Abstract
Low temperature or cold stress is one of the major constraints of rice production and productivity in temperate rice-growing countries and high-altitude areas in the tropics. Even though low temperature affects the rice plant in all stages of growth, the percent seed set is damaged severely by cold and this reduces the yield potential of cultivars significantly. In this study, a new source of cold-tolerant line, IR66160-121-4-4-2, was used as a donor parent with a cold-sensitive cultivar, Geumobyeo, to produce 153 F8 recombinant inbred lines (RILs) for quantitative trait locus (QTL) analysis. QTL analysis with 175 polymorphic simple sequence repeat (SSR) markers and composite interval mapping identified three main-effect QTLs (qPSST-3, qPSST-7, and qPSST-9) on chromosomes 3, 7, and 9. The SSR markers RM569, RM1377, and RM24545 were linked to the identified QTLs for cold tolerance with respect to percent seed set using cold-water (18–19°C) irrigation in the field and controlled air temperature (17°C) in the greenhouse. The total phenotypic variation for cold tolerance contributed by the three QTLs was 27.4%. RILs with high percent seed set under cold stress were validated with linked DNA markers and by haplotype analysis that revealed the contribution of progenitor genomes from the tropical japonica cultivar Jimbrug (Javanica) and temperate japonica cultivar Shen-Nung89-366. Three QTLs contributed by the cold-tolerant parent were identified which showed additive effect on percent seed set under cold treatment. This study demonstrated the utility of a new phenotyping method as well as the identification of SSR markers associated with QTLs for selection of cold-tolerant genotypes to improve temperate rice production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice, the major cereal crop cultivated worldwide can be classified into two major cultivar types. Indica cultivars are grown mostly in the hot and humid tropical low lands; in contrast japonica cultivars are grown in the temperate and sub-temperate regions and the high-altitude areas of the tropics (Mackill and Lei 1997). Even though temperate regions are considered as high yielding in rice production (IRRI 2002), low temperature at the reproductive stage is a major limiting factor for temperate adaptation of cultivars that eventually reduces the yield potential significantly (Xu et al. 2008). The effect of low temperature is manifested at different growth stages such as germination, seedling, vegetative, reproductive, and grain maturity (Andaya and Mackill 2003; Ji et al. 2008; Xu et al. 2008). Low temperature impairs seed germination, reduces seedling vigor, weakens photosynthetic ability by inducing leaf discoloration, reduces plant height, produces degenerated spikes, delays days to heading, reduces spikelet fertility, causes irregular grain maturity, and poor grain quality. Extended low temperature during the reproductive stage of rice in the Republic of Korea caused 17, 78, and 20% damage to the total rice area in 1971, 1980, and 1993, respectively, with maximum yield loss of milled rice of 3.9 t/ha in 1980 (Lee 2001). Yield loss of 1–2 t/ha due to low temperature during the reproductive stage in 1995–96 was also reported in Australia and an annual yield loss of 3–5 million tons was recorded in China (Farrell et al. 2001; Xu et al. 2008).
Japonica cultivars are adapted to low temperature and exhibit better cold tolerance than indica cultivars. Some tropical japonica (javanica) cultivars are reported to also have cold tolerance, and cold tolerance genes from the cultivars Silewah, Lambayque 1, and Padi Labou Alumbis were introduced into many temperate japonica breeding lines in Japan (Abe et al. 1989; Glaszmann et al. 1990; IRRI 1978; Saito et al. 2001). Genetic analysis revealed that two dominant alleles control tolerance of low temperature at the reproductive stage (Nishimura and Hamamura 1993). Nonetheless, this cold tolerance is reported as a complex trait controlled by four or more genes (Nagasawa et al. 1994). Low temperature at the booting stage often causes anther injury, an increase in the number of aborted microspores, eventually resulting in high spikelet sterility and decreased rice yield (Satake 1989). The spikelet sterility type of cold injury is the most serious problem not only at high latitudes but also in high-altitude areas at low latitudes and is characterized by the occurrence of sterile spikelets induced by low temperature during meiosis causing serious yield losses (Murai et al. 1991; Nagasawa et al. 1994). Analysis of mutants from the cultivar Taichung 65 treated with cold water at 19°C revealed that pollen development was inhibited and the embryo sac was malformed in some mutants, resulting in lower spikelet fertility (Nagasawa et al. 1994). The percentage of fertile spikelets has been used as an effective parameter of cold tolerance of rice at the reproductive stage, which includes both booting and flowering (anthesis) stages (Hayase et al. 1969; Saito et al. 1995; Takeuchi et al. 2001).
Simple sequence repeat (SSR) markers (McCouch et al. 2002) were used to identify quantitative trait loci (QTLs) controlling cold tolerance at the reproductive stage (Andaya and Mackill 2003; Saito et al. 2001). QTLs for cold tolerance were identified on chromosomes 3 and 4 derived from a tropical japonica cultivar, Silewah (Saito et al. 1995). Three QTLs for cold tolerance derived from tolerant cultivar Koshihikari were identified on chromosomes 1, 7, and 11 and QTLs for low temperature-induced sterility were mapped on chromosomes 1 and 12 (Li et al. 1997).
QTL analysis has been conducted by using near-isogenic lines (NIL) to tag the genes Ctb1 and Ctb2 for cold tolerance at the booting stage on chromosome 4 (Saito et al. 2004). Eight QTLs for cold tolerance at the reproductive stage were also reported on chromosomes 1, 4, 5, 10, and 11 using NILs (Xu et al. 2008). However, using recombinant inbred lines (RILs) derived from a japonica and indica cross, five QTLs controlling spikelet fertility from a japonica line were located on chromosomes 1, 2, 3, and 9 for booting-stage cold tolerance (Andaya and Mackill 2003). Three QTLs for cold tolerance were also identified on chromosomes 1, 7, and 11 using a doubled haploid (DH) population derived from a cross between two temperate japonica cultivars (Takeuchi et al. 2001). A QTL (qCTB 8) for cold tolerance has been identified on chromosome 8 using an F2 mapping population derived from a cross between two temperate japonica cultivars and by F7 substitution mapping (Kuroki et al. 2007).
The genetics and molecular basis of cold tolerance at the reproductive stage are not completely understood due to unreliable phenotyping methods, non-availability of effective QTLs, and the use of the same genetic sources of cold tolerance. Therefore, it is imperative to use a reproducible phenotyping method to measure tolerant and sensitive traits under cold stress and apply improved QTL analysis procedures to identify effective QTLs for cold tolerance at the reproductive stage. The procedure of screening rice genotypes in the field using cold-water irrigation at 18–19°C from the vegetative stage until grain maturity has been considered as a reliable method to measure cold tolerance (Jena et al. 2004; Xu et al. 2008), although air temperature could not be controlled under field conditions.
The objective of this study was to identify and analyze QTLs derived from a new genetic source (IR66160-121-4-4-2) by using an F7–8 RILs through phenotyping in the field with cold-water irrigation as well as with cool air temperature in the greenhouse with respect to spikelet fertility trait (percent seed set). Here, we report the identification of three reliable QTLs located on chromosomes 3, 7, and 9 responsible for cold tolerance, and haplotype analysis of QTL alleles and their validation using linked SSR marker analysis on some representative cold-tolerant RIL genotypes.
Materials and methods
Plant materials
The breeding line IR66160-121-4-4-2 was used as the donor parent for cold tolerance (Jena et al. 2004). An RIL population (F7 and F8) consisting of 153 plants was produced by single seed descent (SSD) from an F2 population of a cross between the cold-sensitive japonica cultivar Geumobyeo and the cold-tolerant breeding line IR66160-121-4-4-2. These RILs were used to construct a molecular genetic map and identify QTLs controlling cold tolerance. The breeding line IR66160-121-4-4-2 is a new plant type (NPT) line developed at the International Rice Research Institute (IRRI) from a cross between temperate japonica cultivar Shen-Nung89-386 from northern China and tropical japonica cultivar Jimbrug from Indonesia (Peng et al. 1999). Korean japonica cultivars Jinbubyeo, Odaebyeo, and Junganbyeo were used as cold-tolerant checks and Saetbyeolbyeo was used as a cold-sensitive check. Seeds of Shen-Nung89-386, Jimbrug, and IR66160-121-4-4-2 were obtained from the Genetic Resources Center of IRRI, Manila, Philippines, and seeds of Geumobyeo, Saetbyeolbyeo, Jinbubyeo, Odaebyeo, and Junganbyeo were obtained from the Rice Research Division of the National Institute of Crop Science (NICS), Rural Development Administration (RDA), Suwon, Republic of Korea.
Evaluation of cold tolerance
Cold tolerance screening in cold-water irrigation plot and trait measurement
Some 153 RILs along with the parents were planted in a cold-water irrigation plot for phenotypic evaluation during the summer of 2007 and 2008 at Chuncheon substation of NICS, RDA, Republic of Korea. Thirty-day-old seedlings were transplanted in normal-water and cold-water irrigation plots with 25 plants in a single row with 15-/30-cm spacing between plants and rows. The field planting followed a completely randomized block design (CRBD) with two replications. Irrigation-water temperature was normal until the tillering stage (20 days after transplanting). Cold water at 17°C was supplied with water depth of 5 cm during the entire period of rice growth from tillering to grain maturity following standard agronomic practices (NICS 2004). Water temperature showed a gradient from 17°C at the inlet to around 21°C at the outlet (measured by temperature sensors fixed in the plot) in a 6-m-long screening plot. The temperature zone of 17–18°C severely affected the normal development of agronomic traits in most of the RILs. Therefore, we have considered 18–19°C water temperature as the critical temperature zone to measure genotypes as cold tolerant and cold sensitive. Phenotypic data on percent seed set were collected from the first three panicles of five plants at the critical temperature zone in cold-water and normal-water plots. Percent seed set in normal (PSSN) water-irrigated plots and percent seed set in cold water-treated (PSST) plots were measured as the indices for cold tolerance and sensitivity at the reproductive stage, and calculated as the average number of fertile grains in percentage (%) in cold water-irrigated plots and normal water-irrigated plots. Data for percent seed set (PSS) were transformed to measure the effect of the cold-water treatment as follows: a reduction ratio of PSS (PSSR) was calculated as a percentage of the deviation of seed set in the cold water-irrigated plot value from the normal water-irrigated plot value.
Cold tolerance screening in greenhouse with controlled air and water temperature
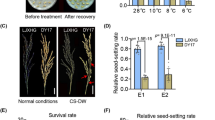
A subset of nine selected cold-tolerant RILs was evaluated for cold tolerance at the booting stage in a controlled cool-environment greenhouse maintained with 17°C air/water temperature following a modified screening procedure of Andaya and Mackill (2003). Five rice seedlings each of the nine RILs and two parents were transplanted in plastic pots containing pulverized dry soil with commercial fertilizer (9–4.5–5.7, N–P2O5–K2O). Extra tillers were removed from each plant in the pot, leaving the main tiller per plant to avoid overcrowding and to promote better growth. Three healthy plants per pot showing uniform development stage were selected and one tiller each from the three plants was tagged. The plants were moved to the controlled-air-and-water-temperature greenhouse maintained at 17°C when the auricle of the flag leaf was approximately 4 cm inside the penultimate leaf of the tillers. After 10 days of cold treatment, the plants were taken back to the normal greenhouse and grown until maturity. The tagged tillers were harvested at maturity and the average number of fertile grains (in %) was measured.
SSR analysis
Genomic DNA was extracted from the fresh leaves of RILs, parents, and progenitor lines of the cold-tolerant parent by a modified CTAB method (Rogers and Bendich 1988). A total of 380 SSR markers distributed at regular intervals (around 3-5 cM) on rice chromosomes were used in a polymorphism survey between the parents. SSR markers that detected polymorphism between the two parents were used for genotype analysis of the RILs. We also searched for additional SSR markers on chromosome regions to locate QTLs for cold tolerance in the SSR map constructed by McCouch et al. (2002) and on the Gramene Web site (http://www.gramene.org/). PCR amplification was done using Taq polymerase (SolGent Co., Ltd., Korea) with the following condition: one cycle at 95°C for 4 min, followed by 35 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, with a final extension at 72°C for 10 min (Bio-Rad, PTC-200 Thermocycler, Germany). PCR products were detected using a 4% denaturing polyacrylamide gel electrophoresis followed by silver staining. SSR analysis was performed following the method of Suh et al. (2009).
Linkage map construction and QTL analysis
The molecular linkage map for the whole genome was constructed using MAPMAKER 3.0 (Lincoln et al. 1992). A Kosambi mapping function was used to convert recombination frequencies to map distances in centiMorgans (cM). The GROUP command was set for logarithm of odds (LOD) at 5.0 to identify initial linkage groupings, and markers that did not belong to the initial group were included in the groupings at LOD = 3.0. Chi-square tests were also performed to examine segregation ratios at the marker loci for deviation from the expected ratio of 1:1, and skewness was determined.
The chromosomal locations of QTLs using RILs were determined by composite interval mapping (CIM). CIM analysis was performed using WinQTL Cartographer v2.5 (Wang et al. 2007). Significance threshold values of LOD scores for QTL detection were determined by using permutation tests (Churchill and Doerge 1994) with 1,000 replicates. For CIM, the critical threshold value of LOD at a genome-wide significance level of P = 0.05 and 0.01 was 2.5 and 4.2, respectively. Correlation analysis between the traits was performed using the correlation procedure of the SAS program (SAS version 8.0, SAS Institute, Cary, NC, USA, 2000).
Haplotype analysis and QTL validation
Sixteen genotypes such as the progenitor lines of IR66160-121-4-4-2 (Shen-Nung89-366 and Jimbrug), parental lines of RILs (IR66160-121-4-4-2 and Geumobyeo), four cold-tolerant RILs, four cold-sensitive RILs, and four check cultivars were used for haplotyping, and to confirm the QTLs controlling cold tolerance. SSR markers closely linked with the three QTLs associated with cold tolerance on chromosomes 3, 7, and 9 were used to compare the genotype patterns of progenitors with parents. The presence or absence of marker allele types corresponding to three QTLs in each of the 16 tested genotypes was determined based on the unique band sizes derived from the PCR products.
Results
Phenotypic variation for cold tolerance
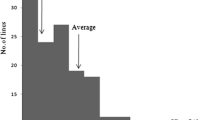
The frequency distribution of PSSN, PSST, and PSSR in the RIL population is shown in Fig. 1. PSSN, PSST, and PSSR showed continuous distribution reflecting quantitative inheritance of the traits. The PSSN of Geumobyeo and IR66160-121-4-4-2 was 91.7 and 93.6%, respectively. The PSSN for RILs ranged from 68.1 to 98.3%, with a mean of 89.1%. The PSST at the critical temperature zone (18–19°C) for Geumobyeo and IR66160-121-4-4-2 was 6.5% and 72.5%, respectively. The PSST for RILs ranged from 0.5 to 80.6%, with a mean of 23.8%. The PSSR of Geumobyeo and IR66160-121-4-4-2 was 92.9 and 24.9%, respectively. However, the PSSR of RILs ranged from 4.9 to 99.5%, with a mean of 73.3%, and it had a left- or right-skewed distribution of the ratings toward cold sensitivity (Fig. 1). A significant negative correlation (−0.995**) was observed between PSST and PSSR at P < 0.01 (Table 1).
Linkage map construction and marker segregation
A genetic linkage map was constructed using 175 polymorphic SSR markers distributed on the 12 rice chromosomes (Fig. 2). The number of surveyed markers per chromosome ranged from 10 to 21 and the average polymorphism between Geumobyeo (temperate japonica) and IR66160-121-4-4-2 (tropical japonica) was 55%. The genetic linkage map covered a total length of 1,641.3 cM, with an average interval size of 9.6 cM between adjacent markers. Some regions on chromosomes 3, 4, 5, 9, and 12 exhibited a limited number of polymorphic markers (Fig. 2). However, additional polymorphic SSR markers derived from a search of the SSR database were inserted on the sparse chromosomal regions.
The SSR linkage map and chromosomal locations of putative QTLs for percent seed set detected in the RIL population. QTLs detected by composite interval mapping analysis (WinQTL cartographer v2.5) are represented to the left of the chromosomes as boxes. qPSSN: QTLs for percent seed set in the normal plot, qPSST: QTLs for percent seed set in cold-water treated plot, qPSSR: QTLs for percent seed set reduction ratio in cold-water treated plot, downward arrow indicates Skewed markers
SSR markers with distorted segregation were detected using the χ2-test for goodness of fit to the expected allelic frequency of 1:1 (Geumobyeo:IR66160-121-4-4-2). Of the 175 mapped loci, 10.3% (18 loci) were skewed toward the Geumobyeo allele type, while 2.9% (5 loci) were skewed toward the IR66160-121-4-4-2 allele type (Fig. 2). Chromosome 11 showed only distorted marker segregation that skewed toward IR66160-121-4-4-2 alleles.
QTL identification
QTL analysis using CIM identified two significant QTLs for PSSN on chromosomes 8 and 11 that explained 9.3 and 7.4% of the phenotypic variation, respectively. These QTLs explained only 16.7% of the total phenotypic variation (Table 2). However, three significant QTLs (qPSST-3, qPSST-7, and qPSST-9) for PSST were detected at the marker intervals (4.6 cM) of RM569-RM231, (1.7 cM) RM3767-RM1377, and (10.8 cM) RM24427-RM24545 on the short arm of chromosome 3, short arm of chromosome 7, and long arm of chromosome 9, respectively. The QTLs qPSST-3, qPSST-7, and qPSST-9 explained 7.1, 10.9, and 9.4% of the phenotypic variation, respectively. The IR66160-121-4-4-2 derived alleles in all three QTL regions increased PSS. Three significant QTLs for PSSR were detected on the same chromosomes (3, 7, and 9), and were associated with PSST, which explained 5.8, 10.9, and 8.7% of the phenotypic variation, respectively. The IR66160-121-4-4-2 allele decreased PSSR at all three QTLs, in contrast to Geumobyeo alleles that increased PSSR at all loci. All the detected QTLs for qPSST and qPSSR totally explained 27.4 and 25.4% of the phenotypic variation, respectively (Table 2).
QTL validation for cold-tolerant RILs
Nine RILs with high PSST in 2007 were evaluated for cold tolerance in a cold-water irrigation plot (18–19°C) and controlled-environment greenhouse with 17°C air/water temperature in 2008. These RILs had the presence of at least one of the three or all QTLs in a homozygous state derived from the cold-tolerant parent for PSST (Table 3). Three of the nine selected cold-tolerant RILs showed higher PSS than even the cold-tolerant parent IR66160-121-4-4-2 in both the screening systems (Table 3). RIL-115 also showed PSS similar to that of IR66160-121-4-4-2. The nine RIL progenies have good phenotypic acceptability for the main agronomic traits and the phenotypes were similar to Geumobyeo. RIL-105 has one QTL (qPSST-3), RIL-110 has two QTLs (qPSST-3 and qPSST-9), RIL-115 has two QTLs (qPSST-7 and qPSST-9), and RIL-138 has all three QTLs (qPSST-3, qPSST-7, and qPSST-9) as detected in this study (Table 3; Fig. 3).
Haplotype analysis at QTLs
A comparative haplotype analysis of 16 genotypes using SSR markers closely linked to the three QTLs for cold tolerance revealed matching alleles of the progenitors present in the cold-tolerant parent (Fig. 4). The closely linked markers, RM1377 for qPSST-7 and RM24545 for qPSST-9, amplified the 145-bp and 152-bp alleles, respectively, in cold-tolerant RILs same as IR66160-121-4-4-2 and the progenitor cultivar Jimbrug. However, the closely linked marker RM231 for qPSST-3 amplified a 186-bp allele in cold-tolerant RILs same as both IR66160-121-4-4-2 and Shen-Nung89-366. The haplotypes with three QTL markers associated with cold tolerance revealed specific banding patterns for the cold-tolerant parent, IR66160-121-4-4-2, as well as cold-tolerant RILs differing from those of japonica check cultivars and cold-sensitive RILs (Fig. 4).
PCR amplification of genotypes using SSR markers linked with QTLs for cold tolerance on chromosomes 3, 7, and 9. Samples in lanes 1, 2, 3, and 4 are Saetbyeolbyeo (cold sensitive), Jinbubyeo, Odaebyeo, and Junganbyeo (cold tolerant), respectively. Samples in lanes 5 and 6 are Shen-Nung89-366 and Jimbrug, the progenitors of IR66160-121-4-4-2. Samples in lanes 7 and 8 are Geumobyeo (P1) and IR66160-121-4-4-2 (P2). Samples in lanes 9, 10, 11, and 12 are cold-tolerant RILs and samples in lanes 13, 14, 15, and 16 are cold-sensitive RILs
Discussion
Low temperature or cold is one of the major abiotic environmental stresses affecting rice plant at different growth stages and it eventually reduces rice yield significantly. Even though molecular and cellular responses to low-temperature stress have been studied extensively at the physiological and biochemical level, the genetic mechanism of cold tolerance in crop plants is not fully understood (Pandey et al. 2009; Yamaguchi-Shinozaki and Shinozaki 2005; Thomashow 1999). Accurate phenotyping of segregating populations and breeding lines for cold tolerance, and their association with molecular markers, is the major limitation to developing cold-tolerant rice cultivars with high spikelet fertility. Therefore, the identification of new sources of cold tolerance followed by the development of appropriate DNA markers associated with a cold tolerance phenotype for percent seed set are important breeding strategies for improving japonica cultivars in temperate regions and high-altitude areas of the tropics.
The main finding of this study is the identification of three QTLs located on chromosomes 3, 7, and 9 and their association with percent seed set, which is the main agronomic trait contributing to yield and which is usually affected during cold stress. We used IR66160-121-4-4-2 as a new source of cold tolerance at the reproductive stage and identified QTLs different from those of other studies in which the sources of cold tolerance were Silewah, Kunmingxiaobaigu, and M-202 (Andaya and Mackill 2003; Kuroki et al. 2007; Takeuchi et al. 2001; Xu et al. 2008). The new QTLs for cold tolerance identified in this study are used for the first time to track early-maturing and fertile cold-tolerant lines in a japonica genetic background.
A molecular marker-based genetic map using 175 polymorphic SSR markers has been used efficiently for QTL analysis in this study. The clustering and distribution of SSR markers on the respective chromosomes coincided with the observation of McCouch et al. (2002). Although the SSR markers were distributed at regular distances (3–5 cM) on all chromosomes and were surveyed to detect polymorphism between the parents, some regions on chromosomes 3, 4, 5, 9, and 12 still have a limited number of polymorphic markers. It suggests that the genetic background of parents may not be very divergent in those chromosomal regions.
Of the 175 SSR markers surveyed, 23 were skewed toward one of the parents. Segregation distortion patterns are frequently observed in the progenies of intervarietal crosses and different types of mapping populations such as F2, RILs, and DH in rice (Andaya and Mackill 2003; Xu et al. 1997; Huang et al. 1997). The segregation distortion of marker alleles observed in this study using RILs from a cross of temperate japonica and an NPT line with a tropical japonica background may be attributed to linkage of the markers with a partially lethal gene as reported by Cheng et al. (1998) or may be due to the presence of gametophytic or sterility genes (Xu et al. 1997). The RILs showed high spikelet fertility although they were developed from a cross between a temperate japonica and an NPT breeding line derived from a temperate japonica and tropical japonica cross. Another cause of segregation distortion for marker alleles in this study may be the selection of fertile progenies developed through the SSD method that excluded sterile plants. Nonetheless, the molecular genetic linkage map constructed in this study using a RIL population is comparable to the maps developed in other studies and should be useful for genetic mapping of different agronomic traits in rice.
Phenotypic variations in PSST and PSSR under cold stress are critical to differentiate cold-tolerant and cold-sensitive individuals in a mapping population. Depending on the origin of the mapping populations, phenotypic indices vary as evident in the progenies from indica/japonica crosses, in which it is difficult to distinguish between floret sterility caused by gametic abortion and that caused by low temperature (Takeuchi et al. 2001). In this study, however, 93% of the RILs showed more than 80% spikelet fertility in the control plot, indicating suitability of the RIL population for QTL analysis for cold tolerance where the progenies could be distinguished for spikelet fertility and sterility. Hence, construction of a molecular linkage map using this RIL population is considered ideal in contrast to an indica/japonica population in which spikelet sterility is often a problem and japonica/japonica population where low DNA polymorphism is normally encountered.
The cold-tolerant donor line used in this study produced high percent seed set (spikelet fertility) under cold-water stress and contributed toward the identification of reliable QTLs. In this study, we used PSST and PSSR to measure the potential of RIL genotypes for cold tolerance or cold sensitivity. These two genetic loci were (PSST and PSSR) mapped on the same genomic regions. Three significant QTLs (qPSST-3, qPSST-7, and qPSST-9) for PSST were identified on chromosomes 3, 7, and 9, respectively. The RILs possessing these QTLs/genes may be involved in reducing the degenerative process of reproductive tissues during cold stress. It is imperative to note that, during cold stress, the RILs tend to reduce plant height and flower either early or late. However, QTLs for plant height and early heading are also putatively located in the different regions of QTLs associated with cold tolerance (Suh et al. unpublished). Reduced panicle nodes of primary tillers might be a critical factor associated with cold tolerance and spikelet fertility (Huang et al. 2009).
QTLs for cold tolerance at the booting and reproductive stages in rice were previously mapped on all the chromosomes (Andaya and Mackill 2003; Dai et al. 2004; Kuroki et al. 2007; Li et al. 1997; Liu et al. 2003; Saito et al. 1995; Saito et al. 2001; Takeuchi et al. 2001; Xu et al. 2008). Saito et al. (1995) identified one QTL for spikelet fertility around the region of qPSST-3 on chromosome 3 as detected in our study. Likewise, Takeuchi et al. (2001) mapped a QTL for percent floret sterility on chromosome 7 in a nearby region similar to that of qPSST-7. These two QTLs may be the reliable indices for cold tolerance. Andaya and Mackill (2003) reported the existence of the QTLs qCTB-7 for undeveloped spikelet (US) and qCTB-9 for spikelet fertility in growth chamber screening, which are also located in the region of qPSST-7 and qPSST-9 for PSST identified in our study. The QTLs detected in our study are very specific and stable because we conducted phenotyping for PSST under long-term cold-water irrigation in the field as well as in controlled environmental conditions in the greenhouse that yielded the same result compared with a short period of cold treatment in the growth chamber. Even though the identified QTLs are relatively minor as indicated by the phenotypic contribution of each QTL, additive values indicate that IR66160-121-4-4-2 is the source contributing to cold tolerance. In a previous study, a QTL (Ctb-1) with minor phenotypic variation for cold tolerance was fine mapped and candidate genes were identified (Saito et al. 2004).
Our results on foreground selection of QTLs associated with cold tolerance are of interest because we could trace the presence of marker alleles in the progenitor cultivars contributing cold tolerance in IR66160-121-4-4-2. Amplification of a 186-bp band at the qPSST-3 locus in the donor, RILs, and progenitor (Shen-Nung89-366) indicated its origin from a temperate japonica genotype of northern China. Similarly, amplification of 145-bp and 152-bp bands at qPSST-7 and qPSST-9 loci, respectively, was attributed to their inheritance from tropical japonica cultivar Jimbrug originated from Indonesia. The qPSST-7 and qPSST-9 might be novel alleles from tropical japonica rice germplasm to improve the cold tolerance of Korean japonica cultivars because the allele sizes of IR66160-121-4-4-2 on the two QTLs were different from those of Korean check cultivars and the QTLs were different from those of Silewah, a tropical japonica (javanica) rice cultivar.
Various methods can be used to create cold injury environments for evaluating cold tolerance at the booting and reproductive stages in rice. The effect of cold-water treatment genetically affected the genotypes of RILs as we have compared the genotype effect in the control plot where there was high percent seed set for all RILs. We studied this effect on RILs for 2 years in two replications. We also used cold tolerant and cold sensitive check cultivars to avoid any biased phenotyping besides testing the RILs in the control plot with normal water irrigation. Because of reproducible results, we used this new method to correctly phenotype RILs for QTL mapping of cold tolerance. Our method of using cold-water irrigation in the field sufficiently induced spikelet sterility and spikelet fertility in RILs although some extreme phenotypes were detected at 17°C water temperature. It is necessary to study the genetics of booting-stage cold tolerance in a growth chamber or greenhouse to minimize the problems imposed by differences in maturity of the test materials (Andaya and Mackill 2003). Interestingly, in our study, the RILs with higher spikelet fertility were observed in both field and greenhouse testings. Similar result was obtained in another study (Xu et al. 2008). We conducted precise experiments to reconfirm cold-tolerant RILs in the greenhouse-controlled air and water temperature. Nine selected cold-tolerant RILs showed cold-tolerant phenotypes under cold-water stress and also produced the same results in greenhouse testing. Spikelet fertility was influenced by both water and air temperature. It is hard to screen many materials in the greenhouse at the critical reproductive stage. From a practical plant breeding point of view, we need to select cold-tolerant breeding lines with high spikelet fertility in the field. Therefore, we confirmed our results in both methods using a subset of cold-tolerant RILs. It is recommended that the cold-water irrigation method with water depth of 5 cm is effective and reproducible to screen and identify cold-tolerant lines in rice. Four of the nine selected RILs have one or two or all three QTLs for PSST and have high percent seed set compared to IR66160-121-4-4-2. However, these QTLs do not show a cumulative effect as a RIL with one QTL (qPSST-3) expressed the same level of percent seed set as a RIL with two or three QTLs. The QTLs on chromosomes 3, 7, and 9 may have a common set of genes controlling spikelet fertility. Even though the effect of QTLs on cold tolerance of the selected RILs does not look like additive based on percent seed set, our results from the analysis of the RIL population in the cold-water irrigated plot revealed additive effect of the QTLs conferring cold tolerance. Additional genetic studies are needed to partition the percent seed set trait to correctly understand the role of the QTLs identified in our study for cold tolerance. We are now developing near-isogenic lines possessing these QTLs using the four cold-tolerant RILs to fine map the QTL genomic regions and determine precisely the effect of each QTL for percent seed set under cold stress.
In this study, marker-QTL association for cold tolerance has been validated using selected RILs for 2 consecutive years in the field and under greenhouse conditions. These QTLs may be useful to facilitate the selection and development of improved cold-tolerant genotypes with high percent seed set for their cultivation in temperate environments and high-altitude areas in the tropics.
References
Abe N, Kotaka S, Toriyama K, Kobayashi M (1989) Development of the “rice Norin-PL8” with high tolerance to cool temperature at the booting stage. Res Bull Hokkaido Natl Agric Exp Stn 152:9–17
Andaya VC, Mackill DJ (2003) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica × indica cross. Theor Appl Genet 106:1084–1090
Cheng R, Kleinhofs A, Ukai Y (1998) Method for mapping a partial lethal-factor locus on a molecular-marker linkage map of a backcross and doubled-haploid population. Theor Appl Genet 97:293–298
Churchill GA, Doerge RW (1994) Empirical threshold value for quantitative trait mapping. Genetics 138:963–971
Dai L, Lin X, Ye C, Ise K, Saito K, Kato A, Xu F, Yu T, Zhang D (2004) Identification of quantitative trait loci controlling cold tolerance at the reproductive stage in Yunnan landrace of rice, Kunmingxiaobaigu. Breed Sci 54:253–258
Farrell TC, Fox KM, Williams RL, Fukai S, Reinke RF, Lewin LG (2001) Temperature constraint to rice production in Australia and Laos: a shared problem. In: Increased lowland rice production in the Mekong region, Proceedings of an international workshop held in Vientiane, Laos. pp 129–137
Glaszmann JC, Kaw RN, Khush GS (1990) Genetic divergence among cold tolerant rices (Oryza sativa L.). Euphytica 45:95–104
Hayase H, Satake T, Nishiyama I, Ito N (1969) Male sterility caused by cooling and the fertilizing ability of pistils. Proc Crop Sci Soc Jpn 38:706–711
Huang N, Parco A, Mew T, Magpantay G, McCouch S, Guiderdoni E, Xu JC, Subudhi P, Angeles ER, Khush GS (1997) RFLP mapping of isozymes, RAPD and QTLs for grain shape, brown planthopper resistance in a doubled-haploid rice population. Mol Breed 3:105–113
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41:494–497
International Rice Research Institute (IRRI) (1978) Screening for cold tolerance. IRRI annual report for 1977, p 142
International Rice Research Institute (IRRI) (2002) Rice production in Europe, Australia, USA, World. http://www.riceweb.org/riceprodleurope.htm
Jena KK, Jeung JU, Yea JD, Choi YH, Hwang HG, Moon HP, Virk PS, Yanagihara S, Mackill D (2004) Evaluation and identification of cold-tolerant rice genotypes by cold-water irrigation stress. Int Rice Res Notes 29(1):54–56
Ji SL, Jiang L, Wang YH, Liu SJ, Liu X, Zhai HQ, Atsushi Y, Wan JM (2008) QTL and epistasis for low temperature germinability in rice. Acta Agron Sin 34(4):551–556
Kuroki M, Saito K, Matsuba S, Yokogami N, Shimizu H, Ando I, Sato Y (2007) A quantitative trait locus for cold tolerance at the booting stage on rice chromosome 8. Theor Appl Genet 115:593–600
Lee MH (2001) Low temperature tolerance in rice: the Korean experience. In: Increased lowland rice production in the Mekong region, Proceedings of an international workshop held in Vientiane, Laos, pp 109–117
Li HB, Wang J, Liu AM, Liu KD, Zhang Q, Zou JS (1997) Genetic basis of low-temperature-sensitive sterility in indica-japonica hybrids of rice as determined by RFLP analysis. Theor Appl Genet 95:1092–1097
Lincoln SE, Daly MJ, Lander E (1992) Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical report, 3rd edn. Whitehead Institute, Cambridge
Liu FX, Sun CQ, Tan LB, Fu YC, Li DJ, Wang XK (2003) Identification and mapping of quantitative trait loci controlling cold-tolerance of Chinese common wild rice (O. rufipogon Griff.) at booting to flowering stages. Chin Sci Bull 48:2068–2071
Mackill DJ, Lei X (1997) Genetic variation for traits related to temperate adaptation of rice cultivars. Crop Sci 37:1340–1346
McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton M, Fu BY, Maghirang R, Li ZK, Xing YZ, Zhang QF, Kono I, Yano M, Fjellstom R, Declerck G, Scheider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers of rice (Oryza sativa L.). DNA Res 9:199–207
Murai M, Hirose S, Sato S, Takebe M (1991) Effects of dwarfing genes from Dee-Geo-Woo-Gen and other varieties on cool temperature tolerance at booting stage in rice. Jpn J Breed 41:241–254
Nagasawa N, Kawamoto T, Matsunaga K, Sasaki T, Nagato Y, Hinata K (1994) Cold-temperature sensitive mutants at the booting stage of rice. Breed Sci 44:53–57
National Institute of Crop Science (NICS) (2004) Stress tolerance breeding of rice in Korea, pp 7–34
Nishimura M, Hamamura K (1993) Diallel analysis of cool tolerance at the booting stage in rice varieties from Hokkaido. Jpn J Breed 43:557–566
Pandey AK, Jain P, Podila GK, Tudzynski B, Davis MR (2009) Cold induced Botrytis cinerea enolase (BcEnol-1) functions as a transcriptional regulator and is controlled by cAMP. Mol Genet Genomics 281:135–146
Peng S, Cassman KG, Virmani SS, Sheehy J, Khush GS (1999) Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop Sci 39:1552–1559
Rogers OS, Bendich AJ (1988) Extraction of DNA from plant tissues. Plant Mol Biol Manual A6:1–10
Saito K, Miura K, Nagano K, Hayano-Saito Y, Saito A, Araki H, Kato A (1995) Chromosomal location of quantitative trait loci for cool tolerance at the booting stage in rice variety Norin-PL8. Breed Sci 45:337–340
Saito K, Miura K, Nagano K, Hayano-Saito Y, Araki H, Kato A (2001) Identification of two closely linked quantitative trait loci for cold tolerance on chromosome 4 of rice and their association with anther length. Theor Appl Genet 103:862–868
Saito K, Hayano-Saito Y, Maruyama-Funatsuki W, Sato Y, Kato A (2004) Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor Appl Genet 109:515–522
Satake T (1989) Male sterility caused by cooling treatment at the young microspore stage in rice plants. XXIX. The mechanism of enhancement in cool tolerance by raising water temperature before the critical stage. Jpn J Crop Sci 58:240–245
Suh JP, Roh JH, Cho YC, Han SS, Kim YG, Jena KK (2009) The Pi40 gene for durable resistance to rice blast and molecular analysis of Pi40-advanced backcross breeding lines. Phytopathology 99(3):243–250
Takeuchi Y, Hayasaka H, Chiba B, Tanaka I, Shimano T, Yamagishi M, Nagano K, Sasaki T, Yano M (2001) Mapping quantitative trait loci controlling cool-temperature tolerance at booting stage in temperate japonica rice. Breed Sci 51:191–197
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Wang SC, Basten CJ, Zeng ZB (2007) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm)
Xu Y, Zhu L, Xiao J, Huang N, McCouch SR (1997) Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled-haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol Gen Genet 253:535–545
Xu LM, Zhou L, Zeng YW, Wang FM, Zhang HL, Shen SQ, Li ZC (2008) Identification and mapping of quantitative trait loci for cold tolerance at the booting stage in a japonica rice near-isogenic line. Plant Sci 174:340–347
Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoter. Trends Plant Sci 10:88–94
Acknowledgments
We thank the Rural Development Administration, Suwon, Korea, for providing necessary facilities and financial support for this study. We are also thankful to Ms. Park So-Hyun for technical assistance. We are grateful to Bill Hardy, Senior science editor, IRRI, for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Wissuwa.
Rights and permissions
About this article
Cite this article
Suh, J.P., Jeung, J.U., Lee, J.I. et al. Identification and analysis of QTLs controlling cold tolerance at the reproductive stage and validation of effective QTLs in cold-tolerant genotypes of rice (Oryza sativa L.). Theor Appl Genet 120, 985–995 (2010). https://doi.org/10.1007/s00122-009-1226-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1226-8