Abstract
Pineapple (Ananas comosus (L.) Merrill) is the second most important tropical fruit in term of international trade. The availability of whole genomic sequences and expressed sequence tags (ESTs) offers an opportunity to identify and characterize microsatellite or simple sequence repeat (SSR) markers in pineapple. A total of 278,245 SSRs and 41,962 SSRs with an overall density of 728.57 SSRs/Mb and 619.37 SSRs/Mb were mined from genomic and ESTs sequences, respectively. 5′-untranslated regions (5′-UTRs) had the greatest amount of SSRs, 3.6–5.2 fold higher SSR density than other regions. For repeat length, 12 bp was the predominant repeat length in both assembled genome and ESTs. Class I SSRs were underrepresented compared with class II SSRs. For motif length, dinucleotide repeats were the most abundant in genomic sequences, whereas trinucleotides were the most common motif in ESTs. Tri- and hexanucleotides of total SSRs were more prevalent in ESTs than in the whole genome. The SSR frequency decreased dramatically as repeat times increased. AT was the most frequent single motif across the entire genome while AG was the most abundant motif in ESTs. Across six examined plant species, the pineapple genome displayed the highest density, substantially more than the second-place cucumber. Annotation and expression analyses were also conducted for genes containing SSRs. This thorough analysis of SSR markers in pineapple provided valuable information on the frequency and distribution of SSRs in the pineapple genome. This genomic resource will expedite genomic research and pineapple improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pineapple (Ananas comosus (L.) Merrill) is the second most important tropical fruit after banana in term of international trade. Although it originated in South America, pineapple is currently cultivated on 1 million hectares in 85 coutries and the global production amounted to 24.79 million tons in 2013, having nearly doubled over the last decade (http://faostat.fao.org). Pineapple is consumed or served fresh, cooked, juiced and canned. In addition to its exceptionally palatable and juicy fruit, it has outstanding nutritional and medicinal properties. The pineapple species is a perennial monocot with a diploid number of 50 chromosomes (2n = 2× = 50); it belongs to the family Bromeliaceae in the order Bromeliales (Sharma and Ghosh 1971). Pineapple is a reasonably close relative of grasses. The pineapple plant uses the Crassulacean Acid Metabolism (CAM) photosynthetic pathway, an evolutionary adaptation that occurred in some plants in response to arid conditions. CAM results in increased photosynthetic efficiency while preventing excessive water loss. Pineapple is a non-climacteric fruit that lacks the ethylene-associated respiratory peak during ripening. In contrast to the self-compatibility of wild species, cultivated pineapple is self-incompatible, thus containing high level of heterozygosity. These unique attributes make pineapple an exceptionally promising system for genetic and genomic studies to address many biological questions such as obligate CAM photosynthesis, parthenocarpic fruit development and non-climacteric ripening process, and the molecular basis of self-incompatibility in monocots as well as offering evidence for cereal genome evolution.
Microsatellites, also referred to as short tandem repeats (STRs) or simple sequence repeats (SSRs), are short tandem repetitive DNA sequences of 2–6 base pairs that are ubiquitously present in the genomes of prokaryotes and eukaryotes (Gur-Arie et al. 2000; Powell et al. 1996; Tautz 1989). DNA replication slipped-strand mispairing (Levinson and Gutman 1987; Tachida and Iizuka 1992) and recombination between DNA strands (Harding et al. 1992) can result in microsatellite instability, a ubiquitous phenomenon in the origin and evolution of microsatellites. SSRs can be categorized into class I (hypervariable markers), and class II (potentially variable markers) based on the length of the repeat motif, and consist of SSRs ≥20 bp and SSRs ≥12 bp < 20 bp, respectively (Temnykh et al. 2001). Owing to their abundance, high polymorphism in the number of repeats, multi-allelic nature, codominant inheritance and low-cost of rapid PCR-based tests, study and detection of microsatellites have found numerous applications allowing important advances in many fields, including the analysis of phylogenetic or genetic diversity (Castillo et al. 2008; Feng et al. 2013; Mian et al. 2005; Sharma et al. 2008), genetic mapping (Chen et al. 2015; Gailing et al. 2013; Yu et al. 2011), marker-assisted selection (MAS) (Steele et al. 2006), population genetics (Innan et al. 1997), comparative genomics (Garza et al. 1995) and gene localization (Molnar et al. 2003).
The computational capabilities of massive genomic sequences along with the completion of sequencing of many genomes have provided insights into genomic distribution, putative functions and mutational mechanisms of microsatellites (Li et al. 2002). The density, distribution and motif composition of SSRs vary unevenly and non-randomly across species as well as among different genomic fractions, i.e. introns, exons, coding sequences (CDS), untranslated regions (UTRs) and intergenic regions (Biswas et al. 2014; Cavagnaro et al. 2010; Liu et al. 2013; Luo et al. 2015; Vásquez and López 2014). A higher overall SSR density was observed in intergenic regions compared genic regions in almost all taxa. SSRs are significantly more frequent in non-coding regions of the genome than in the coding region except for tri- and hexanucleotides (Zhang et al. 2004). However, most SSRs in noncoding regions are close to or linked to expressed genes thus represent functional markers that are of particular interest (Morgante et al. 2002). SSRs from non-coding regions are associated with anonymous genomic sequences and can therefore provide sufficient polymorphisms to discriminate between closely related species or conduct genome comparisons. Expressed sequence tag SSRs (EST-SSRs) are associated with functional genes and usually more conserved in a wide range of species due to the higher selection pressure. In plants, 1.5–7.5 % of ESTs consist of SSRs (Kantety et al. 2002; Thiel et al. 2003). Compared to SSRs from non-coding regions, EST-SSRs are more transferable among related germplasm, enabling genome evolution and comparative mapping studies. Estimates suggest that class II SSRs have a significantly higher density than class I SSRs in both genomic sequences and ESTs in a wide range of plant species (Kantety et al. 2002; Wang et al. 2008). With the rapid increase of the sequencing of cDNA clones and the current availability of many reference genomes, more EST libraries and databases representing the vast majority of the information content of the genome have been established for many organisms, thus providing an avenue for SSR mining in the expressed transcripts.
In pineapple, a number of morphological, biochemical and nucleic acid-based markers such as isozymes, RFLPs (restriction fragment length polymorphisms), AFLPs (amplified fragment length polymorphisms) or RAPDs (randomly amplified polymorphic DNAs) have been employed to characterize pineapple germplasm (Aradhya et al. 1994; Carlier et al. 2012; de Sousa et al. 2013; DeWald et al. 1992; Duval et al. 2001; Kato et al. 2005; Paz et al. 2012; Sripaoraya et al. 2001). However, most genetic markers have failed to provide high-resolution genetic maps in pineapple. Among others, the recently developed user-friendly microsatellite markers have enjoyed much greater success in the field of pineapple genetics due to its rather high polymorphism and genome specificity (Feng et al. 2013; Kinsuat and Kumar 2007; Rodríguez et al. 2013; Shoda et al. 2012). The latest integrated genetic map of pineapple assembles 741 markers including 25 SSRs and 12 EST-SSRs in 28 linkage groups, spanning 2113 cM and covering approximately 86 % of the genome (de Sousa et al. 2013). Those microsatellites are so far the largest set mapped in this species. The most recently reported efforts in the search of pineapple SSRs identified 94 SSRs from pineapple genomic libraries, and 1110 SSRs in 5659 pineapple ESTs (Feng et al. 2013). The small number of SSRs previously found and mapped could be due to the limited number of genome sequences reported at that time. The number of robust and informative SSR markers genome wide and in ESTs publicly available for pineapple is still insufficient for some studies, hindering the development of diversity and phylogenetic studies, as well as the high-resolution genetic maps, which are instrumental for marker-assisted selection, positional gene cloning and comparative mapping. More polymorphic SSR markers and a denser map are needed. Recently, the genome of cultivated pineapple variety F153 was sequenced and assembled using several whole genome sequencing approaches, generating a contig N50 of 126.5 kb and a scaffold N50 of 11.8 Mb (Ming et al. 2015). This genome spans 382 Mb, 72.6 % of the estimated 526 Mb pineapple genome. Based on extensive pineapple RNA-Seq data from the pineapple genome project, a substantial number of novel transcripts that significantly complement current EST databases were identified and collected. The availability of the pineapple draft genome sequence and a large collection of EST sequences are now providing an opportunity for a large scale development of microsatellite markers, which would facilitate the pineapple research community and expedite breeding progress. The present study was conducted for in silico systematic and genome-wide characterization of microsatellite sequences in the pineapple genome for crop improvement. In this study, we 1) mined microsatellites throughout pineapple assembled genome and ESTs, 2) investigated the distribution, density, repeat and motif structure of SSRs in different genomic fractions as well as transcripts, 3) performed comparative analysis among pineapple and other plant species using SSRs from genomic and EST sequence datasets. 4) In addition, to gain some insight into the putative function of SSRs present in the gene regions, Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation and expression pattern analyses were carried out for genes containing SSRs.
Results
SSR Classes and Density Distribution in Different Genome Regions and ESTs
In this study, the class and distribution of pineapple SSRs with a minimum repeat length of 12 bp and a unit size of 2 to 6 bp were analyzed. A total of 278,245 perfect SSRs were identified from 381.91 Mb from the most recently assembled pineapple genome with an overall density of 728.57 SSRs/Mb (i.e., one SSR per 1.37 kb of sequence, excluding mononucleotide SSRs), of which 82,261 (29.6 %) were defined as class I (≥20 bp) SSRs with a density of 215.4 SSRs/Mb, and 195,984 (70.4 %) as class II (≥12 bp and <20 bp) with a higher density of 513.17 SSRs/Mb on average (Table 1). An SSR search in coding, UTRs, intron and exon sequences was also performed to determine the distribution of SSRs on a genic scale. Following the search of SSRs for each of these regions, we found that the densities of SSRs were significantly different in coding and noncoding regions (Table 1 and Fig. 1). The densities of SSRs in noncoding regions were 2839.91 SSRs/Mb for 5′-UTR, 545.88 SSRs/Mb for 3′-UTR, 589.12 SSRs/Mb for introns, and 783.98 SSRs/Mb for intergenic regions. The abundance of SSRs in noncoding regions was much higher than that of coding regions. Only 19,727 SSRs (7 %) were located in the CDS, while 93 % were located in noncoding regions. Class II SSRs were substantially more prevalent than class I SSRs on both genomic and genic scale. The average density of SSRs in CDS (592 SSRs/Mb) was lower than the genome taken as a whole (Fig. 1). In the genic region, 3′-UTR regions were found to have the lowest density of SSRs, whereas 5′-UTR regions contained the greatest amount of SSRs. For instance, we identified 8282 (1718.96 SSRs/Mb) SSRs in 5′-UTRs and only up to 3098 (396.63 SSRs/Mb) SSRs in 3′-UTR regions. 5′-UTR sequences were observed to have between 3.6- to 5.2-fold higher SSR density than other regions and approximately four-fold higher density than in the whole genome. It appeared that the SSRs were denser in the intergenic region (783.98 SSRs/Mb) than in its genic region counterpart (633.7 SSRs/Mb) and genome-wide region. The genome-wide GC content (27.7 %) was lower than the genic GC content (36.4 %) (Table 1).
From the 53.46 Mb EST sequences, 41,962 SSRs were mined with an overall density of 619.37 SSRs/Mb, including 4339 (10.3 %) class I and 37,623(89.7 %) class II SSRs with densities of 64.05 SSRs/Mb and 555.33 SSRs/Mb, respectively. From these two sequence sources, the assembled genome and EST sequences, EST sequences had lower SSR density and a much higher GC content (53.5 %). Class I SSRs had a density of 64.05–215.4 per Mb among genomic, genic and EST sequences, while class II SSRs occurred at a significantly higher density of 446.82–555.33 per Mb.
SSR Repeat Length and Motif Length Frequency in Different Genome Regions and ESTs
The repeat length ranged in size from 12 to 1439 bp in the whole genome and from 12 to 25 bp in EST sequence datasets. 12 bp was the predominant repeat length both in assembled genome and EST, accounting for 37.3 % and 43.3 % of the total SSRs (Fig. 2a), 53 % and 48.3 % of the class II SSRs respectively (Fig. 2b). The second most frequent repeat length was 18 bp. The trend was also similar between the assembled genome and EST, representing 10.5 % and 20.2 % of the total SSRs and 14.9 % and 22.6 % of the class II SSRs. Class I SSRs were extremely underrepresented compared to their class II SSRs counterparts. Within the class I SSRs grouping, the 20 bp repeats with 13,043 SSRs (15.9 %) were the most common SSRsfollowed by 24 bp with 12,369 SSRs (15.0 %) repeats with scarce variance in whole genome sequences (Fig. 2b). In EST sequences, by contrast, the percentage of SSRs with 20 bp repeats (43.2 %) remained the most abundant but followed by 21 bp repeats instead of 24 bp repeats (Fig. 2c).
Regarding the SSR motif lengths, dinucleotide repeats of total SSRs and of each class outnumbered all of the other motif lengths and were the most abundant on a genome wide scale (Fig. 3). The observed SSR frequency decrease was not strictly correlated with the number of nucleotides. For example, in genomic regions, hexa- motifs were slightly more abundant than penta- motifs both in total and of each class of SSRs. In class I SSRs, the number of dimers was significantly higher than other motifs with a high percentage of 62.1 %, followed by tri- motif with 11.4 %, hepta- with 10.9 %, tetra- with 5.1 % (Fig. 3 and Supplementary Table 1). Hepta- to decanucleotide repeats of class II SSRs were totally absent from both sources of genome sequences. In gene models, the density of SSRs differed among exons, CDS, UTRs and introns (Fig. 1). CDS regions differed from other genic regions by their high trinucleotide SSRs percentage (78.4 % vs. 28.7 %, 29.9 %, and 17.7 % in 5′-UTR, 3′-UTR, and intron, respectively) and hexanucleotide SSRs percentage (13.1 % vs. 7.8 %, 6.2 % and 5.2 % in 5′-UTR, 3′-UTR, and intron, respectively) (Fig. 3a and Supplementary Table 1). Across the sequence sources, tri- and hexa- repeats of total SSRs and class II were more prevalent in ESTs than in the whole genome while other types of repeats were slightly reduced in the EST sequences (Fig. 3a and c). In class I SSRs, tetra- and penta- followed the same trend in addition to tri- and hexanucleotide repeats (Fig. 3b). For example, the tri- repeats of total SSRs and of each class were 1.9 to 2.5 times more frequent in ESTs than in whole genome sequences, making these the most dominant motif in class II and in the total SSRs, and the second most dominant motif in class I after dimers. The relative abundance of EST-related motif frequency (tri- and hexa-) was therefore found to be due to their abundance in genomic protein coding regions.
Interspecific Comparison of SSR Distribution in Genomic and EST Sequences
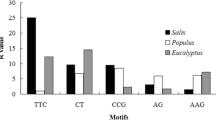
The distribution of SSRs in pineapple genomic sequences and in five other selected species with comparably large genomes was analyzed and summarized in Table 2. We analyzed the genomes from three monocots (Ananas comosus, Oryza sativa, Sorghum bicolor) and three dicots (Arabidopsis thaliana, Cucumis sativus L., Vitis vinifera). These six genomes were studied by applying the same criteria for the identification of SSRs that is a minimum repeat length of 12 bp and a unit size of 2 to 6 bp. The six analyzed species showed a large variance in their microsatellite density ranging from 315.5 SSRs/Mb to 728.57 SSRs/Mb (Table 2). It was an unexpected find that pineapple had the highest density among the species analyzed, significantly ahead of second-place cucumber (536.7 SSRs/Mb). Sorghum was found to have the lowest abundance of SSRs with a density of 315.5 SSRs/Mb, comparable to that of Arabidopsis (364.1 SSRs/Mb). From Table 2, the most ubiquitous SSR motif length were dinucleotides from pineapple genomic sequences, representing 46.8 % of the total SSRs, followed by tri- (23.6 %) and tetranucleotides (18 %). Penta- and hexanucleotides were the least common motif lengths, together covering less than 12 % of all SSRs. Across the six species, the distribution of motif length in the monocot pineapple genome was markedly distinguished from the others, especially from those of dicots cucumber and grapevine, for which tetranucleotides were the most frequent motif type. Trinucleotides were observed to be the most prevalent motif length in the other two monocots, rice and sorghum, together with one dicot, Arabidopsis. Tetranucleotides prevailed in most (two out of three) of the dicot species analyzed. Comparatively, the density of dinucleotide repeats in pineapple was much higher than in the other species. Apart from the dominant motif length, no other significant differences were evident between monocots and dicots.
The overall microsatellite density in pineapple ESTs was lower than that from genomic sequences. Compared to those from genomic sequences, frequencies of tri- and hexanucleotides were much higher in transcripts (51.6 % and 9.7 % of total SSRs), whilst dinucleotide type were greatly decreased (23.1 % in ESTs) (Table 2). In other species, distributions of microsatellites in the expressed fraction of the genomes revealed the similar tendency that tri- and hexanucleotides were relatively more abundant, and dinucleotides were rarer than them at the genomic level. Trinucleotides were by far the most frequent type followed in order by di-, tetra-, hexa- and pentanucleotides, in abundance. In all other species analyzed the relative abundance of SSR types was only slightly different from but rather consistent with pineapple: trinucleotieds were the most abundant, followed by tetra-, di-, hexa- and pentanucleotides. Although there was a trend of trinucleotide frequency being highest across species, the absolute density of trinucleotides varied by nearly 4-fold in magnitude, from 108.4 in grapevine to 485.7 SSRs/Mb in rice. The trinucleotides density in monocots was above 300 SSRs/Mb, while that of dicots was under 250 SSRs/Mb.
Distribution of SSR Motifs in Genome Wide and ESTsequences
We examined the frequencies of pineapple SSR motifs with regard to the repeat times at the genomic and transcript levels (Fig. 4). Consistently, the SSR frequency decreased dramatically as the number of repeat units increased for all analyzed five motif lengths, especially for longer ones (tetra- to hexanucleotides) which showed the most dramatic drop in frequency. As a result, the mean repeat number in dinucleotides was about 1.7–2.2 times the number of repeat units in trinucleotides, and it was nearly 2.5–3.4 times greater than the number of tetra- to hexanucleotides (Table 2). In the genomic region, the cumulative sequence length of dinucleotides was 2906 kb, the longest one compared to any other motif length type. Therefore, dinucleotides (340.58 SSRs/Mb) not only occurred most frequently in the pineapple assembled genome, but also accounted for the greatest contribution to the genome fraction occupied by SSRs (di- to hexanucleotides) due to their highest number of repeat units. In transcript sequences, although dinucleotides had a higher number of repeat units (7.9) than trinucleotides (4.7), trinucleotides, due to their higher density (319.44 SSRs/Mb), made a greater contribution to the transcript fraction occupied by SSRs: the cumulative sequence length of di- and trinucleotides was 152 kb and 307 kb, respectively. The longest SSR in the pineapple genome appeared in dinucleotide repeat patterns, which was (CA)1439, reaching up to 2878 bp, followed by the (TC)1438. Longer repeats in the genome generally tend to have higher mutation rates, which is associated with a high frequency of polymorphisms (Karaoglu et al. 2005). In transcript sequences, tetranucleotide (ATCC)24 at 96 bp and hexanucleotide (CTTCTC)14 at 84 bp were the longest and the second longest SSRs. The frequency of repeat motif varied for each length class from different sequence sources. A more detailed examination of the frequencies of individual repeat motifs is shown in Tables 3 and 4 and in Fig. 5 and Supplementary Table 2.
Dinucleotide Motifs
The analysis of AC, AG, AT and CG repeats revealed that the dominance of dinucleotides was attributed to an overrepresentation of AT motifs in the pineapple whole genome sequences (Table 3), while GC was rather rare, accounting for only 0.15 % of all SSRs. AT was also the overall most common single motif across the entire genome, representing 24.3 % of the total SSRs, followed by the dinucleotide motif AG (20 %). In contrast, the frequency of AT repeat units (4 %) was dramatically lower in the pineapple transcript sequences, with the AG motif (17 %) being the most abundant dinucleotide. In general, the transcript sequences showed a dominance of AT-CG balanced dinucleotide repeats, whereas whole genome sequences displayed a prevalence of AT-rich dinucleotides (Fig. 5). AC and CG repeats were the most rare dinucleotides from both sequence sources.
Trinucleotide Motifs
In terms of trinucleotide motifs, there was prevalence of AAT and AAG motifs represented by approximately 6.4 % and 5.5 % of all SSRs respectively in the entire genome, while the CCG motif was the predominant motif in ESTs, contributing to 14.3 % of the discovered SSR loci (Tables 3 and 4). The abundance of CCG in ESTs was also found in the monocots rice and sorghum. Conversely, an opposite distribution was observed in dicots, where CCG was the most rare trinucleotide in both genomic and EST datasets (Cavagnaro et al. 2010). ACT was the most infrequent trinucleotide repeat pattern in both pineapple genomic and EST datasets, similarly representing less than 1 % from both sources. A strong bias in the distribution of trinucleotides towards AT-rich motifs was found in the genomic sequences, and towards GC-rich motifs in ESTs. Overall, AT-rich motifs accounted for 14.24 % of the identified SSRs, nearly 1.5 times the number of GC-rich motifs (9.4 %) in the assembled genome, whereas GC-rich motifs (36.1 %) were more than twice the AT-rich ones (15.5 %) in ESTs.
Tetranucleotide Motifs
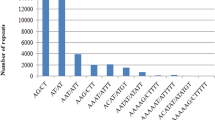
A clear predominance of AT-rich motifs and underrepresentation of GC-rich motifs was observed in genomic and EST sequences (Fig. 5). Compared to ESTs, the shift from AT-rich to GC-rich motifs in genomic sequences was gentler. Overall AT-rich motifs composed up to 80 % in genomic sequences and 62.9 % in ESTs of all tetramer repeats, whereas the GC-rich repeats were the least common with relative frequencies of 4.4 % and 12.7 %, respectively (Fig. 5, Supplementary Table 2a and b). The most prevalent tetramer SSRs in pineapple genomic sequences was ATTT, covering 42.2 % of tetranucleotide repeats, followed by AATT for 18.4 %, AAAG for 14.9 %, AGAT for 9.6 % (Supplementary Table 2a). In relative terms, the GC-rich motifs AGGC, ACGG, ACCG, and AGCC were, in that order, the least prevalent tetramers. A similar distribution was observed in their ESTs counterparts, for which ATTT prevailed, whereas AGGC was the scarcest (Supplementary Table 2b).
Pentanucleotide and Hexanucleotide Motifs
AT-rich motifs were predominant among pentanucleotide repeats in SSRs from the entire assembled genome and ESTs, covering 85 % and 72 %, respectively (Fig. 5, Supplementary Table 2a and b). AAAAT and AAAAG were the most abundant repeats in both genomic and EST sequences, outnumbering the next most common repeats AATAT in genomic sequences or AAAAC in ESTs, by at least two-fold.
As for pentanucleotide repeats, AT-rich hexanucleotides were prominent in the pineapple genomic data (~53 % of total hexanucleotides), such as AAATTT, AAAAAG, AAAAAT, and AAAATT, followed by GC-rich (~28 %) and AT/GC balanced (~19 %) (Fig. 5 and Supplementary Table 2a). On the contrary, there was a high prevalence of GC-rich repeats represented by approximately 53 % of all hexamers in transcript data (Fig. 5 and Supplementary Table 2b), followed by AT/GC balanced (~29 %) and AT-rich (~18 %). Both pineapple genomic and ESTs sequences had a much higher overall density of hexanucleotide motifs compared to other analyzed species, and nearly 3.4–4.5 fold the density found in Arabidopsis (Table 2).
Annotation and Expression Patterns of SSR-Containing Genes
Pineapples genes can be classified as SSR-containing genes and no-SSR genes. In total, 21,631 genes (72.7 %) containing one or more SSRs were identified in this study, only 8115 genes (27.3 %) containing no SSR (Fig. 6a). Of all SSR-containing genes, 5878 genes contained only one SSR and 15,753 genes contained more than one. The amount of SSRs contained in each gene ranged from 0 to 181. A sharply decreasing trend of gene number was observed as the contained SSR number increased (Fig. 6b). Of all SSR-containing genes, genes within one SSR were the most common, 5878, followed by 2-SSR containing genes, 3-SSR containing genes with 4389 and 3230, respectively (Fig. 6b).
The expression of all pineapple genes was quantified using FPKM values, and 16,119 (54.2 %) genes had an FPKM value >0.3 in at least one tissue. The distribution of FPKM values of no-SSR genes (Fig 7 a), genes within one SSR (Fig 7 b) and genes within two or more SSRs (Fig 7 c) in different pineapple tissues including flower, leaf, root and fruit are shown. The heat map results indicate that all tissues from each of the three categories showed identical expression patterns. In general, the greater the number of SSRs in each gene, the more the expression levels of that gene increased. In genes within the two or more SSRs category, most genes had higher expression levels than the other two categories, while genes in the no-SSR category had the lowest expression levels. To further estimate the statistical significance of the correlation between number of SSRs contained in a gene and expression levels, genes with high FPKM values were separated by the number of SSRs contained inside (0 to 181). Graphically, they showed no significant correlation across four tissues (R2 = 0.0002–0.01494, P > 0.2) (Fig. 8).
The GO annotation of SSR-containing genes identified in this study is shown in Fig. 6c. Of the 21,631 genes containing SSRs, 11,700 (54.1 %) were able to be assigned one or more GO annotations, resulting in 14,338, 8668, and 18,110 biological process, cellular component and molecular function terms, respectively (Supplementary Table 3). The molecular function ontology category was comprised of a high portion of protein binding (61.9 %) and catalytic activity (46.5 %), followed by transporter activity (4.8 %) and nucleic acid binding transcription factor activity (2.8 %). With regard to the cellular component, 32 % were assigned to cell or cell part followed by membrane, organelle and macromolecular complex with 12.7, 10.1, and 7.0 %, respectively. In the biological process ontology category, metabolic process (48.2 %), cellular process (40.3 %) and single-organism process (27.6 %) were the top three most dominant groups. The remaining processes were localization, biological regulation, response to stimulus, cellular component biogenesis and signaling, and others. The biological interpretation of the SSR-containing genes was further completed using KEGG pathway analyses. Of all the SSR-containing genes, 1599 (7.4 %) had one or more KEGG annotations, and they belonged to 136 pathways, of which some were consistent with the biological processes identified through GO analyses. Those pathways may represent all KEGG pathways in the pineapple genome.
Discussion
Microsatellites have been of paramount importance for genetics, ecology, taxonomy and evolution studies. Analysis of pineapple microsatellites in coding and non-coding regions, coupled with information of their frequency, distribution and sequence motifs could contribute to the understanding of the pineapple genome architecture and evolution, and provide insights into the possible roles of SSRs in genomic localization and gene regulation. The high abundance of SSRs can be applied to many genetic and genomic studies in pineapple. With the recent pineapple genome release, a global analysis of SSRs is feasible. Genome-wide mining and characterization of pineapple microsatellites was performed and reported for the very first time in this work.
Classes, Frequency and Distribution of SSRs in the Pineapple
In this study, microsatellites in the repeat unit size range of 2 to 6 bp and a minimum length of 12 bp in the pineapple genome were mined and analyzed. In total, the cumulative sequence length of SSRs was 5.2 Mb, which contributed 1.4 % to the estimated 381.91 Mb genome assembly of pineapple and the density was one SSR per 1.37 Kb (728.57 SSRs/Mb). This observed SSR density was higher than those reported from other plant species (Table 2) (Biswas et al. 2014; Cavagnaro et al. 2010; Jena et al. 2012; Liu et al. 2013; Luo et al. 2015; Vásquez and López 2014; Wang et al. 2008). A generally negative correlation between genome size and the density of SSRs or the number of SSRs in plants has been reported (Morgante et al. 2002). Our data obtained applying the same criteria for SSR identification on most selected plant species is consistent with this reported trend. Sorghum with the largest genome size (739 Mb) had a lowest density among others. A similar trend has also been observed in the larger soybean (1115 Mb) (Arumuganathan and Earle 1991) and maize (2365 Mb) genomes (Huo et al. 2008). However, the observation from pineapple deviates from this general trend. Having an estimated genome size larger than that of rice (389 Mb) (Project IRGS, 2005), cucumber (367 Mb) (Arumuganathan and Earle 1991) and grapevine (487 Mb) (Jaillon et al. 2007), the pineapple genome (526 Mb) (Arumuganathan and Earle 1991) harbors an SSR density (728.57 SSRs/Mb) higher than the densities found in these other species. Arabidopsis, the plant with the smallest genome size from our dataset, also deviates from this trend; Arabidopsis had a density substantially smaller than that of those species with 2-fold or 3-fold larger genomes. It has been observed SSRs are preferentially found in non-repetitive or low-copy fractions of the genome (Morgante et al. 2002; Temnykh et al. 2001). The assembled pineapple genome is 381.91 Mb compared to the estimated 526 Mb. The lacking sequences of about ~144 Mb could consist of repetitive DNA that has not been assembled. The high density of SSRs in pineapple genome could be associated with the possibility that the non-assembled sequences are repetitive parts of the plant genome where microsatellites are underrepresented. It has been reported that the frequency of SSRs is considerably higher in dicot species compared to monocots (Sonah et al. 2011). However, this tendency was not observed in this work.
The shorter class II SSRs are more abundant than class I SSRs in genomic and different genic regions of pineapple (Table 1, Fig. 2) and of other plant species (Cavagnaro et al. 2010; Wang et al. 2008). Microsatellites are known to be mutational hotspots in genomes and thus play a significant role in the origin and evolutionary dynamics of genomes (Ellegren 2004; Li et al. 2004b; Luo et al. 2012). Therefore, this tendency might be due to the inherent instability of the longer class I repeats that are prone to mutate to imperfect SSRs by mechanisms of replication slippage, point mutation or recombination (Ellegren 2004), whereas the shorter class II repeats are more tolerant to mutations and retained.
Distribution of SSRs in Different Genome Regions and Transcript Regions
This study represents the first focused analysis of the distribution of SSRs in different genome regions of pineapple. Several lines of evidence have shown that SSRs are nonrandomly distributed across different genomic fractions (Li et al. 2004b; Li et al. 2002). A previous report indicated that UTR regions are SSR rich, and 3′-UTRs were expected to have more SSRs than the 5′-UTRs (Morgante et al. 2002). From our SSR search of each analyzed genome region, 3′-UTRs were found to have the lowest density of SSRs (545.9 SSR/Mb), whereas 5′-UTRs (2839.9 SSR/Mb) had the greatest amount of SSRs, at least 3.6-fold higher than other regions in general (Fig. 1). Higher SSR densities in 5′-UTR were also observed in Arabidopsis, rice, sweet orange and cassava (Biswas et al. 2014; Lawson and Zhang 2006; Vásquez and López 2014). The SSR density varies between genomic and transcribed sequences of pineapple and other organisms (Table 2). In the intergenic or genomic regions of pineapple, microsatellites were present at a higher density than in transcribed regions of genomes, including protein-coding genes and ESTs. This trend was also found in several other plant species, such as papaya, cucumber and grapevine, whereas the opposite was found for rice, sorghum, soybean and citrus. These findings partially contradict a previous study by Morgante et al. (Morgante et al. 2002), although these discrepancies may be due to limited data resources at that time. In addition, inherent genome variations between different species as well as different software used for SSR detection with different set of parameters and algorithms may account for the observed differences among species. In general, SSRs were less abundant in coding regions than in non-coding regions. This is consistent with the report that a considerable amount of SSRs are embedded in non-coding regions either in the intergenic sequences or introns (Ellegren 2004; Tóth et al. 2000). Although SSRs are scarcer in coding regions, the majority of SSRs in noncoding regions are actually close to or linked to expressed genes, making SSRs attractive potential markers for gene localization (Morgante et al. 2002). Furthermore, SSRs from non-coding regions are associated with anonymous genomic sequences, and can therefore provide sufficient polymorphisms to discriminate between closely related species or be useful when conducting genome comparisons.
In pineapple genic regions, UTRs harbor more SSRs than the coding regions. In the untranslated portion of transcripts, SSRs were observed to be densest in 5′-UTRs, followed by introns and 3′-UTRs. The observations of SSR densities in different genomic regions were congruent with previous reports from various plants (Biswas et al. 2014; Cavagnaro et al. 2010; Mun et al. 2006; Wang et al. 2008) and fungi (Labbé et al. 2011; Li et al. 2014; Murat et al. 2011). The transcript regions had a higher G + C content than the whole genomic regions (Table 1). SSRs from different genic locations (CDS, UTRs and introns) may play various roles in development, adaptation, survival and evolution. Mutations in SSRs in the genic regions could affect the corresponding gene products. For example, SSR deletions/insertions in coding regions could lead to a gain or loss of gene function via frameshift mutation or expanded toxic mRNA (Li et al. 2004b; Li et al. 2002); the presence of certain polymorphic SSRs in UTRs or introns could affect gene expression levels; SSRs in 5′-UTRs could be responsible for regulating transcription/translation, gene regulation adaptation, mRNA stabilization as well as short-time phenotypic changes; SSR expansions in the 3′-UTRs could cause transcription slippage, slicing disruption and cellular function damage; SSR variations in introns can affect gene transcription, mRNA slicing or export to the cytoplasm (Li et al. 2004a; Li et al. 2004b; Zhang et al. 2006). In light of this, the observed high density of SSRs in the 5′-UTRs of the pineapple genome provides a good opportunity to gain insight into the influence of SSRs on pineapple gene expression and regulation. In total, 8282 SSRs were found in the 5′-UTRs of genes coding for proteins (Table 1). These gene models are good candidates for future studies.
Character of SSR Motifs, Repeat Number and Repeat Length
SSRs motifs show species and genome fraction or region specificity in eukaryotic and prokaryotic organisms (Mrázek et al. 2007; Tóth et al. 2000). In pineapple, most SSRs are di-, tri- and tetranucleotide repeats, which together account for 88 % of all SSRs. The dinucleotide motifs (46.8 %) outnumbered all other microsatellite repeat categories and were the most prevalent motif length type in pineapple genomic regions (Fig. 3). Especially in the genomic class I (long) microsatellites, dimers accounted for up to 62 %. The prevalent motif length varied across species. Similarly to the trend observed in pineapple, dimers prevailed in sweet orange, cranberry, and cassava. These findings are in agreement with previous reports of highly abundant dinucleotides in the genomic DNA of many evaluated species (Kalia et al. 2011). However, some exceptions exists such as the prevalence of trinucleotides in Arabidopsis, rice, sorghum and flax, whereas tetranucleotide repeats are found to be the most common type in cucumber and grapevine. Comparison of transcript regions (i.e., CDs, exons, ESTs) and whole pineapple genomic regions showed that all repeat types except tri- and hexanucleotide repeats were comparatively less abundant in the transcript regions (Fig. 3 and Table 2). This tendency was also observed in other species. Trinucleotide repeats were found to be the most prevalent repeat type in protein-coding sequences or ESTs of pineapple genome and all other taxa including plants, insects and human (Biswas et al. 2014). In transcript regions, trinucleotide SSRs were the most abundantly found SSRs, followed by di-, tetra-, hexa- and tetranucleotides. This trend was consistent with the most recent study on pineapple EST-SSRs mining (Ong et al. 2012). This predominance of tri- and hexanucleotides over other repeat types has been attributed to negative selection against frameshift mutations. Tri- and hexanucleotides are an integration of multiple of codons, therefore their mutations probably will not disrupt the reading frame, a process that may be associated with genetic conservation. Although a similar situation was observed across species, trinucleotides being the most frequent motif, the absolute density of triplets varied extensively depending on the species. In all examined species, we found that the density of trinucleotides in monocots was at least 1.4-fold higher than that of dicots in transcripts regions.
For all SSR motif types, as the number of repeat unit increased their occurrence strikingly decreased at genomic and transcript levels. The longer motifs, tetra- penta- and hexanucleotides, showed a more dramatic reduction in frequency as the number of repeats increased (Fig. 4). The higher mutation rates of longer repeats may account for this trend. Dinucleotides with a cumulative length of nearly 3 Mb were the greatest contributors to the total percentage of SSRs in the genome fraction due to their highest number of repeat units and highest frequency (Table 2 and Fig. 4). In transcript regions, trinucleotides with a cumulative length of 307 kb represented the largest proportion of microsatellites due to their highest density. The longest SSR in pineapple genome was (CA)1439. It has been proposed that repetitive sequences are recombinogenic elements in eukaryotic chromosomes (Treco and Arnheim 1986; Wahls et al. 1990), dinucleotides especially are preferential sites for recombination because of their high affinity for recombination enzymes (Biet et al. 1999). As molecular markers, the lower stability, higher frequency and longer repeats indicates that the region will be richer in polymorphisms, making dinucleotides more important than other types of nucleotide as the most sought-after markers for practical applications in pineapple population genetics.
There may be striking differences in the frequency of SSRs within certain nucleotide compositions among eukaryotic genomes or between sequence datasets (genomic and EST) of a species. Overall, the base composition of the SSR motifs showed a strong bias to AT-rich in the pineapple genome, and an increased GC-rich or AT-CG balanced motifs in the transcript regions (Fig. 5). Similarly to many other understudied plant species (Cavagnaro et al. 2010; Morgante et al. 2002; Tangphatsornruang et al. 2009), AT were the densest dinucleotide motifs, whereas the least frequent was the GC motifs accounting for only 1 % of dinucleotides, and other genomic SSRs with GC-rich repeats were also rare. This result in pineapple contradicts previous reports indicating that AT-rich repeats prevail in dicot species but not in monocots by virtue of the relative GC content of their genomes (average 34.6 % in dicots vs. 43.7 % in monocots) (Cavagnaro et al. 2010; Wang et al. 2008). This dicot-like trend in pineapple could be explained by its relatively low GC content compared to other monocots, only 38.3 % genome-wide (Ming et al. 2015). ESTs showed higher frequencies of AG repeats than AT repeats in pineapple, in agreement with previous findings from pineapple ESTs (Wöhrmann and Weising 2011) and from many other vascular plants, e.g. Arabidopsis thaliana, coffee, kiwifruit, cassava and cereals (Fraser et al. 2004; Katti et al. 2001; Morgante et al. 2002; Poncet et al. 2006; Vásquez and López 2014). Homopurine-homopyrimidine stretches like AG in the 5′-UTR have been reported to take part in gene regulation (Varshney et al. 2005) and are preferentially associated with genes involved in transcription, nucleic acid metabolism and the regulation of gene expression (Scaglione et al. 2009). The length polymorphism of a (AG)n in the 5′-UTR of the waxy gene also proved to correlate to the amylose content in rice (Ayres et al. 1997). Likewise in many dicots such as legume species but not in Arabidopsis, the trinucleotide repeat AAT was overrepresented in genomic sequences of pineapple, nearly 1.16 times more than the second most trinucleotide repeats. By contrast, GGC repeats were typically predominant in the monocots (Mun et al. 2006). The representation level of AAT in the pineapple genome was midway between the levels found in the legume species and those of rice. Typically, GC-rich tri- and hexa-motifs dominate in the transcribed regions and are less pronounced in non-transcribed regions (Mun et al. 2006; Tóth et al. 2000), in agreement with the observations from our study. CCG as the most common triplet in transcript regions is a known feature of monocots, including pineapple in this study and all cereal species (Li et al. 2004b), and seems to correlate with the increase of GC content in monocot genomes (Morgante et al. 2002). The CCG motif in 5′-UTR of ribosomal protein genes is involved in the regulation of fertilization in maize (Dresselhaus et al. 1999). The taxon-specific accumulation of certain repeat motifs could be explained by strand slippage or a positive selection pressure, such as a preference of codon usage in exons or a regulatory effect of particular repeats in non-coding regions (Mun et al. 2006), which may drive specialization and divergence of genomes.
Expression and Functional Annotation of SSR-Containing Genes
It is important to note that SSRs identified in genic regions are informative and potential powerful molecular markers for the plant breeding community. A better understanding of SSRs could reduce the effort and resources required in the early stages of development of markers closely linked to particular genes due to their location inside genes. Additionally, gene-related SSR markers can be employed in association with mapping studies to help map the particular genes in which they reside. However, the functional significance of SSRs in plant genes remains poorly understood. Putative functional annotation and categorization of pineapple genes containing SSRs from this study revealed that those genes have a range of functions such as protein binding, catalytic activity, metabolic enzymes, disease signaling, structural and storage proteins, and transcription factors, suggesting a biological significance of SSRs in plant metabolism and gene evolution. In the molecular function category, the majority of genes containing SSRs were homologous to proteins with binding and catalytic activities, mostly associated with cell, membrane and organelle according to the cellular component category. Cellular and metabolic processes were associated with most SSR-containing genes, while a small number were involved in reproduction, biological adhesion and growth processes. Similar results were found from date palm (Zhao et al. 2012) and citrus (Liu et al. 2013) transcript sequences containing SSRs, suggesting that genes containing SSRs involved in protein metabolism and biosynthesis are well conserved in plants. Genes containing SSRs are nearly 3 times more likely than no-SSR genes to appear in the pineapple genome (Fig. 6a). In general, a positive correlation between gene expression level and the number of SSRs present was found (Fig. 7). This evidence suggests that SSRs may play an important role in the regulation of gene expression and many other associated functions. The particular role of SSRs and preferred motifs for the function of related genes needs to be further investigated in pineapple.
Conclusions
The current work contributed to a detailed characterization of microsatellites in pineapple and the comparison of these microsatellites to related species. We reported the identification of 278,245 SSRs and 41,962 SSRs with an overall density of 728.57 SSRs/Mb and 619.37 SSRs/Mb in genomic and EST sequences, respectively. This was unexpectedly high given the moderate size of the pineapple genome. Class II SSRs were more abundant than class I SSRs in all genome fractions. ESTs of pineapple were less abundant in microsatellites compared to genomic sequences. Dinucleotide repeats were the most frequent SSRs in the genome with AT being the overall most common single motif, whereas trinucleotides strongly predominated in EST sequences. AT-rich motifs prevailed in the pineapple genome and an increased GC-rich or AT-CG balanced motif in the transcription regions were observed. The putative functional annotation and categorization of genes containing SSRs revealed that those genes are involved in various aspects of pineapple development. Our transcriptome analysis reflected a positive relationship between expression levels and SSR number contained in a gene. Based on this preliminary study, primer design and laboratory validation for genomic and ESTs SSR marker development will be necessary to develop this line of research. These potential SSR markers, especially SSR loci with GO terms, may facilitate a number of genetic and genomic studies in Ananas comosus, such as functional genomics, genetic mapping, discrimination of genotypes, diversity analysis, transferability, as well as positional gene-cloning and QTL analysis.
Materials and Methods
RNA Extraction and Library Construction
Pineapple transcripts were sampled from the major tissues including flower, leaf, root, and fruit. Total RNA was extracted from ground leaves using the Qiagen RNeasy Plant Mini Kit (Qiagen, #74,904) and following the manufacturer’s instructions. DNA was removed with the DNA-free™ DNA Removal Kit (Life Technologies, #AM1906M). A single indexed RNAseq library was constructed using the Illumina TruSeq stranded RNA Sample Preparation Kit (Illumina, #RS-122-2001), and then sequenced by Illumina HiSeq2500 in paired-end 100 nt mode. Three biological replicates were studied for each time point.
Source of Genomic and EST Sequences
The pineapple genome sequencing project yielded 381.91 Mb genome sequences from A. comosus F153, accounting for 72.6 % of the estimated 526 Mb pineapple genome. The contig N50 is 126.5 kb and scaffold N50 is 11.8 Mb (Ming et al. 2015). Pineapple ESTs were downloaded from Genbank (access date: 10/27/2014). Transcripts were assembled using TRINITY (Grabherr et al. 2011), from several RNAseq libraries including flower, leaf, root, and fruit S1 to S8. Finally, we combined the Genbank ESTs and TRINITY transcripts, which were further assembled into 61,522 unigenes using CDHIT (percent identity cutoff at 98 %). These two sources of pineapple sequences were used for SSR mining.
SSR Mining
A large-scale, genome-wide SSR search was performed in pineapple genome using a Perl program MISA, MicroSAtellite identification tool (Thiel et al. 2003) available at http://pgrc.ipk-gatersleben.de/misa/. Both perfect and compound repeats were considered, with a basic motifs ranging from 2 to 6 bp and a minimum repeat length of 12 (for di- to tetra-), 15 (for penta-), 18 (for hexa-). Mononucleotide repeats were not considered due to possible confusion between bona fide SSRs and errors during sequencing, assembly process or polyadenylation tracks. With respect to compound repeats (distinct and adjacent SSRs), the maximum difference between two SSRs was set as 100 bp. 3′-UTR, 5′-UTR, CDS, exon and intron were extracted from gff3 annotation file using an on-line gff2bed python script (http://bedops.readthedocs.org/en/latest/content/reference/file-management/conversion/gff2bed.html ), ‘bedtools getfasta’ to convert bed format to fasta and some in-house python script. Altogether we obtained ~41.95 Mb of exon sequences, ~100.49 Mb of intron sequences, ~33.32 Mb of coding sequences, and ~2.92 Mb and ~5.68 Mb of 5′ and 3′-UTR sequences, respectively. SSR density, GC content, motif, repeat length and repeat times distribution in pineapple whole genome, genic region and EST sequences were estimated, analyzed and compared with each other via windows Excel 2010 and Linux python scripts. The repeats motifs on a complementary strand were considered equivalents and grouped into one motif. For instance, the motif AG is equivalent to GA, TC, CT, and so forth.
SSR-Containing Gene and No-SSR Gene GO Annotation and Expression Estimation
The trimmed paired-end reads of each sample were aligned to repeat-masked pineapple assembly version 3 using TopHat v2.0.9 default settings (Trapnell et al. 2009). The normalized FPKM value (Number of fragments per kilobase of exon per million fragments mapped) of each sample were estimated by Cufflinks v2.2.1, followed by Cuffnorm v2.2.1 using default setting with pineapple gene model annotation provided (−g option). Several in-house python scripts (available upon request) were used to extract ID of genes within no SSR, one SSR, and two or more SSRs. The log2 FPKM value for each class of genes was used to generate heatmaps via pheatmap in R (edgeR) ver. 3.2.1 statistical package (www.CRAN.R-project.org). To further identify the relationship between the SSR number contained in a gene and its expression levels, genes with high FPKM values were separated by the number of SSR contained inside. The genes with a log2 FPKM value >5 were considered to derive from active transcribed regions and defined as highly expressed genes. GO terms and KEGG pathway information associated with each protein were computed using INTERPROSCAN (Zdobnov and Apweiler 2001).
Abbreviations
- AFLP:
-
amplified fragment length polymorphism
- bp:
-
Base pair
- CAM:
-
Crassulacean Acid Metabolism
- CDS:
-
Coding sequences
- EST:
-
Expressed sequence tag
- FPKM:
-
Number of fragments per kilobase of exon per million fragments mapped
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- kb:
-
Kilo base pairs
- MAS:
-
Marker-assisted selection
- Mb:
-
Mega base pairs
- RAPD:
-
Randomly amplified polymorphic DNA
- RFLP:
-
Restriction fragment length polymorphism
- SSR:
-
Simple sequence repeat
- STR:
-
Short tandem repeat
- UTR:
-
Untranslated regions
References
Aradhya MK, Zee F, Manshardt RM (1994) Isozyme variation in cultivated and wild pineapple. Euphytica 79:87–99
Arumuganathan K, Earle E (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Report 9:229–241
Ayres N, McClung A, Larkin P, Bligh H, Jones C, Park W (1997) Microsatellites and a single-nucleotide polymorphism differentiate apparentamylose classes in an extended pedigree of US rice germ plasm. Theor Appl Genet 94:773–781
Biet E, Sun J-S, Dutreix M (1999) Conserved sequence preference in DNA binding among recombination proteins: an effect of ssDNA secondary structure. Nucleic Acids Res 27:596–600
Biswas MK, Xu Q, Mayer C, Deng X (2014) Genome wide characterization of short tandem repeat markers in sweet Orange (Citrus sinensis). PLoS One 9. doi:10.1371/journal.pone.0104182
Carlier JD, Sousa NH, Santo TE, d’Eeckenbrugge GC, Leitão JM (2012) A genetic map of pineapple (Ananas comosus (L.) merr.) including SCAR, CAPS, SSR and EST-SSR markers. Mol Breed 29:245–260
Castillo A, Budak H, Varshney RK, Dorado G, Graner A, Hernandez P (2008) Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis in Hordeum chilense. BMC Plant Biol 8:97
Cavagnaro PF et al. (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genomics 11:569
Chen H et al. (2015) A high-density SSR genetic map constructed from a F2 population of Gossypium hirsutum and Gossypium darwinii. Gene 574:273–286
de Sousa N, Carlier J, Santo T, Leitão J (2013) An integrated genetic map of pineapple (Ananas comosus (L.) merr.). Sci Hortic-Amsterdam 157:113–118
DeWald M, Moore G, Sherman W (1992) Isozymes in Ananas (pineapple): genetics and usefulness in taxonomy. J Am Soc Hortic Sci 117:491–496
Dresselhaus T, Cordts S, Heuer S, Sauter M, Lörz H, Kranz E (1999) Novel ribosomal genes from maize are differentially expressed in the zygotic and somatic cell cycles. Mol Gen Genet 261:416–427
Duval M, Noyer J, Perrier X, d’Eeckenbrugge C, Hamon P (2001) Molecular diversity in pineapple assessed by RFLP markers. Theor Appl Genet 102:83–90
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genet 5:435–445
Feng S et al. (2013) Development of pineapple microsatellite markers and germplasm genetic diversity analysis. Biomed Res Int 2013:11
Fraser L, Harvey C, Crowhurst R, De Silva H (2004) EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108:1010–1016
Gailing O, Bodénès C, Finkeldey R, Kremer A, Plomion C (2013) Genetic mapping of EST-derived simple sequence repeats (EST-SSRs) to identify QTL for leaf morphological characters in a Quercus robur full-sib family. Tree Genet Genomes 9:1361–1367
Garza JC, Slatkin M, Freimer NB (1995) Microsatellite allele frequencies in humans and chimpanzees, with implications for constraints on allele size. Mol Biol Evol 12:594–603
Grabherr MG et al. (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–652
Gur-Arie R, Cohen CJ, Eitan Y, Shelef L, Hallerman EM, Kashi Y (2000) Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res 10:62–71
Harding RM, Boyce A, Clegg J (1992) The evolution of tandemly repetitive DNA: recombination rules. Genetics 132:847–859
Huo N et al. (2008) The nuclear genome of Brachypodium distachyon: analysis of BAC end sequences. Funct Integr Genomics 8:135–147
Innan H, Terauchi R, Miyashita NT (1997) Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics 146:1441–1452
Jaillon O et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jena SN et al. (2012) Development and characterization of genomic and expressed SSRs for levant cotton (Gossypium herbaceum L.). Theor Appl Genet 124:565–576
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan A (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Karaoglu H, Lee CMY, Meyer W (2005) Survey of simple sequence repeats in completed fungal genomes. Mol Biol Evol 22:639–649
Kato CY, Nagai C, Moore PH, Zee F, Kim MS, Steiger DL, Ming R (2005) Intra-specific DNA polymorphism in pineapple (Ananas comosus (L.) merr.) assessed by AFLP markers. Genet Resour Crop Ev 51:815–825
Katti MV, Ranjekar PK, Gupta VS (2001) Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol 18:1161–1167
Kinsuat M, Kumar S (2007) Polymorphic microsatellite and cryptic simple repeat sequence markers in pineapples (Ananas comosus var. comosus). Mol Ecol Notes 7:1032–1035
Labbé J, Murat C, Morin E, Le Tacon F, Martin F (2011) Survey and analysis of simple sequence repeats in the Laccaria bicolor genome, with development of microsatellite markers. Curr Genet 57:75–88
Lawson MJ, Zhang L (2006) Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol 7:R14
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221
Li YC, Korol AB, Fahima T, Beiles A, Nevo E (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11:2453–2465
Li B, Xia Q, Lu C, Zhou Z, Xiang Z (2004a) Analysis on frequency and density of microsatellites in coding sequences of several eukaryotic genomes. Genomics Proteomics Bioinform 2:24–31
Li Y-C, Korol AB, Fahima T, Nevo E (2004b) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007
Li W, Feng Y, Sun H, Deng Y, Yu H, Chen H (2014) Analysis of simple sequence repeats in the Gaeumannomyces graminis var. tritici genome and the development of microsatellite markers. Curr Genet 60:237–245
Liu S-R, Li W-Y, Long D, Hu C-G, Zhang J-Z (2013) Development and characterization of genomic and expressed ssrs in citrus by genome-wide analysis. PLoS One:8. doi:10.1371/journal.pone.0075149
Luo J et al. (2012) Microsatellite mutation rate during allohexaploidization of newly resynthesized wheat. Int J Mol Sci 13:12533–12543
Luo H et al. (2015) Genome-Wide Analysis of Simple Sequence Repeats and Efficient Development of Polymorphic SSR Markers Based on Whole Genome Re-Sequencing of Multiple Isolates of the Wheat Stripe Rust Fungus. PLoS One:10. doi:10.1371/journal.pone.0130362
Mian MAR, Saha MC, Hopkins AA, Wang Z-Y (2005) Use of tall fescue EST-SSR markers in phylogenetic analysis of cool-season forage grasses. Genome 48:637–647
Ming R et al. (2015) The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47:1435–1442
Molnar SJ, Rai S, Charette M, Cober ER (2003) Simple sequence repeat (SSR) markers linked to E1, E3, E4, and E7 maturity genes in soybean. Genome 46:1024–1036
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Mrázek J, Guo X, Shah A (2007) Simple sequence repeats in prokaryotic genomes. P Natl Acad Sci 104:8472–8477
Mun J-H et al. (2006) Distribution of microsatellites in the genome of Medicago truncatula: a resource of genetic markers that integrate genetic and physical maps. Genetics 172:2541–2555
Murat C et al. (2011) Distribution and localization of microsatellites in the perigord black truffle genome and identification of new molecular markers. Fungal Genet Biol 48:592–601
Ong W, Voo CLY, Kumar SV (2012) Development of ESTs and data mining of pineapple EST-SSRs. Mol Biol Rep 39:5889–5896
Paz EY et al. (2012) Genetic diversity of Cuban pineapple germplasm assessed by AFLP Markers. Crop Breed Appl Biot 12:104–110
Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, Hamon P (2006) SSR mining in coffee tree EST databases: potential use of EST–SSRs as markers for the Coffea genus. Mol Gen Genomics 276:436–449
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Project IRGS (2005) The map-based sequence of the rice genome. Nature 436:793–800
Rodríguez D, Grajal-Martín M, Isidrón M, Petit S, Hormaza J (2013) Polymorphic microsatellite markers in pineapple (Ananas comosus (L.) Merrill). Sci Hortic-Amsterdam 156:127–130
Scaglione D, Acquadro A, Portis E, Taylor CA, Lanteri S, Knapp SJ (2009) Ontology and diversity of transcript-associated microsatellites mined from a globe artichoke EST database. BMC Genomics 10:454
Sharma AK, Ghosh I (1971) Cytotaxonomy of the family bromeliaceae. Cytologia 36:237–247
Sharma R, Gupta P, Sharma V, Sood A, Mohapatra T, Ahuja PS (2008) Evaluation of rice and sugarcane SSR markers for phylogenetic and genetic diversity analyses in bamboo. Genome 51:91–103
Shoda M et al. (2012) DNA profiling of pineapple cultivars in Japan discriminated by SSR markers. Breed Sci 62:352
Sonah H et al. (2011) Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS One:6. doi:10.1371/journal.pone.0021298
Sripaoraya S, Blackhall N, Marchant R, Power J, Lowe K, Davey M (2001) Relationships in pineapple by random amplified polymorphic DNA (RAPD) analysis. Plant Breed 120:265–267
Steele K, Price A, Shashidhar H, Witcombe J (2006) Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor Appl Genet 112:208–221
Tachida H, Iizuka M (1992) Persistence of repeated sequences that evolve by replication slippage. Genetics 131:471–478
Tangphatsornruang S et al. (2009) Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean (Vigna radiata (L.) Wilczek). BMC Plant Bio 9:137
Tautz D (1989) Hypervariabflity of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Tóth G, Gáspári Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25:1105–1111
Treco D, Arnheim N (1986) The evolutionarily conserved repetitive sequence d(TG. AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol Cell Biol 6:3934–3947
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Vásquez A, López C (2014) In silico genome comparison and distribution analysis of simple sequences repeats in cassava. Int J Genomics 2014
Wahls WP, Wallace LJ, Moore PD (1990) Hypervariable minisatellite DNA is a hotspot for homologous recombination in human cells. Cell 60:95–103
Wang J et al. (2008) Genome-wide comparative analyses of microsatellites in papaya. Trop Plant Biol 1:278–292
Wöhrmann T, Weising K (2011) In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across bromeliaceae. Theor Appl Genet 123:635–647
Yu Y, Yuan D, Liang S, Li X, Wang X, Lin Z, Zhang X (2011) Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genomics 12:15
Zdobnov EM, Apweiler R (2001) InterProScan – An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848
Zhang L et al. (2004) Preference of simple sequence repeats in coding and non-coding regions of Arabidopsis thaliana. Bioinformatics 20:1081–1086
Zhang L et al. (2006) Conservation of noncoding microsatellites in plants: implication for gene regulation. BMC Genomics 7:323
Zhao Y, Williams R, Prakash C, He G (2012) Identification and characterization of gene-based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol 12:237
Acknowledgments
This work was supported startup fund from Fujian Agriculture and Forestry University to R.M.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Paulo Arruda
Electronic supplementary material
ESM 1
(DOCX 85 kb)
Rights and permissions
About this article
Cite this article
Fang, J., Miao, C., Chen, R. et al. Genome-Wide Comparative Analysis of Microsatellites in Pineapple. Tropical Plant Biol. 9, 117–135 (2016). https://doi.org/10.1007/s12042-016-9163-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-016-9163-6