Abstract
A collection of 5,659 expressed sequence tags (ESTs) from pineapple [Ananas comosus (L.) Merr.] was screened for simple sequence repeats (EST-SSRs) with motif lengths between 1 and 6 bp. Lower thresholds of 15, 7 and 5 repeat units were used to define microsatellites of the mono-, di-, and tri- to hexanucleotide repeat type, respectively. Based on these criteria, 696 SSRs were identified among 3,389 EST unigenes, together representing 2,840 kb. This corresponds to an average density of one SSR every 4.1 kb of non-redundant EST sequences. Dinucleotide repeats were most abundant (38.4% of all SSRs) followed by trinucleotide repeats (38.1%). Flanking primer pairs were designed for 537 EST-SSR loci, and 49 of these were screened for their functionality in 12 accessions of A. comosus, 14 accessions of 5 additional Ananas species and 1 species of Pseudananas. Distinct PCR products of the expected size range were obtained with 36 primer pairs. Eighteen loci analyzed in more detail were all polymorphic in pineapple, and primer pairs flanking these loci also generated PCR products from a wide range of genera and species from six subfamilies of the Bromeliaceae. The potential to reveal polymorphism in a heterologous target species was demonstrated in Deuterocohnia brevifolia (subfamily Pitcairnioideae).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microsatellites or simple sequence repeats (SSRs) are short, tandemly repeated DNA regions that are ubiquitously present in the genomes of pro- and eukaryotes. Because of their abundance, multiallelic behavior, high polymorphism, codominant inheritance and excellent reproducibility, PCR-amplified SSRs have become the markers of choice for many applications, including genetic mapping and population genetics (Powell et al. 1996). Among the few disadvantages of SSR markers are the often tedious and costly cloning and enrichment procedures required for their generation (Zane et al. 2002; Squirrell et al. 2003; Weising et al. 2005).
Since about a decade, computational methods have been applied to search for microsatellites in the vast body of sequence data generated by high-throughput genomics. Expressed sequence tag (EST) databases have become particularly attractive resources for such in silico mining, as was demonstrated in, e.g., citrus (Chen et al. 2006), coffee (Poncet et al. 2006; Aggarwal et al. 2007), sugarcane (Pinto et al. 2004), sunflower (Pashley et al. 2006; Heesacker et al. 2008) and particularly in the cereals (Kantety et al. 2002; Thiel et al. 2003; Yu et al. 2004). Several reasons account for the high popularity of EST-derived microsatellite markers (EST-SSRs). First, marker development from already existing sequence data is fast, easy and economical. Any type of SSR will be detected using an appropriate search program, whereas only SSRs with predefined motifs are captured by enrichment cloning. Second, SSRs are a common component of ESTs, which is due to the preferential association of SSRs with the non-repetitive portion of plant genomes (Morgante et al. 2002). Third, EST-SSRs are physically linked to expressed genes and therefore represent so-called functional markers that are of particular interest for, e.g., marker-assisted selection (Andersen and Lübberstedt 2003). Finally, primer target sequences residing in the expressed DNA regions are expected to be relatively well conserved; thus, enhancing the chance of marker transferability across taxonomic boundaries (Varshney et al. 2005).
The research of our group aims at elucidating evolutionary processes in Bromeliaceae, which comprise more than 3,100 almost exclusively neotropical species (Luther 2008). A well-known representative of the family is Ananas comosus (L.) Merr. (pineapple), the third most important fruit crop worldwide after banana and citrus (Botella and Smith 2008). Bromeliads are famous for their ecological plasticity that allows them to occupy a great variety of terrestrial and epiphytic habitats. Their great versatility is associated with several key innovations, like CAM photosynthesis, succulence, and the formation of foliar trichomes capable of water absorption (Benzing 2000). Despite the ecological importance of the family and the economic importance of pineapple, only a few genomic resources have yet been developed for Bromeliaceae (Botella and Smith 2008). Thus, only about 3,000 nucleotide sequence entries of bromeliads (except ESTs) have been deposited in GenBank by December 2010, a single, medium-sized EST library is available for pineapple (Moyle et al. 2005), and comparatively a few SSR markers have so far been published for any bromeliad species (Boneh et al. 2003; Sarthou et al. 2003; Palma-Silva et al. 2007; Kinsuat and Kumar 2007; Paggi et al. 2008; Carlier et al. 2011).
The present study aims at expanding the genomic resources for Ananas and other Bromeliaceae by developing a large set of EST-SSR markers. To achieve this, we screened the publicly available collection of 5,659 pineapple ESTs for the presence of SSRs, designed flanking primer pairs to amplify SSR-containing ESTs, evaluated a subset of these primer pairs for the extent of polymorphism that they reveal in A. comosus, and tested marker portability across the genus Ananas as well as to more distant members of the Bromeliaceae family.
Materials and methods
EST data mining
By December 2010 the PineappleDB EST database developed by Moyle et al. (2005) contained 5,659 EST sequences that are also available via NCBI (GenBank). These ESTs were derived from five cDNA libraries constructed from uninfected root tips, dissected vascular cylinders of galls from early and late infection, mature green fruit and mature yellow fruit, respectively. We used the Geneious 5.0 software (Drummond et al. 2010) to assemble the pineapple ESTs into non-redundant unigenes (contigs and singletons).
Primer design and Basal Local Alignment Search Tool (BLAST) searches
The EST unigenes were screened for the presence of perfect SSRs using a combination of SciRoKo (Kofler et al. 2007) and BatchPrimer3 (You et al. 2008). In the present study, a microsatellite was defined as a DNA sequence containing at least 15 repeat units for mononucleotide repeats, seven units for dinucleotide repeats, and five units for tri-, tetra-, penta-, and hexanucleotide repeats. Primer pairs flanking the EST-SSRs were designed using the BatchPrimer3 interface which employs the primer design features of Primer3 (Rozen and Skaletsky 2000). Primer construction was based on the following criteria: length ranging from 18 to 23 nucleotides (20 as the optimum), PCR product size ranging from 100 to 300 bp, annealing temperature from 50 to 70°C (55°C as the optimum), and GC content between 30 and 70% (50% as the optimum). All other parameters were set to default values. To determine putative gene functions, a BLASTx search was performed against the Oryza sativa reference protein database in NCBI (GenBank). The OrfPredictor software (Min et al. 2005) was used to determine whether the repeat motifs were located in the 5′ untranslated region (5′UTR), the protein-coding region (CDS) or the 3′ untranslated region (3′UTR).
Plant materials and genomic DNA isolation
The plant materials used in the present study comprised 26 accessions from 6 of the 7 Ananas species recognized by Smith and Downs (1979), 1 accession of Pseudananas sagenarius, 9 accessions of Deuterocohnia brevifolia and 1 accession each of 73 species from 30 genera within 6 subfamilies of Bromeliaceae. These plants were supplied by various organizations and Botanical Gardens (see compilation in supplementary Table S1). Some species and accessions of Deuterocohnia and Fosterella (subfamily Pitcairnioideae) were collected from the wild in various regions of Bolivia. The total genomic DNA from fresh or silica-dried leaf tissue was extracted using a variant of the CTAB method (Weising et al. 2005). Final DNA concentrations were determined electrophoretically versus known amounts of λ-DNA as standards.
SSR amplification and detection
Polymerase chain reactions (PCR) were performed in total volumes of 12.5 μL containing 1× PCR Mango-buffer (provided by the manufacturer, Bioline, Luckenwalde, Germany), 5 μg BSA, 1.5 mmol/L MgCl2, 0.1 mmol/L of each dNTP, 10 pmol of each primer, ~10 ng of genomic DNA and 0.1 U of Taq DNA polymerase (Mango-Taq, Bioline, Luckenwalde, Germany). Primers were used either unlabeled, or with the forward or reverse primer carrying a fluorescent IRDye700 or IRDye800 label at its 5′-end. After initial denaturation at 94°C for 6 min, a touchdown PCR protocol was followed for 30 cycles, each consisting of 94°C for 45 s, 65–54°C for 30 s, and 72°C for 45 s. Starting at 65°C, the annealing temperature was reduced by 1°C per cycle during the first 11 cycles and then left constant at 54°C for the remaining 19 cycles. Final extension was at 72°C for 8 min.
For the initial functionality screens, unlabeled PCR products were separated by electrophoresis on 1.5% agarose gels (NEOO Ultra Quality, Roth, Karlsruhe, Germany) in 0.5× TBE buffer (Sambrook and Russell 2001) at 10 V/cm, stained with ethidium bromide (1 μg/mL) and visualized under UV-light. A 100 bp DNA ladder (Roth, Karlsruhe, Germany) was used as the molecular size standard. Using the same cycling conditions, fluorescently labeled PCR fragments used in the cross-taxon transferability tests were mixed with an equal volume of loading buffer [10 mmol/L EDTA, 0.1% (w/v) basic fuchsin, 98% (v/v) formamide], and denatured at 85°C for 5 min. Half-μL samples of each reaction were electrophoresed on high resolution, denaturing 41 cm × 0.2 mm polyacrylamide gels [6% Sequagel XR (National Diagnostics, Atlanta, GA, USA) in 1× TBE buffer (Sambrook and Russell 2001)] using an automated sequencer (Li-Cor 4200 IR2, Li-Cor Inc., Lincoln, NE, USA). Gels were run at constant power of 25 mA and constant temperature of 45°C. Fragment mobility was measured by real-time laser scanning.
Fragment sizing and assessment of genetic variability
Fragment sizing was performed manually with the help of a known sequencing reaction that served as an external size standard. Reference samples were included in all gels. The extent of SSR marker variability within a species was estimated using the following parameters: observed number of alleles (NA), expected heterozygosity (H e) and observed heterozygosity (H o). Expected heterozygosity was estimated using the formula:
where n is the number of gene copies in the sample, k is the number of alleles, and p i is the sample frequency of the ith allele. All calculations were performed using Arlequin version 3.1 (Excoffier et al. 2005). The NTSYSpc version 2.1 software package (Rohlf 2000) was used to perform principal coordinate analyses (PCoA) based on a square-root transformed distance matrix calculated with the index of Bray and Curtis (1957).
Results
Identification, frequency and characterization of EST-SSRs
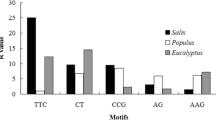
The PineappleDB database generated by Moyle et al. (2005) comprises 5,659 ESTs with an average length of 730 bp. Of these, 3,389 sequences (824 contigs and 2,565 singletons, together representing 2,840 kb) were identified as unigenes by the Geneious 5.0 software (Drummond et al. 2010). Using the minimal length criteria outlined in Materials and methods, a total of 696 unique, perfect SSRs were retrieved from 565 unigenes, corresponding to an average frequency of one SSR per 4.1 kb. A single SSR each was present in 457 unigenes, whereas 108 unigenes contained two or more SSRs. The frequencies and sequence characteristics of the 696 SSRs are summarized in Tables 1, 2. Forty-two different SSR motif types were represented. Repeat numbers ranged from 5 to 92, and the total lengths of the repeats ranged from 14 to 92 bp (average 37.8 bp). Dinucleotide repeats were most abundant (38.4%), closely followed by trinucleotide repeats (38.1%). Tetra-, penta- and hexanucleotide repeats together represented <10% of the total EST-SSRs. With 231 loci, (AG) n /(CT) n was by far the most frequent dinucleotide repeat motif, whereas only two (CG) n repeats were detected. (AGC) n /(GCT) n proved to be the most abundant trinucleotide repeat motif, followed by (AGG) n and (AAG) n .
Marker development, and polymorphism of pineapple EST-SSRs within the genus Ananas
As mononucleotide repeat polymorphisms are often difficult to interpret, only di-, tri-, tetra-, penta- and hexanucleotide repeats were considered as potential candidates for EST-SSR marker development (Table 3). Sufficient flanking sequence of appropriate quality for primer design was present in 421 unigenes, which together contained 537 perfect microsatellites (i.e., 92.4% of all di- to hexanucleotide repeats present in the database). Forward and reverse sequences of the 537 PCR primer pairs designed for these loci are compiled in the supplementary Table S2, along with predicted allele lengths and microsatellite repeat motifs. Forty-nine primer pairs, all specifying single, perfect di-, tri- or tetranucleotide repeats, were randomly selected and used to amplify a test panel consisting of 12 accessions of Ananas comosus, 5 accessions each of Ananas nanus and Ananas bracteatus, 1 Ananas accession with unknown species assignment, 2 accessions of Ananas ananassoides, and 1 accession each of Ananas fritzmuelleri, Ananas lucidus and Pseudananas sagenarius (for a detailed list of taxa see supplementary Table S1). As evidenced by electrophoresis on 1.5% agarose gels and ethidium bromide staining, 40 of the 49 primer pairs (81.6%) generated one or two distinct PCR products of the expected size range for all A. comosus accessions. Thirty-six primer pairs (73.5%) also performed well in most other Ananas species and in Pseudananas.
Eighteen selected primer pairs were fluorescently labeled and used to amplify the same Ananas test panel followed by PCR fragment separation on high-resolution sequencing gels. Primer sequences, locus characteristics and results of genetic diversity analyses are summarized in Table 4. High quality, easy-to-type banding patterns were generated throughout, as exemplified for two loci in Fig. 1. All 18 SSR loci were polymorphic at the intraspecific level in A. comosus. The number of alleles ranged from 2 to 6 (mean 4.3) with an average expected heterozygosity of 0.58, and an average observed heterozygosity of 0.49 over all loci. Larger bands than expected were observed at two loci (Acom_22.22 and Acom_91.2) suggesting that an intron may be present between the primer binding sites. 16 of the 18 primer pairs also worked in all other Ananas and Pseudananas species tested, whereas Acom_39.5 and Acom_67.2 failed in all A. nanus accessions and Acom_39.5 failed in one accession of A. ananassoides. Typically one or two bands were found in each sample, consistent with the homozygous or heterozygous state of the respective locus. In P. sagenarius which is a tetraploid species (Collins 1960; Duval et al. 2001), three to four bands were generated by six of the primer pairs tested (Acom_9.9, Acom_12.12, Acom_78.4, Acom_86.4, Acom_101.1, Acom_109.6; see Fig. 1).
To illustrate the genetic divergence and relationships among the 28 accessions of Ananas and Pseudananas, the data set was subjected to a PCoA (Fig. 2). The first 3 axes explained approximately 61% of the overall genetic variation. Identical multilocus SSR genotypes each were found for (1) A. comosus BT_77 and BT_81, (2) A. comosus BT_79 and BT_85, (3) A. bracteatus BT_15 and BT_28, (4) A. bracteatus BT_73, A. bracteatus BT_80 and A. comosus BT_75 and (5) A. nanus BT_84, BT_86 and BT_90. All other accessions had a unique multilocus genotype. Three loosely defined groups were formed. One group is made up of all but one individual of A. comosus, a second group combines all individuals of A. nanus, and a third one harbors all but one accessions of A. bracteatus together with one aberrant A. comosus individual. No obvious groups were formed by the remaining samples.
Principal coordinates analysis (PCoA) based on microsatellite data for 18 EST-SSR markers and 28 Ananas accessions, including 11 accessions of A. comosus, one accession each of A. comosus var. porteanus and A. comosus var. erectifolius, five accessions of A. bracteatus, two accessions of A. ananassoides, five accessions of A. nanus, and one accession each of A. fritzmuelleri, A. lucidus, an Ananas accession of unknown species affiliation and Pseudananas sagenarius. Each species is represented by a unique symbol. The first three axes together represent 61.4% of the total genetic variation (axis 1 = 28.4%, axis 2 = 20.6%, axis 3 = 12.4%). See Fig. 1 and supplementary Table S2 for explanation of plant codes
Cross-species amplification of pineapple EST-SSRs in Bromeliaceae
The 18 primer pairs listed in Table 4 were also used to assess the extent of cross-species amplification within a wide range of Bromeliaceae outside the genera Ananas and Pseudananas, encompassing 74 species from 30 genera within 6 subfamilies. PCR products were visualized by polyacrylamide gel electrophoresis and fluorescence detection on an automated sequencer. The results are summarized in Table 5. Cross-species amplification was considered successful when either one or two distinct bands were observed on the respective lane of the sequencing gel. Under this criterium, success rates of marker transferability ranged from 17.9% (Acom_65.1) to 96.4% (Acom_12.12) with a mean value of 58.6%. Only one primer pair (Acom_39.5) failed with all samples outside Ananas. Eleven loci were particularly well conserved and showed consistent amplification in more than 60% of all species included in the test panel. The highest values of primer cross-species transportability were detected among Bromelioideae (mean 74.8%) followed by Puyoideae (mean 60.0%), Pitcairnioideae (mean 51.6%), Hechtioideae (mean 42.6%), Tillandsioideae (mean 27.3%) and Brocchinioideae (mean 19.5%).
The extent of polymorphism of the 18 selected pineapple EST-SSR markers in a distantly related, heterologous target species was tested on 9 individuals of Deuterocohnia brevifolia (subfamily Pitcairnioideae). Genetic diversity parameters calculated for these samples are summarized in Table 4, along with the respective values obtained for the 12 A. comosus accessions. 10 of the 18 primers pairs generated PCR products from all D. brevifolia accessions. Eight loci were polymorphic, with allele numbers ranging from three to five. Two loci (Acom_93.4 and Acom_109.6) were monomorphic and produced a single band only. The average values of expected and observed heterozygosity were 0.51 and 0.42, respectively.
Sequence homology search
BLAST searches were performed for the ESTs from which the 18 selected primer pairs were designed. The results are summarized in Table 4. 15 of the 18 ESTs showed significant homology with sequences in the Oryza sativa protein database in GenBank. As determined by OrfPredictor (Min et al. 2005), seven of these SSRs were located within coding regions, and four each were located in the 5′ and 3′UTR, respectively. There was no obvious correlation between the extent of marker polymorphism and the location of the SSR in coding versus untranslated regions.
Discussion
Pineapple ESTs are a rich source of SSRs
ESTs have become attractive and valuable resources for microsatellite markers in plants, including horticultural crops like grape (Scott et al. 2000), kiwifruit (Fraser et al. 2004), apricot (Decroocq et al. 2003) and citrus (Chen et al. 2006). A single, medium-sized EST database is also available for pineapple (Moyle et al. 2005). In the present work, we identified a total of 696 unique SSRs among the 3,389 unigenes currently present in the NCBI database, with one SSR every 4.1 kb. This frequency is at the upper limit of the ranges usually reported from other plant species (typically one SSR per 2–10 kb; see Morgante et al. 2002; Kantety et al. 2002; Kumpatla and Mukhopadhyay 2005; Varshney et al. 2005; Chen et al. 2006; Poncet et al. 2006; Scaglione et al. 2009). Although it is difficult to compare relative estimates of the frequency and abundance of EST-SSRs in different species because of the different cut-off criteria and search tools used, our findings clearly demonstrate that pineapple ESTs are a rich source of SSRs. During the preparation of the present manuscript, the NCBI database was also searched for pineapple microsatellites in an independent study that was focussed on genetic mapping (Carlier et al. 2011). Primer pairs were constructed for 45 EST-SSR loci, of which 13 were successfully included in a genetic map of A. comosus, comprising RAPD, ISSR, AFLP, SCAR, SSR and CAPS markers.
Di- and trinucleotides are the predominant type of repeat present in the pineapple EST database of Moyle et al. (2005) each comprising approximately 38% of all EST-SSRs (Tables 2, 3). This reflects the situation observed in other plants, where either dinucleotide repeats (e.g., Pinto et al. 2004; Kumpatla and Mukhopadhyay 2005) or trinucleotide repeats (e.g., Scott et al. 2000; Kantety et al. 2002; Chen et al. 2006) were the most common type of repeat depending on the report. Trinucleotide repeats generally prevail in coding regions, which is usually attributed to selection against frameshift mutations caused by length variation in non-trimeric repeats (Metzgar et al. 2000). Dinucleotide repeats are typically more frequent in 5′- and/or 3′UTR regions, but occasionally occur in coding regions as well. With 33.2% of all SSRs and 86.5% of all dinucleotide repeats, (AG) n /(CT) n was by far the most abundant individual SSR motif in the pineapple EST database (Table 1). High proportions of AG/CT repeats in ESTs were also found in, e.g., kiwifruit (70% of all EST-SSRs; Fraser et al. 2004), in coffee (62% of all dinucleotide EST-SSRs; Poncet et al. 2006), and in almost all ESTs of various dicotyledonous species analyzed by Kumpatla and Mukhopadhyay (2005). A large abundance of AG/CT repeats seems to be a general property of plant genomes, whereas AC/GT repeats are more common in animal genomes. The functional significance of SSRs in genic regions of plants is unclear, but homopurine–homopyrimidine stretches like (AG) n /(CT) n in the 5′UTR have been reported to be involved in gene regulation (reviewed by Varshney et al. 2005). For example, the length polymorphism of a (CT) n microsatellite in the 5′UTR of the waxy gene in rice proved to be correlated with amylose content (Ayers et al. 1997). In agreement with these findings, (AG) n repeats in the 5′UTR of globe artichoke ESTs were preferentially associated with genes involved in transcription, nucleic acid metabolism and the regulation of gene expression (Scaglione et al. 2009).
Polymorphism of pineapple EST-SSRs in Ananas
Because of their association with transcribed and hence conserved regions of the genome, EST-SSR markers are often assumed to be less polymorphic than their genomic counterparts (reviewed by Varshney et al. 2005; Ellis and Burke 2007). Comparative studies using both types of markers showed that this is not necessarily the case, and equivalent levels of polymorphism revealed by genomic versus EST-SSRs were, e.g., reported for kiwifruit (Fraser et al. 2004) and sunflower (Pashley et al. 2006). In the present study, all 18 tested EST-SSR markers were reasonably polymorphic with two to six alleles and an average H e over all loci of 0.58 among 12 A. comosus accessions (see Fig. 1). Although a comparative assay was not the intention of our investigation, these values compare quite favorably with the two to four alleles, and an average H e of 0.509 revealed by 19 genomic SSRs among 180 individuals of A. comosus grown in six fields in Malaysia (Kinsuat and Kumar 2007). We are therefore confident that the level of polymorphism exhibited by EST-SSR markers in pineapple will be sufficient for many putative applications, including genetic mapping (Carlier et al. 2004, 2011).
The quality of the banding patterns was high throughout, which appears to be a general property of EST-SSR markers (Varshney et al. 2005; Pashley et al. 2006). Whereas single or double bands were observed in all Ananas species, three or four bands were occasionally produced in Pseudananas sagenarius, in accordance with the presumed tetraploid status of this species (2n = 4× = 100; Collins 1960). Three accessions of A. nanus, one pair of accessions of A. bracteatus and two pairs of accessions of A. comosus each had identical multilocus genotypes, which can be attributed to vegetative propagation and/or exchange between Botanical Gardens. One A. comosus individual (BT_75) was probably mislabeled, as it harbored the same genotype as two A. bracteatus individuals (BT_73 and BT_80) and formed a group in the PCoA with most other A. bracteatus accessions (Fig. 2). Heterozygosity values became slightly lower when this aberrant accession was pruned from the A. comosus data set (average H e over all loci of 0.56; average H o over all loci of 0.47). Loosely defined groups were formed by all A. nanus accessions and most A. comosus accessions, respectively, but groups were not distinct and no obvious relationships between species became apparent in the PCoA.
The taxonomy of Ananas and Pseudananas is not fully settled yet. In their comprehensive monograph on the Bromeliaceae, Smith and Downs (1979) recognized seven species within the genus Ananas (i.e., A. ananassoides, A. bracteatus, A. comosus, A. fritzmuelleri, A. lucidus, A. nanus, and A. parguazensis), with the monotypic, tetraploid P. sagenarius being the closest relative. All seven Ananas species are interfertile (Collins 1960), and clear distinction between taxa has been difficult because of environmental plasticity of morphological characters. A more or less continuous variation among the seven Ananas species also became apparent with molecular data sets, including nuclear and chloroplast RFLPs (Duval et al. 2001; 2003). In an attempt to simplify the taxonomy, Coppens d′Eeckenbrugge and Leal (2003) therefore proposed to have the seven species of Ananas downgraded to the level of five botanical varieties of A. comosus. Under their new classification, the tetraploid P. sagenarius would then become A. macrodontes Morren. Our PCoA analysis of microsatellite data seems to be in agreement with the view that there is no clear separation between the seven species of Ananas, but a much broader sampling would of course be required to substantiate these findings.
Transportability of pineapple EST-SSR markers across Bromeliaceae
The development of nuclear microsatellite marker sets by enrichment and cloning is a tedious and costly affair (Zane et al. 2002). In silico mining is a feasible and attractive alternative, but sufficiently large EST databases are only available for a limited number of plant species, mainly for crop and/or model species. Cross-species transfer of microsatellite markers therefore remains an important issue (Peakall et al. 1998; Barbará et al. 2007), and a major purpose of our study was the development of genic SSR markers for population genetic studies in heterologous species of Bromeliaceae (see Ellis and Burke 2007). On a theoretical basis, EST-SSRs are expected to be more transferable across taxa than genomic SSRs, because they originate from more conserved coding regions. In accordance with this assumption, the portability of genomic SSRs is usually restricted to congeners (e.g., in Glycine; Peakall et al. 1998), whereas EST-SSRs are frequently transportable across genera (reviewed by Ellis and Burke 2007) and sometimes also across subfamilies (Decroocq et al. 2003; Zhang et al. 2005; Heesacker et al. 2008) or even families (Gupta and Prasad 2009).
The results obtained in the present study fit well into this picture. Almost all of the pineapple EST-SSR markers showed good transferability within the genus Ananas and a remarkable proportion of primer pairs also generated PCR products of expected size in distantly related species from various genera and subfamilies of the Bromeliaceae. In agreement with taxonomic expectations, the average percentage of successfully amplifying primer pairs was the highest (74.8%) in subfamily Bromelioideae (to which Ananas belongs), and decreased with increasing evolutionary distance down to an average of 60.0% in Puyoideae, 51.6% in Pitcairnioideae, 42.6% in Hechtioideae, 27.3% in Tillandsioideae and 19.5% in the most distant subfamily Brocchinioideae (Table 5).
A marker that is coined “transferable” among species should of course be present and polymorphic in the target species, which is quite frequently the case in congeners (Ellis and Burke 2007). In the present study, marker transferability was also tested across a larger taxonomic distance by genotyping nine individuals of D. brevifolia (subfamily Pitcairnioideae) with the 18 EST-SSR markers developed from pineapple. Eight loci proved to be polymorphic in the target species with allele numbers ranging from three to five, and average expected and observed heterozygosity values of 0.51 and 0.42, respectively. Obviously, the extent of marker polymorphism will have to be tested in each individual target species before embarking onto any large-scale study.
Conclusions
The EST-SSR markers developed in the present study increase the numbers of SSR markers that are currently available for Bromeliaceae several fold. Many of them show good cross-species amplification and are suitable marker candidates for population genetic analyses, and other genotyping applications in a wide range of bromeliad species from various subfamilies. Within Ananas, EST-SSRs could be exploited to assess the extent of interspecific gene flow and problems of species delimitation, the origin of putative hybrids, and the origin and parent(s) of the tetraploid P. sagenarius. Finally, functional markers like EST-SSRs are supposed to play a pivotal role in the context of marker-assisted selection and genetic mapping projects in pineapple (Carlier et al. 2011).
References
Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet 114:359–372. doi:10.1007/s00122-006-0440-x
Andersen JR, Lübberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560. doi:10.1016/j.tplants.2003.09.010
Ayers NM, McClung AM, Larkin PD, Bligh HFJ, Jones CA, Park WD (1997) Microsatellites and a single nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germplasm. Theor Appl Genet 94:773–781. doi:10.1007/s001220050477
Barbará T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C (2007) Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol 16:3759–3767. doi:10.1111/j.1365-294X.2007.03439.x
Benzing DH (2000) Bromeliaceae: profile of an adaptive radiation. Cambridge University, Cambridge
Boneh L, Kuperus P, van Tienderen PH (2003) Microsatellites in the bromeliads Tillandsia fasciculata and Guzmania monostachya. Mol Ecol Notes 3:302–303. doi:10.1046/j.1471-8286.2003.00432.x
Botella JR, Smith M (2008) Genomics of pineapple, crowning the king of tropical fruits. In: Moore PH, Ming R (eds) Plant genetics and genomics vol. 1: genomics of tropical crop plants. Springer, New York, pp 441–450. doi:10.1007/978-0-387-71219-2_18
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349
Carlier JD, Reis A, Duval MF, Coppens d`Eeckenbrugge G, Leitão JM (2004) Genetic maps of RAPD, AFLP and ISSR markers in Ananas bracteatus and A. comosus using the pseudo-testcross strategy. Plant Breed 123:186–192. doi:10.1046/j.1439-0523.2003.00924.x
Carlier JD, Horta Sousa N, Espírito Santo T, Coppens d`Eeckenbrugge G, Leitão JM (2011) A genetic map of pineapple (Ananas comosus (L.) Merr.) including SCAR, CAPS, SSR and EST-SSR markers. Mol Breed. doi 10.1007/s11032-010-9543-9
Chen C, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257. doi:10.1007/s00122-006-0226-1
Collins JL (1960) The pineapple. Interscience Publishers, New York
Coppens d′Eeckenbrugge G, Leal F (2003) Morphology, anatomy and taxonomy. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 13–32
Decroocq V, Favé MG, Hagen L, Bordenave L, Decroocq S (2003) Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet 106:912–922. doi:10.1007/s00122-002-1158-z
Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v5.0, available from http://www.geneious.com [accessed 3 December 2010]
Duval M-F, Noyer JL, Perrier X, Coppens d′Eckenbrugge G, Hamon P (2001) Molecular diversity in pineapple assessed by RFLP markers. Theor Appl Genet 102:83–90
Duval MF, Buso GS, Ferreira FR, Noyer JL, Coppens d′Eeckenbrugge G, Hamon P, Ferreira ME (2003) Relationships in Ananas and other related genera using chloroplast DNA restriction site variation. Genome 46:990–1004. doi:10.1139/G03-074
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132. doi:10.1038/sj.hdy.6801001
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.1: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fraser LG, Harvey CF, Crowhurst RN, De Silva HN (2004) EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108:1010–1016. doi:10.1007/s00122-003-1517-4
Gupta S, Prasad M (2009) Development and characterization of genic SSR markers in Medicago truncatula and their transferability in leguminous and non-leguminous species. Genome 52:761–771. doi:10.1139/G09-051
Heesacker A, Kishore VK, Gao W, Tang S, Kolkman JM, Gingle A, Matvienko M, Kozik A, Michelmore RM, Lai Z, Rieseberg LH, Knapp SJ (2008) SSRs and INDELs mined from the sunflower EST database: abundance, polymorphisms, and cross-taxa utility. Theor Appl Genet 117:1021–1029. doi:10.1007/s00122-008-0841-0
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rize, sorghum and wheat. Plant Mol Biol 48:501–510. doi:10.1139/G04-055
Kinsuat MJ, Kumar SV (2007) Polymorphic microsatellite and cryptic simple repeat sequence markers in pineapples (Ananas comosus var. comosus). Mol Ecol Notes 7:1032–1035. doi:10.1111/j.1471-8286.2007.01764.x
Kofler R, Schlötterer C, Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685. doi:10.1093/bioinformatics/btm157
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48:985–998. doi:10.1139/G05-060
Luther HE (2008) An alphabetical list of bromeliad binomials, 11th edn. Bromeliad Society International, Mary Selby Botanical Gardens, Sarasota
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80. doi:10.1101/gr.10.1.72
Min XJ, Butler G, Storms R, Tsang A (2005) OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res 33:W677–W680. doi:10.1093/nar/gki394
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nature Genet 30:194–200. doi:10.1038/ng822
Moyle RL, Crowe ML, Ripi-Koia J, Fairbairn DJ, Botella JR (2005) PineappleDB: an online pineapple bioinformatics resource. BMC Plant Biol 5:21. doi:10.1186/1471-2229-5-21
Paggi GM, Palma-Silva T, Bered F, Cidade FW, Sousa ACB, Souza AP, Wendt T, Lexer C (2008) Isolation and characterization of microsatellite loci in Pitcairnia albiflos (Bromeliaceae), an endemic bromeliad from the Atlantic Rainforest, and cross-amplification in other species. Mol Ecol Resour 8:980–982. doi:10.1111/j.1755-0998.2008.02126.x
Palma-Silva C, Cavallari MM, Barbará T, Lexer C, Gimenes MA, Bered F, Bodanese-Zanettini MH (2007) A set of polymorphic microsatellite loci for Vriesea gigantea and Alcantarea imperialis (Bromeliaceae) and cross-amplification in other bromeliad species. Mol Ecol Notes 7:654–657. doi:10.1111/j.1471-8286.2006.01665.x
Pashley CH, Ellis JR, McCauley DE, Burke JM (2006) EST databases as a source for molecular markers: lessons from Helianthus. J Hered 97:381–388. doi:10.1093/jhered/es1013
Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A (1998) Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: implications for the transferability of SSRs in plants. Mol Biol Evol 15:1275–1287
Pinto LR, Oliveira KM, Ulian EC, Garcia AAF, de Souza AP (2004) Survey in the sugarcane expressed sequence tag database (SUCEST) for simple sequence repeats. Genome 47:795–804. doi:10.1139/G04-055
Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, Hamon P (2006) SSR mining in coffee tree EST databases: potential use of EST-SSRs as markers for the Coffea genus. Mol Gen Genomics 276:436–449. doi:10.1007/s00438-006-0153-5
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222. doi:10.1016/1360-1385(96)86898-1
Rohlf FJ (2000) NTSYSpc: numerical taxonomy and multivariate analysis system, ver. 2.1. Applied Biostatistic Inc, Port Jefferson
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: Methods in molecular biology. Humana Press, Totowa, pp 365–386
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sarthou C, Boisselier-Dubayle MC, Lambourdière J, Samadi S (2003) Polymorphic microsatellites for the study of fragmented populations of Pitcairnia geyskesii L.B. Smith (Bromeliaceae), a specific saxicolous species of inselbergs in French Guiana. Mol Ecol Notes 3:221–223. doi:10.1046/j.1471-8286.2003.00404.x
Scaglione D, Acuadro A, Portis E, Taylor CA, Lanteri S, Knapp SJ (2009) Ontology and diversity of transcript-associated microsatellites mined from a globe artichoke EST database. BMC Genomics 10:454. doi:10.1186/1471-2164-10-454
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726. doi:10.1007/s001220051344
Smith L, Downs RJ (1979) Bromelioideae (Bromeliaceae). Monograph 14, part 3, Flora Neotropica. New York Botanical Garden, New York
Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W (2003) How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol 12:1339–1348. doi:10.1046/j.1365-294X.2003.01825.x
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422. doi:10.1007/s00122-002-1031-0
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55. doi:10.1016/j.tibtech.2004.11.005
Weising K, Nybom H, Wolff K, Kahl G (2005) DNA fingerprinting in plants: principles, methods, and applications, 2nd edn. CRC Press, Taylor & Francis Group, Boca Raton
You FM, Huo N, Gu YQ, Luo M-C, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD (2008) BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9:253. doi:10.1186/1471-2105-9-253
Yu J-K, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME (2004) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47:805–818. doi:10.1139/G04-057
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation; a review. Mol Ecol 11:1–16. doi:10.1046/j.0962-1083.2001.01418.x
Zhang LY, Bernard M, Leroy P, Feuillet C, Sourdille P (2005) High transferability of bread wheat EST-derived SSRs to other cereals. Theor Appl Genet 111:677–687. doi:10.1007/s00122-005-2041-5
Acknowledgments
The plant material used in the present study was kindly provided by the Botanical Gardens of Berlin, Frankfurt, Göttingen, Hannover, Heidelberg, Kassel, Witzenhausen (all Germany) and Vienna (Austria), Prof. Dr. Ana Maria Benko Iseppon (University of Pernambuco, Recife, Brazil), Dr. Rafael B. Louzada (University of Sao Paulo, Brazil), Dipl.-Biol. Daniele Silvestro and Prof. Dr. Georg Zizka (Senckenberg Research Institute and Biodiversity and Climate Research Centre [BiK-F], Frankfurt), Dipl.-Biol. Nicole Schütz and Dipl.-Biol. Natascha Wagner (University of Kassel). We thank the ZFF (Zentrale Forschungsförderung) of the University of Kassel for funding the project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Nybom.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wöhrmann, T., Weising, K. In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across Bromeliaceae. Theor Appl Genet 123, 635–647 (2011). https://doi.org/10.1007/s00122-011-1613-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1613-9