (<50 %), no intermediate type was found. There are 2452 individuals with low spikelet fertility (<50 %) in this F2 population and the spikelet fertilities of the rest individuals were normal (>70 %). The χ 2-test result showed that the proportion of sterile and normal plants in the F2 population was well fitted to 1:3 ratio (χ 2 = 1.80 < \(\chi_{0.05}^{2}\) = 3.84), implying that qSF12 is a single recessive sterile locus on chromosome 12.
In order to finely map qSF-12, based on the information about the donor segment from Habataki on the chromosome 12, seven polymorphic SSR markers were initially used for genotyping the 192 F2 individuals with low spikelet fertility (<50 %), and qSF-12 was delimited to a region between RM6905 and RM270 (Fig. 3a). To finely map qSF-12, more newly-developed polymorphic STS markers (Supplemental Table S3) were used to genotype the another 1420 F2 individuals with low spikelet fertility. Based on the number of recombinant events (individuals with Sasanishiki type homozygous or heterozygous genotypes) at each marker locus, the qSF-12 gene was finally restricted to a 137-kb DNA fragment in AL928774 on chromosome 12 between STS marker M12–16 and M12–13, which was co-segregated with M12–y22 (Fig. 3b, c).
Genetic mapping of qSF-12. a Location of qSF-12 on chromosome 12. b qSF-12 was located between M12–16 and M12–13. c qSF-12 was delimited to a 137-kb long region on PAC AL731884 and AL928774. d The nine candidate genes with full length cDNA support in the 137-kb long region. e A 3-bp insertion (GAT) was found at the end of exon 10 in Habataki compared to that of Sasanishiki. f The sequence different can be revealed by STS marker M12–y22
Candidate gene analysis
Gene prediction analysis (http://rice.plantbiology.msu.edu) suggests that there are 19 putative open reading frames (ORF) on the 137-kb DNA fragment covering the qSF-12 locus, Of which, 11 ORFs had full length cDNA supports. Based on the information from RGAP (http://rice.plantbiology.msu.edu), nine ORFs were confirmed to express in different organs with various FPKM (Reads Per Kilobase of exon model per Million mapped reads), while for LOC_Os12g39020 and LOC_Os12g39030, their RNA-Seq FPKM values were 0, suggesting that the two predicted ORFs may be ghost (Supplemental Table S4). Furthermore, the CDS re-sequencing results of the nine candidate genes showed that sequence differences between Sasanishiki and Habataki were only found in LOC_Os12g38850, LOC_Os12g38900, LOC_Os12g38940, LOC_Os12g38950 and LOC_Os12g38970. Taken together, the five ORFs mentioned above were considered to be the putative candidate gene of qSF-12 (Supplemental Table S5).
Discussion
In the present study, we conducted the genetic analysis on the hybrid breakdown in the progeny of a cross between Sasanishiki and Habataki, two typical indica/japonica varieties. Based on the CSSL population, we identified a major QTL, qSF-12, responsible for spikelet sterility (embryo sac sterility). Moreover, another QTL, qSF-8, was detected to interact with qSF-12 in BIL population. Our results provided a direct evidence that hybrid breakdown is caused by the complementary gene interaction, consistent with the hypothesis proposed by Stebbins (1950). In addition, the interaction analysis between qSF-12 and qSF-8 further suggested that at the two loci, either of allele replaced by its opposite one would lead to the decrease of spikelet fertility in one genetic background, which fits to the Dobzhansky–Muller model (Dobzhansky 1937). Moreover, we found that the SH or HS genotypes exhibited similar fertility level with ~60 % (Fig. 1c), suggesting that both qSF-12 and qSF-8 seem to have similar genetic effects. Of course, the CSSL covering qSF-8 in Sasanishiki genetic background is needed to further confirm our speculation. It should be noted that, the SL438 belongs to the SH class, when cultivated in Yangzhou (2008, 2012), it showed very low spikelet fertility (30.6 ± 8.3 %; Fig. 1a), but the BILs carrying the genotype classes of SH and HS showed moderate spikelet fertility (~60 %) (Fig. 1c). The reason underlying the phenomenon may be due to the recombination events existing between the linked marker and QTL. When we analyzed the interaction between qSF-8 and qSF-12, the two closely linked markers were used to divide the population into four classes. Due to the recombination between C1121 and qSF-8, as well as the recombination between R1709 and qSF-12, the average fertility of SH class seems to be relatively higher than that in CSSL. In fact, in the BIL population, some lines with low spikelet fertility (20–30 %) were observed, which belong to SH class.
In addition, qSF-8 was only identified in BIL population, not in CSSL population. According to the interaction analysis between qSF-8 and qSF-12, qSF-8 also should be detected in CSSL population theoretically. It seems to be paradoxical. To address the issue, we carefully analyzed the genotypes of four CSSLs (SL425, SL426, SL427 and SL428), in which, the substituted segments were from donor parent Habataki, covering chromosome 8. Interestingly, we found that the substituted segment in all four CSSLs did not cover the qSF-8 anchored region governed by RM8018 and RM6838, instead, they were all the segments from Sasanishiki (Supplemental Fig. S1B). Hence, we considered it may be the reason why we failed to detect the qSF-8 in the CSSL population.
Although the hybrid breakdown is a difficult problem to solve due to its complex mode of inheritance, many genes responsible for it have been identified and mapped (Fukuoka et al. 1998; Kubo and Yoshimura 2002, 2005). In our study, two complementary QTL, qSF-12 (chromosome 12) and qSF-8 (chromosome 8), were found to control the spikelet sterility (embryo sac sterility) in the cross of Sasanishiki/Habataki. It seems that the genes responsible for hybrid breakdown were different depending on the specific crosses although the same genetic mechanism, epistatic interaction, overruled in all reported crosses. It should be mentioned that hsa1 (Kubo and Yoshimura 2005) and qSF-12 are both located on the long arm of chromosome 12 between the marker R1709 and C1069. hsa1 is closely linked to RFLP marker G148 located on chromosome 12 from 24,487,487 to 24,487,785 bp, and the co-segregated marker M12–y22 for qSF-12 is from 23,879,858 to 23,884,982 bp on chromosome 12 in the present study. Because the locations of hsa1 and qSF-12 are very close and both of them have not been cloned and functional characterized, more studies are needed to clarify whether they are allelic. On the contrary, according to the marker information, hsa2 identified in the cross of Asominori/IR24 is closely linked to RFLP marker G104 with 2.1 cM on chromosome 8 (Kubo and Yoshimura 2005), but qSF-8 detected in the present study is far from G104, suggesting they are possibly non-allelic. Of course, the exact allelic relationship between hsa2 and qSF-8 should be elucidated through the fine mapping and cloning of both of them. Taken together, these results suggest that the complicated mechanisms have been evolved for hybrid breakdown during the rice speciationprocess.
Up to date, although many genes/QTL have been identified and assigned to the corresponding chromosomes by using molecular markers, which shown to be responsible for hybrid breakdown (Fukuoka et al. 1998, Kubo and Yoshimura 2002, 2005, Zhang et al. 2011a), few genes have been cloned and functional characterized except for Hwi1 and Hwi2 (Chen et al. 2014). In the present study, we carried out fine mapping of qSF-12, controlling embryo sac fertility, and successfully delimited it to a 137-kb DNA fragment. Based on the RNA-seq data and the DNA sequence diversity, five ORFs (LOC_Os12g38850, LOC_Os12g38900, LOC_Os12g38940, LOC_Os12g38950 and LOC_Os12g38970) are considered to be candidates of qSF-12. Because qSF-12 is believed to play an important role in embryo sac development, not in other organs, so we consider that the candidate of qSF-12 may possibly have the highest FPKM value in the reproductive organs, including the anther and pistil. Among five ORFs, it is obvious that LOC_Os12g38850 has the highest FPKM value in the pistil and anther (Supplemental Table S4), thus, it was preliminarily considered as the putative candidate of qSF-12. In order to verify our hypothesis, we sequenced the genomic DNA of LOC_Os12g38850, and a 3-bp insertion (GAT, a codon of aspartic acid) was found at the end of exon 10 in Habataki when compared to Sasanishiki (Fig. 3e), which can be revealed by STS marker M12–y22 (Fig. 3f).
We also analyzed the nucleotide sequences of LOC_Os12g38850 in six typical indica varieties and six typical japonica varieties. Interestingly, the 3-bp deletion in the coding region was detected in most japonica varieties except for Nongken 57 (Supplemental Table S6). In contrast, the 3-bp is observed to exist in most of indica varieties, except for Minghui 63. The 3-bp insertion/deletion is basically consistent with the indica/japonica differentiation, that is, most of indica varieties have the additional 3-bp in exon 10, whereas most of japonica ones do not have the 3-bp insertion.
On the other hand, the bioinformatics analysis showed that LOC_Os12g38850 encodes a DUF1336 containing protein. Up to date, only two genes were cloned, encoding a DUF1336 domain containing protein. One is EDR2 acting as a negative regulator of cell death, specifically the cell death elicited by pathogen attack and mediated by the salicylic acid defense pathway (Vorwerk et al. 2007). The other one is identified to interact with PtELF5, which was proven to play an important role during the early flowering of precocious trifoliate orange (Zhang et al. 2011b). The accumulated knowledge implies that the protein containing DUF1336 domain may function in cell growth. So, it is plausible to deduce that LOC_Os12g38850 is the candidate of qSF-12; of course, the complementary test is needed to confirm it.
In the follow-up study, we will put efforts on functionally characterization of LOC_Os12g38850. And we believe that the cloning and characterization of qSF-12 will be helpful not only for dissecting the mechanism of hybrid breakdown and better understanding of rice evolution, but also for the direct application of the co-segregated marker m12–y22 in rice breeding program.
References
Chen J, Ding J, Ouyang Y, Du H, Yang J, Cheng K, Zhao J, Qiu S, Zhang X, Yao J, Liu K, Wang L, Xu C, Li X, Xue Y, Xia M, Ji Q, Lu J, Xu M, Zhang Q (2008) A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica–japonica hybrids in rice. Proc Natl Acad Sci 105:11436–11441
Chen C, Chen H, Lin YS, Shen JB, Shan JX, Oi P, Shi M, Zhu MZ, Huang XH, Feng Q, Han B, Jiang LW, Gao JP, Lin HX (2014) A two-locus interaction causes interspecific hybrid weakness in rice. Nat Commun. doi:10.1038/ncomms4357
Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Inc., Sunderland
Dobzhansky T (1937) Genetics and the origin of species. Columbia University Press, New York
Fukuoka S, Namai H, Okuno K (1998) RFLP mapping of the genes controlling hybrid breakdown in rice. Theor Appl Genet 97:446–449
Garavito A, Guyot R, Lozano J, Gavory F, Samain S, Panaud O, Tohme J, Ghesquière A, Lorieux M (2010) A genetic model for the female sterility barrier between Asian and African cultivated rice species. Genetics 185(4):1425–1440
Ji Q, Zhang MJ, Lu JF, Wang HM, Lin B, Liu QQ, Chao Q, Zhang Y, Liu CX, Gu MH, Xu ML (2012) Molecular basis underlying the S5-dependent reproductive isolation and compatibility of indica/japonica rice hybrids. Plant Physiol 158:1319–1328
Kato S (1930) On the affinity of the cultivated varieties of rice plants, Oryza sativa L. J Dept Agric Kyushu Imp Univ 2:241–275
Kitamura E (1962) Genetic studies on sterility observed in hybrids between distantly related varieties of rice, Oryza sativa L. Bull Chugoku Agric Exp Station A 8:141–205
Kubo T, Yoshimura A (2002) Genetic basis of hybrid breakdown in a japonica/indica cross of rice, Oryza sativa L. Theor Appl Genet 105:906–911
Kubo T, Yoshimura A (2005) Epistasis underlying female sterility detected in hybrid breakdown in a Japonica–Indica cross of rice (Oryza sativa L.). Theor Appl Genet 110:346–355
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
Long Y, Zhao L, Niu B, Su J, Wu H, Chen Y, Zhang Q, Guo J, Zhuang C, Mei M, Xia J, Wang L, Wu H, Liu Y-G (2008) Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc Natl Acad Sci 105:18871–18876
McCouch SR (2008) CGSNL (Committee on Gene Symbolization, Nomenclature and Linkage, Rice Genetics Cooperative) Gene nomenclature system for rice. Rice 1:72–84
Oka HI (1978) Phylogenetic differentiation of cultivated rice. XXI. The sporophytic pollen sterility: its genetic basis and interval relationship as shown by F2 sterility. Jpn J Genet 53:397–410
Ouyang Y, Liu YG, Zhang Q (2010) Hybrid sterility in plant: stories from rice. Curr Opin Plant Biol 13:186–192
Rogers SO, Bendch AJ (1988) Extraction of DNA from plant tissues. Plant Mol Biol Man 6:1–10
Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10:516–522
Stebbins GL Jr (1950) Isolation and the origin of species. In: Stebbins GL Jr (ed) Variation and evolution in plants. Columbia University Press, New York, pp 189–250
Vorwerk S, Schiff C, Santamaria M, Koh S, Nishimura M, Vogel J, Somerville C, Somerville S (2007) EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol 7:35
Wang J (2009) Inclusive composite interval mapping of quantitative trait genes. Acta Agron Sin 35(2):239–245
Yamamoto E, Takashi T, Morinaka Y, Lin SY, Kitano H, Matsuoka M, Ashikari M (2007) Interaction of two recessive genes, hbd2 and hbd3, induces hybrid breakdown in rice. Theor Appl Genet 115:187–194
Yamamoto E, Takashi T, Morinaka Y, Lin SY, Wu JZ, Matsumoto T, Kitano H, Matsuoka M, Ashikari M (2010) Hain of deleterious function causes an autoimmune response and Bateson–Dobzhansky–Muller incompatibility in rice. Mol Genet Genomics 283:305–315
Yang HY (1986) The use of a whole stain-clearing technique for observations on embryo sac, embryo, endosperm and embryoid. Acta Bot Sin 28:575–581
Yokoo M (1984) Female sterility in an indica–japonica cross of rice. Jpn J Breed 34:219–227
Zhang H, Zhang CQ, Sun ZZ, Yu W, Gu MH, Liu QQ, Li YS (2011a) A major locus qS12, located in a duplicated segment of chromosome 12, causes spikelet sterility in an indica–japonica rice hybrid. Theor Appl Genet 123:1247–1256
Zhang JZ, Ai XY, Sun LM, Zhang DL, Guo WW, Deng XX, Hu CG (2011b) Molecular cloning and functional characterization of genes associated with flowering in citrus using an early-flowering trifoliate orange (Poncirus trifoliata L. Raf.) mutant. Plant Mol Biol 76:187–204
Acknowledgments
We wish to thank the Rice Genome Project of the National Institute of Agrobiological Sciences of Japan, for kindly providing the BIL and CSSL population. This study was financially supported by the National Natural Science Foundation (31171158), the Ministry of Science and Technology (2011ZX08009-003-005), the Natural Science Foundation of Jiangsu Province (BK2012684), the project of Six Talent Peaks in Jiangsu Province, and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Rongde Li and Min Guo have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, R., Guo, M., Lu, Y. et al. Genetic dissection of hybrid breakdown in an indica/japonica cross and fine mapping of a quantitative trait locus qSF-12 in rice (Oryza sativa L.). Mol Breeding 35, 144 (2015). https://doi.org/10.1007/s11032-015-0331-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0331-4