Abstract
A series of expressed sequence tags-derived polymerase chain reaction (EST-PCR) markers specific to chromosome 2Ai#2 from Thinopyrum intermedium were developed in this study using a new integrative approach. The target alien chromosome confers high resistance to barley yellow dwarf virus (BYDV), which is a severe virus disease in wheat. To generate markers evenly distributed on 2Ai#2, a total of 105 primer pairs were designed based on mapped ESTs from 8 bins of wheat chromosome 2B with intron-prediction by aligning ESTs with genomic sequences of the new model plant Brachypodium distachyon. Eight and seven polymorphic markers on the short arm and the long arm of chromosome 2Ai#2, respectively, were obtained with a polymorphism rate of 14.3%. These chromosome 2Ai#2-specific EST-PCR markers were then used in tracing and exploring the structural variation of the alien chromosome in the population derived from the immature embryo culture of the cross between N452, a 2Ai#2(2D) substitution line, and common wheat CB037. Two centric fusion of translocations involving 2Ai#2 short or long arm with wheat chromosome 2D and some new genetic stocks including telosomes with the alien chromosome short or long arm were identified in the SC3 generations, which provided basic materials to further study the mechanism of the BYDV resistance. BYDV tests in two field seasons suggest that the BYDV resistance was mainly conferred by the short arm, gene interaction on both arms of the alien chromosome was discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L, ABD) is one of the most important food crops in the world, yet, its production is greatly hampered by biotic and abiotic stresses. Barley yellow dwarf virus (BYDV) is a major wheat disease in many wheat growing regions, typically causing a yield loss of between 13 and 25 kg/ha for each 1% of its increase in incidence (McKirdy et al. 2002). Thinopyrum intermedium (Host) Barkworth & D.R. Dewey (E1E2St), a close relative of wheat, was evaluated to contain several resistant genes to BYDV (Chen et al. 1998; Zhang et al. 2009). It has been reported that the 2Ai#2 chromosome in Th. intermedium, which is homoeologous to wheat group 2 chromosomes, carries a BYDV resistant gene, or genes (Xin et al. 1993; Larkin et al. 1995). Incorporating resistant genes into wheat through introgression from wild relatives has been regarded as the most effective way of BYDV control. To achieve this, stable chromosome translocation lines were developed by homoeologous pairing induction with ph1b mutant (Xin et al. 1991), radiation (Crasta et al. 2000), or tissue culture (Singh 1986; Xin et al. 1991; Banks et al. 1995).
Detecting the alien chromosomes carrying BYDV resistant genes in wheat background is a key step in the generation of translocation lines. Traditionally, the hybrid populations of common wheat and its alien chromosome addition or substitution lines are exposed to suitable stress environment, screened by phenotype for BYDV resistance, confirmed by cytological examination such as genomic in situ hybridization (GISH). This is a rather time-consuming process. Since phenotype is controlled by genotype, and affected by environmental factors, such an approach is also often results in inaccurate selection. New and more efficient, less time-consuming molecular selection methods are required to trace the BYDV resistant genes in wheat background, especially for screening large segregating populations in early generations.
In recent years, marker assisted selection (MAS) techniques for BYDV resistance have been applied to detect the alien chromosome from Th. intermedium, such as SSR (simple sequence repeat) (Ayala-Navarrete et al. 2001), random amplified polymorphism DNA (RAPD), and sequence tagged sites (STS) (Lin et al. 2006, 2007; Cui et al. 2006). However, many limitations exist for these techniques. For instance, the transferability of wheat SSR markers to Th. intermedium is rather low, and RAPD markers usually lack locus information.
Expressed sequence tags-derived polymerase chain reaction (EST-PCR) is a novel and rich-in-source functional marker system, and it is usually generated simply by direct primer design based on EST sequences. Unlike other PCR-based markers such as RAPD, amplified fragment length polymorphism (AFLP) and SSR that often target non-coding regions, EST-PCR markers directly target coding regions, and may directly represent the phenotype-related genes, making EST-PCR markers more convenient for screening desirable breeding materials (Schubert et al. 2001; Hagras et al. 2005; Ayala-Navarrete et al. 2007, 2009; Shen and Ohm 2007; Lu et al. 2006). Furthermore, the coding regions of a gene are often more highly conserved between species or genera, making the EST-derived markers more likely are transportable across taxonomic boundaries (Rowland et al. 2003; Sargent et al. 2007). This transportability is especially useful for marker development in species where whole genome sequences are not yet available. In contrast, the intron regions of a gene are usually less conserved than exons, hence designing primers which anneal to exons flanking introns is more likely to produce polymorphic markers (Plomion et al. 1999; Sargent et al. 2007).
Currently, 1,050,717 wheat EST sequences are registered in the NCBI EST database(dbEST), among which more than 16,000 EST loci have been physically mapped onto specific regions of wheat chromosomes using deletion lines (Qi et al. 2004). To date, 2,600 confirmed loci have been mapped onto group 2 homoeologous chromosomes (Conley et al. 2004), including 959 loci (36.9%) from 728 probes mapped onto 2B which contains most deletion bins in this group.
Brachypodium distachyon has been considered as a new potential model plant due to its small genome and DNA sequences from Brachypodium have been shown to be conserved with those from many grass species including wheat (Foote et al. 2004). Moreover, the 8× draft genome assemblies of B. distachyon have been completed (The International Brachypodium Initiative 2010) and a BLAST server is available at (http://blast.brachybase.org/). It is therefore possible to develop useful EST-PCR markers for tracing chromosome 2Ai#2 by combining the mapped EST database of wheat and the genome sequences of the new model plant in grass family (Draper et al. 2001; Opanowicz et al. 2008).
In this study, we initially developed a set of chromosome 2Ai#2-specific EST-PCR markers in wheat background by combining the mapped wheat EST information and the genomic sequences of B. distachyon. These newly developed markers were then used to screen a somatic culture-derived population and two centric fusion lines and three ditelosomic lines were generated and characterized. The ditelosomic lines were tested by BYDV-GAV response in field for the resistance genes localization. BYDV tests in two field seasons suggest that the resistance gene(s) mainly locate on the short arm.
Materials and methods
Plant materials, tissue culture and DNA extraction
Four wheat varieties Zhong8601, Zhong8423, Chinese Spring (CS) and Wan7107, Th. intermedium and seven wheat–Th. intermedium derivatives with 2Ai#2 chromosomes were used to screen 2Ai#2-specific EST-PCR markers. Th. intermedium (RM001941) was kindly provided by Professor Shancheng Sun, Heilongjiang Academy of Agriculture Science and preserved at the Institute of Crop Science (ICS), Chinese Academy of Agricultural Sciences (CAAS). The wheat–Th. intermedium derivatives used in this study were developed by our research group, including the wheat-2Ai#2 disomic addition line Z6 (2n = 44, Xin et al. 1993; Larkin et al. 1995), 2Ai#2(2D) substitution lines N431 (2n = 42) and N452 (2n = 42), 2Ai#2(2B) substitution lines N439 (2n = 42) and N420 (2n = 42), double ditelosomic substitution line N530 [2n = 40 + 4t, with two short arms of alien telosome 2Ai#2S and two long arms of wheat acrotelosomes 2AL (Lin et al. 2006, 2007)], and long arm ditelosomic addition line T980.
Immature embryo culture of hybrids technique was adopted to induce alien gene introgression into wheat genomes. The hybrids were obtained from a cross of N452 × CB037. CB037 is a common wheat line with good tissue culture ability. Callus induced from 14 to 16-day-old hybrid embryos were subsequently used for two further subcultures (4 weeks each). An SC2 population with 222 plants raised from 5 regenerated plantlets was used for MAS. Seeds with expected key chromosome structural variation were harvested from the selected SC2 plants and grown into the next generations (SC3), which were used for further marker detection and GISH analysis.
DNA was extracted from 2 g fresh leaves at three-leaf-stage with CTAB method described by Doyle and Doyle (1990) and purified for further elimination of RNA, amylose and other unwanted components. The purity and concentration of DNA was assessed by comparison with standard DNA samples in 0.8% agarose gel. The DNA was finally diluted to approximately 50 ng/μl each and stored at −20°C.
EST mining and sequence extraction
A total of 7,104 unigene ESTs that mapped into a chromosome bin map of CS were available at GrainGenes-SQL server (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi). ESTs (≥200 bp each) with no paralogous gene on other homologous groups on each bin of chromosome 2B were obtained.
Intron prediction and primer design
Both 5′ and 3′ end wheat ESTs were aligned with the genomic sequence of B. distachyon. Alignment revealed intron positions and readily confirmed whether two ends could be joined together. Primer premier 5 (PREMIER Biosoft International, USA) was subsequently used for primer design. The primer pairs derived from mapped ESTs were designed to flank at least one intron, with a primer length ranging from 19 to 23 bp, Tm from 50 to 62°C and rating more than 90. Primers were synthesized by AuGCT Co. Ltd, Beijing China. 75 of the total 105 primer pairs were designed without intron prediction (see “Discussion”).
PCR amplification and products visualization
PCR reactions were carried out in a total volume of 25 μl containing 100 ng DNA as template, 0.2 mM each primer, 1 U Taq polymerase (Takara, Japan), 0.2 mM of each dNTP, 10 mM Tris–HCl (pH 8.3), 50 mM KCl and 1.5 mM MgCl2. PCR program was set as following: an initial 5 min at 94°C for denaturation followed by 35 cycles of 94°C 45 s, 45 s at melting temperature of each primer and 72°C 1 min, and a final extension at 72°C for 10 min. The annealing temperature could be altered a few degrees if necessary.
A triple-level separation system was set up for the separation and visualization of PCR products, including 2–4% agarose gel electrophoresis (AGE), 6% polyacrylamide gel electrophoresis (PAGE) and single strand conformation polymorphism (SSCP or n-PAGE). AGE was first used for separating all PCR products. Depending on the separation results, the PCR products were further separated by PAGE and SSCP in order to achieve optimal and clear separation and polymorphism results.
Cloning, sequencing and sequence alignment
To confirm that specific fragments on both 2Ai#2 and wheat chromosomes were homoeologous to the original wheat EST from which the primer pairs were derived, these fragments were excised following AGE using Agarose Gel Extraction Kit (TIANGEN, Beijing) and directly cloned into pMD18-T vector. The protocol for fragment recovery from PA (polyacrylamide) gel was done using a simplified “Crush and Soak” method (Maxam and Gilbert 1977). Recovered fragments were amplified by further PCR. These products were purified and cloned into pMD18-T vector and transferred into competent cells following the manufacturer’s protocol (TIANGEN, Beijing). Colony PCR was used to identify successfully transformed cell colonies. Sequencing was conducted in the Sequencing Laboratory of CAAS. To ensure reliability and accuracy of sequencing results, each recovered fragment was sequenced three times from three different samples. Sequence comparison was conducted using DNAMAN software. 2Ai#2-specific sequences were compared to the nr nucleotide database (NCBI) using BLAST algorithms for repetition. New genomic DNA sequences of Th. intermedium were submitted to GenBank.
Marker assisted selection in SC2 populations
The SC2 population was used to detect the alien chromosome in structural variance by seven markers with relatively high discriminative power, including P4, P31, P36, and P97 (Fig. 2a, e, f, b) on the short arm of chromosome 2Ai#2, and P68, P41, P79 (Fig. 2c, g, d) on the long arm, which were distributed on different regions according to their physical sites on wheat chromosome 2B. PCR and separation of PCR products was conducted as described above with at least two repetitions.
Genomic in situ hybridization
Genomic in situ hybridization was performed as described by Wei et al. (2002). Total genomic DNA of Th. intermedium was used as probe, which was labeled with digoxigenin-11-dUTP using the DIG-Nick Translation Mix (Roche Germany). Total genomic DNA of CS was used as blocker at a ratio of 1:50 (probe:blocker). Hybridization signal was detected with FITC-conjugated anti-digoxigenin-fluorescein (Roche, Germany). Preparations were analyzed and photographed using an Olympus BX-51 fluorescence microscope.
BYDV-resistance test in field
To initially locate the BYDV-resistance gene(s) on the alien chromosome in detail, three ditelosomic lines, T997 with the short arm, T980-2 and T980-14 with the long arm of 2Ai#2, were sown in the field at CAAS in the spring of 2009, along with Zhong8601, CB037 and N452 as negative and positive controls. Besides the ditelosomic lines mentioned above, three other short arm ditelosomic homozygous lines T595, T597 and T598 were planted on the same field. Those lines were originated from another cross between common wheat Zhong8601 and the double ditelosomic substitution line N530, and identified by MAS and GISH (result not show). In 2010, the BYDV test was carried out in the field for replication. More test lines were added and the population was increased. Fourteen lines in total, including an addition line Z6, four substitution lines and two short arm ditelosomic lines N530 and N523 previously generated by our group, three short arm ditelosomic lines T595, T597 and T598 mentioned above, two ditelosomic lines T997, T980 obtained in this study were tested. Each line was seeded into nine rows (120 cm × 30 cm each) and followed by two rows of controls. 35–38 seeds were planted in each row. BYDV-GAV was artificially inoculated at the three-leaf stage using viruliferous aphids as a vector. Approximately ten aphids were deposited at the base of each seedling. After a 20 days’ infestation, all plots were sprayed with pesticide to kill aphids. Investigation was finally carried out after 6 weeks when the controls displayed a discrimination reaction to BYDV. According to the yellowing area on the leaves, the reactions were simply described using three levels: high resistance (HR), moderate resistance (MR) and susceptible (S). HR represents no yellowing area observable, MR for yellowing area only observed on leaf tips and plant growth moderately harmed by the virus; and S for leaves turned mostly yellow and a great decline in plant growth.
Results
Amplification patterns with different EST-derived primers
One hundred and five primer pairs in total were designed according to the ESTs located on eight bins of wheat chromosome 2B. These primers were divided into five categories based on PCR products amplified from genomic DNA of wheat and Th. intermedium as templates (see supplementary material 1). The majority (81.9% of the primers) resulted in successful amplification in both wheat and Th. intermedium. This is consistent with the fact that both Thinopyrum and Triticum genus belong to subtribe Triticinae in tribe Triticeae. Only 15 primer pairs (14.3%) produced diverse fragments in Th. intermedium which could be assigned to 2Ai#2, indicating that the 2Ai#2 chromosome is highly homoeologous to wheat group 2 chromosomes (Zhang et al. 2001). These primer pairs were designated as 2Ai#2-specific EST-PCR markers. In addition, 7 primer pairs only amplified products from the wheat template, and the remaining 12 pairs (11.4%) failed to amplify products both in wheat and Th. intermedium .
Marker allocation analysis by using different cytogenetics materials
As mentioned above, 15 2Ai#2-specific EST-PCR markers were obtained using a set of alien addition/substitution lines and relevant parental lines (supplementary material 2). With 2Ai#2 short arm ditelosomic line N530, eight markers were further mapped onto the short arm of 2Ai#2, the seven remaining ones amplified the alien-specific fragments only from ditelosomic line T980 with the long arm but not from line N530 (Fig. 1). It was inferred that at least one marker existed in each bin, with some of the bins containing up to four markers.
Physical linkage map of group 2 chromosomes. The map was drawn with 2Ai#2-specific EST-PCR markers. Wheat chromosomes was divided into several regions (bins) measured by fraction length (FL, defined by Endo and Gill 1996) at right. Note that the order of markers within an identical bin (wheat chromosomes) or arm (2Ai#2) is unknown; however, the colinearity of markers among different subgenomes is presented by dashed lines. Markers were emboldened to indicate that chromosome assignment information revealed in the present study was consistent with that in the mapped EST database (except P80)
Four and eleven markers revealed their polymorphism after AGE and PAGE, respectively. Figure 2 shows some typical 2Ai#2-specific markers. These markers could be divided into two types: type I is defined as the simple polymorphic 2Ai#2-specific marker, which produced less fragments (Fig. 2a, b, e, g). Type II produced several fragments assigned to more than one chromosome (Fig. 2c, d, f, h), indicating a complex polymorphic marker. For example, P36 (Fig. 2f) was a typical complex polymorphic marker on short arm that amplified 2Ai#2-, 2B- and 2D-specific fragments simultaneously. The AGE markers are much more convenient than PAGE markers since they are easy to run without laborious procedures. No 2Ai#2-specific markers were obtained using SSCP.
Electrophoresis patterns of 2Ai#2-specific EST-PCR markers in different polymorphism level and polymorphism type. PCR products were loaded and separated in the order labeled on the top. Each pattern represents markers as follows: a P4, b P97, c P68, d P79, e P31, f P36, g P41, h P77. The fragments produced by certain primer pairs could be easily assigned to chromosomes by the presence or absence of PCR products from 2Ai#2 addition/substitution lines. The arrow indicates the fragment which can be assigned to a certain chromosome
The complex polymorphic markers are useful to simultaneously trace the alien 2Ai#2 chromosome and wheat corresponding chromosomes. In most cases, the wheat chromosome locations revealed by certain EST-PCR markers in the present study are slightly different (usually with less mapped loci) from the mapped EST database. For example, P68 (BQ170567) is considered to exist on both 2B and 2D, but was only found to be present on 2D (in addition to 2Ai#2) in this study. It is worth noting that one marker, P80, is specific to 2A in the current study, rather than to 2B or 2D as the mapped EST database shows. This may be due to the different mapping methods used, i.e. intron-flanking primer design followed by PCR amplification in the current study might be different in polymorphism from the restriction enzymes cutting followed by Southern blot for the EST mapping in the database (Lazo et al. 2004).
Besides alien chromosome-specific markers, a set of primer pairs were mapped onto wheat group 2 chromosomes 2A, 2B and 2D. Moreover, the specificities of these markers were confirmed by amplification with another set of barley–wheat chromosome 2H addition/substitution lines and their parents (unpublished data). All information on wheat chromosome-specific markers is available in supplementary material 3.
Sequence analysis of recovered fragments
Eleven fragments from five 2Ai#2-specific EST-PCR markers P4, P31, P36, P77 and P97 were recovered and sequenced. Alignments of PCR product sequences from 2Ai#2 and wheat corresponding chromosomes with original ESTs were carried out (Fig. 3). In most cases, both wheat and the grass fragments are similar to or the same as the original EST (Fig. 3a, Ta31; b, Ta36-2D), suggesting that the PCR amplifications had good specificity. Conservation among sequences from 2Ai#2 and wheat chromosomes was also observed. This conservation mostly occurred in exons (Fig. 3a, b), indicating a close relationship between the four group 2 chromosomes, while in intron regions, the sequences seem to be more variable than that in exons (Fig. 3c). The six genomic sequences of Th. intermedium chromosome 2Ai#2 were submitted to GenBank (accession numbers: FI187346, FI187347, FI187348, FI187349, FI187350 and FI187351).
Alignments of sequences from recovered fragments of three markers. a P31, b P36, c P77. EST-PX indicates the original ESTs from which the primer pairs were designed. TiX indicates sequence of Th. intermedium. TaX indicates sequence of wheat (T. aestivum). Bd indicates genomic sequence of B. distachyon. Note that the intron position of EST-P77 which was predicted by Bd sequence is consistent with that of actual sequences Ta77-2B and Ti77
Tracing 2Ai#2 chromosome by developed markers
222 SC2 plants were assessed with 7 markers distributing on different regions of chromosome 2B. In most cases alien chromosome 2Ai#2 either was inherited as a whole (48.6%, 108 plants including reciprocal translocations) or was completely lost (43.7%, 97 plants) and therefore could not be detected by any of the seven markers. Chromosome aberrations were detected only in a small part of the population (7.7%, 17 plants). Three kinds of chromosome structural variation were observed in the SC2 population. The first kind of variation was detected in the plants with four short arm-specific markers, but without the three long arm-specific markers (P4+, P31+, P36+, P97+, P68−, P4−, P79−), suggesting that only the short arm of the alien chromosome was present (2Ai#2S+) in these plants. In contrast, the second had the long arm-specific markers (2Ai#2L+), but lost the short arm-specific ones. In the third, whole alien chromosomes remained but the markers on the short and long arms of chromosome 2D detached (2DS+2DL− or 2DS−2DL+).
SC3 plants derived from selected SC2 plants with the key chromosome structural variation were again identified by MAS (Fig. 4) and finally confirmed by GISH (Fig. 5). Table 1 summarized major chromosome abnormality of SC3 plants detected by EST-PCR markers. Like their parents, the inheritance and segregation of different chromosome arms could be easily traced by our markers and this allowed us to have a general idea of the possible karyotype of tested lines. However, the copy number and relationship of these dissociated arms remained unknown. Figure 5 presented further details of these lines. Eventually three translocation lines and four telosomic lines were obtained. Translocation events generally occurred between chromosome 2Ai#2 and 2D, including 2Ai#2S · 2DL homozygous translocation lines T982-2 and T982-7 (Fig. 5a, b) and a 2DS · 2Ai#2L heterozygous translocation line T993-3 (Fig. 5i). Telosomic lines were most frequency recovered chromosome structural change in the long somatic culture procedure. Both the short arm and the long arm could be added to wheat background (2Ai#2S ditelosomic addition line T997-11, Fig. 5c; 2Ai#2L ditelosomic addition line T980-14, Fig. 5f). Occasionally, only one copy of the alien chromosome arm remained (2Ai#2L mono-telosomic addition line T990-5, Fig. 5g, h), in such a case, plants of this kind tended to lose this telosome in next generation. Another 2Ai#2L ditelosomic line T980-2 was different from others because cytological observation showed that there was a pair of telosomic bivalent and an isochromosome besides twenty bivalents in PMC MI of T980-2 (Fig. 5e). The telosomic bivalent could be detected by the DNA probe of Th. intermedium labeled with digoxigenin-11-dUTP and showed strong hybridization signal, while the small ‘o’ shape isochromosome exhibited the wheat background signal of PI (Fig. 5d).
Seven SC3 plants showing structural variation with alien chromosome detected by developed markers. Each plant (including control lines in last 4 lanes) was tested using seven EST-PCR markers, which from different bins of chromosome 2B, hoping that they could also represent different parts of 2Ai#2. Note that among seven markers, complex marker P36 could detect the existence of 2DS and 2BS, while P68 and P79 mapped on the long arm of 2Ai#2 could also represent the long arm of 2D and 2B, respectively
Variations verified by GISH. Using total genomic DNA of Th. intermedium as probe and Chinese Spring as blocker, GISH was carried out for seven previously selected SC3 plants. Hybridization signal was detected with FITC-conjugated anti-digoxigenin and represented as greenish yellow fluorescence (indicated with arrows). Wheat background was counterstained by PI and fluoresced red. a T982-2 is a homozygous translocation line (2n = 21II), with its 2Ai#2S linked to a wheat chromosome long arm. Considering the result in Fig. 4, the translocated long arm would be 2DL (2Ai#2S · 2DL). b T982-7 (root tip cells) is another translocation line, a sibling line of T982-2. c T997-11 (root tip cells) is a ditelosomic addition line with 2Ai#2S in the wheat background. d, e (fluorescence, brightfield) T980-2 is a 2Ai#2L ditelosomic line. The interesting thing is that a wheat isochromosome (indicated by arrowhead) is observed besides alien telosomes (indicated by arrow). In brightfield, this isochromosome clearly forms an ‘O’ shape, which indicates self-pairing. Judging by the lack of 2DL-specific markers and the presence of 2DS in Fig. 4, we believe that this is a 2DS isochromosome. The karyotype is 2n = 20II + 2Ai#2Ltt + iso2DStt. f T980-14 is a 2Ai#2L ditelosomic addition line with 2n = 21II + 2Ai#2Ltt. g, h (pollen mother cells and root tip cells) T990-5 is a 2Ai#2L monotelosomic addition line with 2n = 42 + 1t = 2n = 21II + 2Ai#2Lt. i T993-3 shows a strong hybridization signal on one arm of a rod bivalent. And according to Fig. 4, the hybridized chromosome arm would be 2DS · 2Ai#2L, thus line T993-3 is a heterozygous translocation line with 2n = 21II
Characterization of ditelosomic lines for BYDV resistance
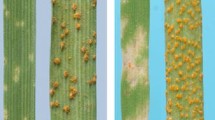
In the field, common wheat line CB037 and Zhong8601 were completely susceptible to BYDV, while substitution lines N452 have good resistance against this virus (Fig. 6). Unlike their resistant ancestor N452, ditelosomic line T980-2, T980-14 and 15 offspring lines of T980-2 presented limited resistance ability to the virus although the long arms of chromosome 2Ai#2 still remain. The short arm ditelosomic homozygous line T997, along with previously generated short arm ditelosomic line N530, N523, T595, T597 and T598, showed moderate BYDV-resistance level among these two. This may possibly suggest that the short arm has a major contribution to the resistance ability of chromosome 2Ai#2. However, this ability could not be fully developed without assistance from the long arm.
BYDV-GAV reaction of ditelosomic lines in field. The resistance ability of test lines: N452 (R) > 2Ai#2S(MR) > 2Ai#2L(S) ≥ CB037, Zhong8601(S). Note that in our study the main symptom of BYDV disease is yellowish leaves. Dwarfing was seldom observed. Thus yellow leaf number (indicated by arrows) and area are indicated in the figure as a brief index of resistance ability of test lines
Discussion
Comparative genomics in this research: application and evidence
In recent years, the greatly accumulated genetic markers and a large number of DNA sequences have made the study of comparative genomics in the grass family possible. It was found that the linear order (colinearity) of genetic markers is well conserved and is retained at the molecular level (microcolinearity) in most cases among the grass genomes (Feuillet and Keller 2002; Li et al. 2004). In this study, we designed 105 wheat EST-derived primer pairs. Most were successful used in PCR amplifications in both wheat and its close relative Th. intermedium, of which 15 were mapped onto the alien chromosome 2Ai#2. Although the linear order of the markers within the alien chromosome arm need to be validated, the markers from different arms of the wheat group 2 chromosome were found to be also located on the corresponding arm of 2Ai#2, which was confirmed by two ditelosomic lines.
Polymorphic rate of EST-PCR markers in wheat background
EST-PCR markers are based on gene sequences that are usually conserved but vary in some degree among species or between genera. Different strategies for EST-PCR marker development yield different levels of efficiency. Randomly designed primer pairs usually results in low levels of polymorphism. Cleaved amplified polymorphic sequences (CAPs) were adopted for further improvement of polymorphism (Gilpin et al. 1997; Schubert et al. 2001; Chee et al. 2004). Some researchers (Cato et al. 2001; Perry and Bousquet 1998; McCallum et al. 2001; Rowland et al. 2003) designed primer pairs by the 5′ or 3′ ends of ESTs for greater polymorphism levels. However, this method often results in failure of primers to pair because of much more divergence in these regions between distanting-related genus. Plomion et al. (1999) and Sargent et al. (2007) introduced the intron-flanking strategy with relatively high efficiency for marker development in genus Pinus and Fragaria. This strategy was based on the fact that introns of a gene are usually less conserved than exons, hence there is an increased likelihood of producing polymorphic markers by designing primers annealing on exons flanking introns. The key step of this method is to accurately predict introns, especially when dealing with ESTs or cDNAs for which whole gene sequences are unknown. A feasible way is to carry out sequence alignments between ESTs or cDNAs with related genomic sequences. This approach is especially useful when the target species has not yet been sequenced (Choi et al. 2004; Wei et al. 2005; Panjabi et al. 2008; Ishikawa et al. 2007; Ayala-Navarrete et al. 2007, 2009). In the present study, primer pairs (P1–P75) were initially designed without considering the intron location, which had a low polymorphic rate of 9.3% (7/75). Of the 75 primer pairs, 10 failed to PCR both in wheat and Th. intermedium. This accounts for 13.3% of the total, and might be largely due to primers annealing at the intron–exon junction site. The later 30 pairs (P76–P105) were designed by intron- flanking way predicted by alignment of wheat EST with related Brachypodium genomic sequences. This approach resulted in a higher polymorphic rate of 26.7% (8/30), threefold over the previous designed primers. Of these, only two primer pairs failed in PCR, accounting for 6.7%, which might be due to inaccurate EST sequences. So, the intron-flanking method is more efficient for the development of EST-markers among different genera, including some diploid (Wei et al. 2005; Hagras et al. 2005) or polyploid plants (Ishikawa et al. 2007; Ayala-Navarrete et al. 2007; Shen and Ohm 2007) compared with conventional methods. However, unlike as with the relatively high polymorphic rate of EST-PCR markers in other plants (Sato et al. 2004; Levi et al. 2008, 2009), it is still difficult to develop EST-PCR markers in wheat and its close relatives (Ishikawa et al. 2007; Ayala-Navarrete et al. 2009), even with the intron-flanking method. This may be largely due to the huge size of the hexaploid wheat genome (17,300 Mb, Bennett and Leitch 1995) and similarity among wheat and its grass relatives such as Th. intermedium. Consequently there are only a few development and application cases of EST-PCR markers in wheat.
Potential of EST-PCR markers in tracing the alien chromatin in wheat background
Seven markers on different physical loci were effectively exploited in the current study according to the corresponding bins on 2B chromosome to track different regions of the alien chromosome. These markers were proved to be very helpful to eliminate most of the unchanged plants and retain plants of interest in the large population of each generation. Detection procedure can be completed in a week instead of weeks of laborious and tedious cytological examinations. After quick MAS, we can have a general idea of each tested lines. GISH results well accorded with the deduction and provided more details. We believe that newly developed EST-PCR markers are reliable and robust. MAS with these markers plus GISH visualization can identify unknown materials quickly, conveniently and accurately.
Unlike chemical and physical mutation-inducing strategies, the overall aberrance rate often stays low during the long somatic culture procedures. But the latter is less severe hence a higher survival rate of regenerated plants. Descended plants tend to grow stronger and thus is beneficial to further development of these plants. Only a few of chromosome abnormality types were detected in this study. Chromosomes tended to break and reconnect at centromere point. Little structural variance within the alien chromosome arm was found in the offspring. This phenomenon was also observed by Zhang et al. (2009) who inferred that the 2Ai#2 chromosome was much more frequently broken in this region than other sites. All of obtained lines will be great helpful to understand more details of BYDV-resistance ability of chromosome 2Ai#2.
The integrity of chromosome 2Ai#2 and its resistance ability against BYDV
Resistance ability of chromosome 2Ai#2 to BYDV has been of great interest since the mid-1990s when it was first recognized as a new resistance source different from that of 7Ai#1 chromosome (Xin et al. 1993; Larkin et al. 1995), and a lot of effort has been put into breeding varieties containing this resistance. In this study, ditelosomic lines, which were carefully selected by the EST-PCR markers and confirmed by GISH, were used to carry out the BYDV test in the filed for two field seasons. Interestingly, the BYDV test results showed that neither of the arms alone confers the full level of resistance associated with the full chromosome substitution and addition lines. It seems that full development of this ability depends on the integrity of this chromosome and the short arm appears to contribute more to this character than the long one does. When the alien chromosome is inherited as a whole, the plants tend to be highly immune to BYDV. We suppose that this ability is fully developed perhaps due to coexistence of resistance genes on the short arm and the long arm, hence possible interaction among these genes. However, things change when this chromosome breaks at the centromere. In this study, tested ditelosomic lines more or less lose this ability. Short arm ditelosomic lines tend to be less sensitive to BYDV compared with long arm ditelosomic lines. This provides possible evidences for our supposition. Another question of interest is that whether the resistance ability will be fully recovered if we combine these two detached arms of chromosome 2Ai#2 into one nucleus together again? If the answer is positive then an interaction among genes of these two arms can be reasonably confirmed.
In conclusion, the mechanism of the BYDV resistance is more complicated than we expected. To make a clear understanding to these genes and gene relations is of great value to our further breeding projects and thereby becoming our new research priority in the future. In current study, effect of the resistance was estimated depend on the symptom of the plants, quantitative analysis of virus in plant (ELISA) is necessary to accurately evaluate this character. The powerful EST-PCR markers along with the well described varieties will undoubtedly increase the opportunity to gain a deeper insight into the BYDV-resistance characteristics of chromosome 2Ai#2 for the first time and finally lead the way to the ultimate program goal.
References
Ayala-Navarrete L, Henry M, Gonz N, Ginkel M, Mujeeb-Kazi A, Keller B, Khairallah M (2001) A diagnostic molecular marker allowing the study of Th. intermedium-derived resistance to BYDV in bread wheat segregating populations. Theor Appl Genet 102:942–949
Ayala-Navarrete L, Bariana HS, Singh RP, Gibson JM, Mechanicos AA, Larkin PJ (2007) Trigenomic chromosomes by recombination of Thinopyrum intermedium and Th. ponticum translocations in wheat. Theor Appl Genet 116:63–75
Ayala-Navarrete L, Tourton E, Mechanicos A, Larkin P (2009) Comparison of Thinopyrum intermedium derivatives carrying barley yellow dwarf virus resistance in wheat. Genome 52:537–546
Banks PM, Larkin PJ, Bariana HS, Lagudah ES, Appels R, Waterhouse PM, Brettell RI, Chen X, Xu HJ, Xin ZY, Qian YT, Zhou XM, Cheng ZM, Zhou GH (1995) The use of cell culture for subchromosomal introgressions of barley yellow dwarf virus resistance from Thinopyrum intermedium to wheat. Genome 38:395–405
Bennett MD, Leitch IJ (1995) Nuclear DNA amounts in angiosperms. Ann Bot 76:113–176
Cato SA, Gardner RC, Kent J, Richardson TE (2001) A rapid PCR-based method for genetically mapping ESTs. Theor Appl Genet 102:296–306
Chee PW, Rong JK, Williams-Coplin D, Schulze SR, Paterson AH (2004) EST derived PCR-based markers for functional gene homologues in cotton. Genome 47:449–462
Chen Q, Conner RL, Laroche A, Thomas JB (1998) Genome analysis of Thinopyrum intermedium and Thinopyrum ponticum using genomic in situ hybridization. Genome 41:580–586
Choi HK, Kim D, Uhm T, Limpens E, Hyunju Lim, Mun JH, Kalo P, Penmetsa RV, Seres A, Kulikova O, Roe BA, Bisseling T, Kiss GB, Cook DR (2004) A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa. Genetics 166:1463–1502
Conley EJ, Nduati V, Gonzalez-Hernandez JL, Mesfin A, Trudeau-Spanjers M, Chao S, Lazo GR, Hummel DD, Anderson OD, Qi LL, Gill BS, Echalier B, Linkiewicz AM, Dubcovsky J, Akhunov ED, Dvořák J, Peng JH, Lapitan NLV, Pathan MS, Nguyen HT, Ma XF, Miftahudin, Gustafson JP, Greene RA, Sorrells ME, Hossain KG, Kalavacharla V, Kianian SF, Sidhu D, Dilbirligi M, Gill KS, Choi DW, Fenton RD, Close TJ, McGuire PE, Qualset CO, Anderson JA (2004) A 2600-locus chromosome bin map of wheat homoeologous group 2 reveals interstitial gene-rich islands and colinearity with rice. Genetics 168:625–637
Crasta OR, Francki MG, Bucholtz DB, Sharma HC, Zhang J, Wang RC, Ohm HW, Anderson JM (2000) Identification and characterization of wheat-wheatgrass translocation lines and localization of barley yellow dwarf virus resistance. Genome 43:698–706
Cui ZF, Lin ZS, Xin ZY, Tang YM, Zhang ZY, Lu Q (2006) Identification of wheat-Thinopyrum intermedium telosomic lines resistant to barley yellow dwarf virus by GISH and STS markers converted from RFLP. Acta Agron Sin 32:1855–1859 (article in Chinese with English abstract)
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Draper J, Mur LAJ, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge APM (2001) Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol 127:1539–1555
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307
Feuillet C, Keller B (2002) Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann Bot 89:3–10
Foote TN, Griffiths S, Allouis S, Moore G (2004) Construction and analysis of a BAC library in the grass Brachypodium sylvaticum: its use as a tool to bridge the gap between rice and wheat in elucidating gene content. Funct Integr Genomics 4:26–33
Gilpin BJ, McCallum JA, Frew TJ, Timmerman-Vaughan GM (1997) A linkage map of the pea (Pisum sativum L.) genome containing cloned sequences of known function and expressed sequence tags (ESTs). Theor Appl Genet 95:1289–1299
Hagras AA, Kishii M, Sato K, Tanaka H, Tsujimoto H (2005) Extended application of barley EST markers for the analysis of alien chromosomes added to wheat genetic background. Breed Sci 55:335–341
Ishikawa G, Yonemaru J, Saito M, Nakamura T (2007) PCR-based landmark unique gene (PLUG) markers effectively assign homoeologous wheat genes to A, B and D genomes. BMC Genomics 8:135. http://www.biomedcentral.com/1471-2164/8/135
Larkin PJ, Banks PM, Lagudah ES, Appels R, Xiao C, Xin ZY, Ohm HW, McIntosh RA (1995) Disomic Thinopyrum intermedium addition lines in wheat with barley yellow dwarf virus resistance and with rust resistances. Genome 38:385–394
Lazo GR, Chao S, Hummel DD, Edwards H, Crossman CC, Lui N, Matthews DE, Carollo VL, Hane DL, You FM, Butler GE, Miller RE, Close TJ, Peng JH, Lapitan NLV, Gustafson JP, Qi LL, Echalier B, Gill BS, Dilbirligi M, Randhawa HS, Gill KS, Greene RA, Sorrells ME, Akhunov ED, Dvoák J, Linkiewicz AM, Dubcovsky J, Hossain KG, Kalavacharla V, Kianian SF, Mahmoud AA, Miftahudin, Ma X-F, Conley EJ, Anderson JA, Pathan MS, Nguyen HT, McGuire PE, Qualset CO, Anderson OD (2004) Development of an expressed sequence tag (EST) resource for wheat (Triticum aestivum L.): EST generation, unigene analysis, probe selection and bioinformatics for a 16,000-locus bin-delineated map. Genetics 168:585–593
Levi A, Wechter WP, Harris KR, Davis AR, Fei Z, Giovannoni JJ (2008) Expression and polymorphism of watermelon fruit ESTs. In: Pitrat M (ed) Proceedings of the IXth EUCARPIA meeting on genetics and breeding of Cucurbitaceae. INRA, Avignon, France, 21–24 May 2008
Levi A, Wechter P, Davis A (2009) EST-PCR markers representing watermelon fruit genes are polymorphic among watermelon heirloom cultivars sharing a narrow genetic base. Plant Genet Resour 7:16–32
Li WL, Peng Zhang, Fellers JP, Friebe B, Gill BS (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40:500–511
Lin ZS, Huang DH, Du LP, Ye XG, Xin ZY (2006) Identification of wheat–Thinopyrum intermedium 2Ai-2 ditelosomic addition and substitution lines with resistance to barley yellow dwarf virus. Plant Breed 125:114–119
Lin ZS, Cui ZF, Zeng XY, Ma YZ, Zhang ZY, Nakamura T, Ishikawa G, Nakamura K, Yoshida H, Xin ZY (2007) Analysis of wheat-Thinopyrum intermedium derivatives with BYDV-resistance. Euphytica 158:109–118
Lu HJ, Fellers JP, Friesen TL, Meinhardt SW, Faris JD (2006) Genomic analysis and marker development for the Tsn1 locus in wheat using bin-mapped ESTs and flanking BAC contigs. Theor Appl Genet 112:1132–1142
Maxam AM, Gilbert W (1977) A new method for sequencing DNA. Proc Natl Acad Sci 74:560–564
McCallum J, Leite D, Pither-Joyce M, Havey MJ (2001) Expressed sequence markers for genetic analysis of bulb onion (Allium cepa L.). Theor Appl Genet 103:979–991
McKirdy SJ, Jones RAC, Nutter FW Jr (2002) Quantification of yield losses caused by barley yellow dwarf virus in wheat and oats. Plant Dis 86:769–773
Opanowicz M, Vain P, Draper J, Parker D, Doonan JH (2008) Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci 13:172–177
Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, Gupta V, Pradhan AK, Pental D (2008) Comparative mapping of Brassica juncea and Arabidopsis thaliana using intron polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9:113. http://www.biomedcentral.com/1471-2164/9/113
Perry DJ, Bousquet J (1998) Sequence-tagged-site (STS) markers of arbitrary genes: development, characterization and analysis of linkage in black spruce. Genetics 149:1089–1098
Plomion C, Hurme P, Frigerio J-M, Ridolfi M, Pot D, Pionneau C, Avila C (1999) Developing SSCP markers in two Pinus species. Mol Breed 5:21–31
Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, Akhunov ED, Dvořák J, Linkiewicz AM, Ratnasiri A, Dubcovsky J, Bermudez-Kandianis CE, Greene RA, Kantety R, La Rota CM, Munkvold JD, Sorrells SF, Sorrells ME, Dilbirligi M, Sidhu D, Erayman M, Randhawa HS, Sandhu D, Bondareva SN, Gill KS, Mahmoud AA, Ma XF, Miftahudin, Gustafson JP, Conley EJ, Nduati V, Gonzalez-Hernandez JL, Anderson JA, Peng JH, Lapitan NLV, Hossain KG, Kalavacharla V, Kianian SF, Pathan MS, Zhang DS, Nguyen HT, Choi DW, Fenton RD, Close TJ, McGuire PE, Qualset CO, Gill BS (2004) A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168:701–712
Rowland LJ, Mehra S, Dhanaraj AL, Ogden EL, Slovin JP (2003) Development of EST-PCR markers for DNA fingerprinting and genetic relationship studies in blueberry (Vaccinium, section Cyanococcus). J Am Soc Hortic Sci 128:682–690
Sargent DJ, Rys A, Nier S, Simpson DW, Tobutt KR (2007) The development and mapping of functional markers in Fragaria and their transferability and potential for mapping in other genera. Theor Appl Genet 114:373–384
Sato K, Nankaku N, Motoi Y, Takeda K (2004) A large scale mapping of ESTs on barley genome. In: Spunar J, Janikova J (eds) Proceedings of the 9th international barley genetics symposium. Committee of 9th international barley genetics symposium, vol 1, Brno, Czech Republic, pp 79–85
Schubert R, Mueller-Starck G, Riegel R (2001) Development of EST-PCR markers and monitoring their intrapopulational genetic variation in Picea abies (L.) Karst. Theor Appl Genet 103:1223–1231
Shen XR, Ohm H (2007) Molecular mapping of Thinopyrum-derived Fusarium head blight resistance in common wheat. Mol Breed 20:131–140
Singh RJ (1986) Chromosomal variation in immature embryo derived calluses of barley (Hordeum vulgate L.). Theor Appl Genet 72:710–716
The International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
Wei WH, Qin R, Song YC, Ning SB, Guo LQ, Gu MG (2002) Location and analysis of introgressed segments in the parthenogenetic progenies of Zea mays × Z. diploperennis by GISH. Acta Bot Sin 44:373–376
Wei H, Fu Y, Arora R (2005) Intron-flanking EST-PCR markers: from genetic marker development to gene structure analysis in Rhododendron. Theor Appl Genet 111:1347–1356
Xin ZY, Xu HJ, Chen X, Lin ZS, Zhou GH, Qian YT, Cheng ZM, Larkin PJ, Banks P, Appels R, Glarke B, Brettell RIS (1991) Development of common wheat germplasm resistant to barley yellow dwarf virus by biotechnology. Sci China (Ser B) 21:36–42
Xin ZY, Chen X, Xu HJ, Lin ZS, Larkin PJ, Banks P, Clarke B, Appels R, Qian YT (1993) Development of addition lines resistant to barley yellow dwarf virus from wheat-Th. intermedium. In: Li ZS, Xin ZY (eds) Proceedings of the 8th international wheat genet symposium. China Scientech Press, Beijing, pp 485–488
Zhang ZY, Xin ZY, Larkin PJ (2001) Molecular characterization of a Thinopyrum intermedium group 2 chromosome (2Ai-2) conferring resistance to barley yellow dwarf virus. Genome 44:1129–1135
Zhang Z, Lin Z, Xin Z (2009) Research progress in BYDV resistance genes derived from wheat and its wild relatives. J Genet Genomics 36:567–573
Acknowledgments
The project is supported by the National Natural Science Foundation of China (30571159).We thank Plant Protection Institute of CAAS for the supply of viruliferous aphids.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by P. Heslop-Harrison.
M. J. Wang and Y. Zhang contribute equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, M.J., Zhang, Y., Lin, Z.S. et al. Development of EST-PCR markers for Thinopyrum intermedium chromosome 2Ai#2 and their application in characterization of novel wheat-grass recombinants. Theor Appl Genet 121, 1369–1380 (2010). https://doi.org/10.1007/s00122-010-1394-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1394-6