Abstract
Rice growth and productivity are greatly affected by cold stress, which is likely to become more of a hindrance for high and stable rice yields. To identify cold tolerance at the booting and flowering stages in rice, a recombinant inbred line was developed by crossing a cold-tolerant japonica cultivated variety, Jileng1, with a cold-sensitive indica cultivated variety, Milyang23. The seed setting rate (SST) of the parents and RIL population were investigated under different temperature environments, and the SST and cold stress tolerance index under natural low temperature were used to evaluate cold tolerance and quantitative trait locus (QTL) mapping. Fifteen QTLs were detected on chromosomes 1, 2, 3, 5, 7, 11 and 12, with log-likelihood values ranging from 2.64 to 4.76. These QTLs account for 3.34% to 12.02% of the phenotypic variance explained. Three QTLs, qCtb1, qCtb5 and qCtb12, were repeatedly detected in different conditions, and they were considered stably expressed QTLs. qCtb5 was localized to chromosome 5 marker between CMB0526.3 and ID5014265. In this interval or nearby, many cold-resistance QTLs have been identified in previous studies, so qCtb5 is considered a major cold-tolerance QTL. Thirteen QTLs with environmental interactions were also detected, and QTLs detected in single environment were all found to be involved in environmental interactions. These results show that environmental interactions have a significant effect on cold tolerance in rice. Stable expression of major QTLs will help to fine mapping cold-tolerance genes and provide gene resources to cultivate cold-tolerance rice varieties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is the main staple food crop worldwide, feeding more than half the world's population (Sasaki and Burr 2000). Rice is a cold-sensitive crop, and low-temperature stress has a negative influence on the vegetative and reproductive stages. The booting and flowering stages are the most sensitive period and encounter cold stress that will be a fatal effect on production (Xu et al. 2008a, b; Cruz et al. 2013). At the booting and heading stages, cold stress affects panicle growth, including pollen activity, seed fertility and seed size, which ultimately results in decreased yield (Li et al. 2017). Therefore, improving cold tolerance is one of the most important methods to maintain high and stable yields for rice cultivation areas vulnerable to cold stress.
Cold tolerance is a complex trait in a quantitative manner and has a complicated genetic basis controlled by a large number of QTLs and affected simultaneously by the environment (Andaya and Mackill 2003a; Zeng et al. 2009; Zhou et al. 2010). Many QTLs related to cold tolerance at the booting stage have been reported, and these QTLs have been identified on chromosomes 1, 2, 3, 5, 6, 7, 9 and 12 (Andaya and Mackill 2003b). Ctb1 and Ctb2 are related to cold resistance, and locate on chromosome 4 (Saito et al. 2001, 2004). Ctb1 has been fine mapped to a 17-kb region that contains two candidate genes, and encodes an F-box protein and a ser/thr protein kinase; the F-box protein has a significant correlation with cold tolerance (Saito et al. 2010). qCTB8 for cold tolerance was detected on the short arm of chromosome 8 (Kuroki et al. 2007). Xu et al. (2008a, b) evaluated cold tolerance via spikelet fertility of the main panicles using BC5F3 populatin, and eight QTLs were identified on chromosomes 1, 4, 5, 10 and 11. CTB4a and qCTB10-2 were fine mapped in subsequent research. CTB4a encodes a conserved leucine-rich repeat receptor-like kinase (Zhang et al. 2017). qCTB10-2 was delimited to a 132.5-kb region containing 17 candidate genes, and four genes are related to cold treatment inducibility (Li et al. 2017). A major QTL, qPSST6, related to cold tolerance is located on chromosome 6 (Sun et al. 2018). Tang et al. (2019) identified cold-tolerance QTLs at the reproductive stage in rice using two RIL populations, and 17 QTLs were detected on chromosomes 1, 3–6, 8, 11 and 12.
In recent years, numerous cold-resistance QTLs have been mapped at the booting stage, and several QTLs have been stably detected in different environments. However, only a few QTLs have been cloned and applied in rice breeding. Previous researchers, to detect stable cold-tolerance QTLs, the F2 and F2:3 populations were developed by crossing JL1 (cold-tolerant japonica cultivated variety) with MY23 (a cold-sensitive indica cultivated variety). Some major QTLs have been identified and have a positive effect towards increasing cold tolerance in the seedling stage and booting stage, respectively (Han et al. 2005a, 2005b). In this study, the RIL population developed by crossing these parents were employed to evaluate cold tolerance at the booting and flowering stages in rice. QTL mapping was carried out to detect stable expression cold-tolerance QTLs for natural low-temperature tolerance. The research results will provide a basis for fine mapping of these major QTLs and successfully cultivating strong cold-resistance varieties in rice.
Materials and methods
Experimental materials
An RIL population containing 253 lines was developed by crossing the cold-tolerant japonica cultivated variety JL1, as the donor, and the cold-sensitive indica cultivated variety MY23, as the receptor, using the single seed descent (SSD) method. The F2 generation from JL1 × MY23 was subjected to more than ten rounds of self-pollination to generate the RIL population.
Field experiment
The natural low-temperature test was carried out in the field of Yunnan Academy of Agricultural Sciences, Songming (SM), Yunnan Province, China (25°05'N, 102°72'E, 2136 m). Between early June and late September, the daily minimum temperature is approximately 15–19 °C (Fig. 1), which is a suitable temperature to evaluate cold tolerance at the booting and flowering stages in rice. The RIL population and their parents were sown in Songming in 2016 (E1), 2017 (E2) and 2018 (E3). Moreover, all lines were sown in the Changping Test Field of the Institute of Crop Science, Chinese Academy of Agricultural Sciences (Beijing, BJ) in 2018 (Eb1) and 2019 (Eb2). All the field experiments were randomized complete block design with two replications. Each line in the RIL population was grown in three rows, 12 holes per row, and a single plant per hole. The transplanting standard was 25 cm × 15 cm. Fertilizer of pure nitrogen was directly applied to transplants at 120 kg/hm2 in the field.
The heading date was investigated for three years under natural low-temperature conditions. The parents and RIL population were sown in early April 2016, and the heading and flowering periods were mainly concentrated from July 5 to August 27. All lines were planted in late April in 2017 and 2018, and the heading and flowering periods occurred from July 15 to September 29 (Fig. 1). Therefore, the varieties experienced the critical temperature cold stress at the booting and flowering stages. We estimated the SST of parents and RIL population using the mean values of five main panicles in each line when plants reached the maturity stage. The phenotypic value of each line was the mean of two replicates. The CSTI was defined as the ratio of the aSSM to the average SST in Beijing (aSBJ).
DNA extraction and genotyping
Genomic DNA was extracted using the CTAB method. A total of 291 polymorphic markers, including 114 single nucleotide polymorphisms (SNP) (Supplementary Table 1), 62 sequence-tagged sites (STS) (Supplementary Table 2) and 115 simple sequence repeats (SSR) markers, evenly distributed throughout the entire genome of rice, were used to genotype the RIL population. The genotyping of SNP markers was performed by SNP chips. The same genotype as JL1 was “2”, and the same genotype as MY23 was “0”. The PCR products of SSR and STS were separated using 8% polyacrylamide gel electrophoresis. At the same migration rate position, the amplified polymorphic DNA segments that were the same as those of JL1 recorded as “2”, and those that were the same as those of MY23 recorded as “0”. In addition, missing segments were recorded as “−1”.
Linkage map construction and QTL analysis
The genetic linkage map was constructed using the JoinMap4 software (Ooijen 2006) by maximum likelihood mapping, and the other parameters were default values, which covered 12 chromosomes, with 114 SNP, 62 STS and 115 SSR markers. The genetic linkage map was drawn by the Mapchart software (Voorrips 2002). Inclusive composite interval mapping (ICIM) method was used to determine the QTL by QTL IciMapping 4.2 (Li et al. 2007) with scanning step was 1 cM, probabilities of adding and removing variables in stepwise regression were set at 0.001 and 0.002, respectively. The minimal logarithm of the LOD score was 2.50. The SST in SM (E1, E2 and E3), aSSM and CSTI under natural low-temperature conditions were used for QTL mapping. QTL mapping was performed in biparental populations and QTL by environment interaction analysis was also conducted by IciMapping 4.2 (Meng et al. 2015; Li et al. 2015). The confidence interval was calculated by one-LOD drop from the estimated QTL position. The QTL naming convention was as described by McCouch et al. (1997).
Results
Phenotypic variation of cold tolerance in the RIL population
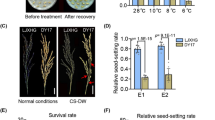
The SST was assessed for the parents and RIL population in Beijing and Songming. The climate of Songming was natural low-temperature conditions from June to september when the RIL population was in booting and flowering stages. Due to the incompatibility of the indica and japonica crossings, varieties with SST less than 70% were eliminated in Eb1 and Eb2. Finally, 219 lines were used for this research. Phenotypic evaluations and comparisons of SST were performed for the parents JL1 and MY23 (Fig. 2a). JL1 showed a higher SST than MY23 under different environments. The average SST of JL1 was 93.39%, which was higher than that of MY23 (78.90%) under Beijing environmental conditions, but there was no significantly different (p < 0.05). In Songming, the average SST of JL1 was 80.70%, which was significantly higher than that of MY23 (p < 0.01), which was 27.34% (Fig. 2a and Table 1). MY23 showed cold sensitive with the SST largely reducing under natural low-temperature conditions. In contrast, JL1 showed stronger cold tolerance than MY23 at the booting and flowering stages.
Violin diagram of SST for the parents and RIL population in Beijing and Songming. A Statistical comparison of the mean SST of parents planted in Beijing (BJ) and Songming (SM). B Violin plots of the distributions of SST for the RIL population grown in Beijing in 2018 (Eb1) and 2019 (Eb2) and Songming in 2016 (E1), 2017 (E2) and 2018 (E3). C Violin plots of the distributions of the average SST in Beijing (aSBJ), the average SST of in Songming (aSSM) and cold stress tolerance index (CSTI). *p < 0.05; **p < 0.01; ns:not significant
There was great variation for the SST (Eb1, Eb2, E1, E2 and E3), aSBJ, aSSM and CSTI under different environments (Fig. 2b–c, Table 1, Supplementary Fig. 1 and Supplementary Table 3). Overall, the SST of BJ environments was higher than that of SM under low-temperature conditions. The variation of E1 (0–85.11%), E2 (0–88.84%) and E3 (0–90.81%) were more rich than that of Eb1 (32.69–93.97%) and Eb2 (49.33–97.50%). The ranges of aSBJ were 54.25–94.85% and 0.83–76.78% for aSSM, and the range of CSTI was 1.00–88.85%. The SST in SM (E1, E2 and E3), aSSM and CSTI were used for QTL mapping for cold tolerance.
Genetic map construction
In total, 295 molecular markers showed polymorphisms between the parents and even distribution across the 12 chromosomes. Finally, a genetic linkage map was constructed based on 291 polymorphic markers, which covered of the rice genome (2619.10 cM) at an average interval of 9.00 cM (Fig. 3). The longest chromosome is chromosome 1, which is 353.48 cM, with an average interval genetic distance of 12.62 cM per marker. The shortest chromosome is chromosome 12, with a length of 137.48 cM and an average interval of 9.82 cM.
QTL mapping
To estimate cold tolerance, the SST in SM (E1, E2 and E3), aSSM and CSTI under natural low-temperature conditions were used for QTL mapping. QTL detected in different single environments were considered as same QTL if their estimated map position was within a 20 cM interval. A total of fifteen cold-tolerance QTLs were detected under different environments (Table 2 and Fig. 3). These QTLs were distributed on chromosomes 1, 2, 3, 5, 7, 11 and 12, with LOD scores ranging from 2.64 to 4.76, and PVE rangeing from 3.34% to 12.02%. Twelve QTLs, including qSST2.1, qSST2.2, qSST3, qSST5, qSST11, qSST12, qSSM2, qSSM5, qSSM7, qSSM12, qCSTI5 and qCSTI12, theirs positive additive effect were from JL1, and three QTL, qSST1, qSSM1 and qCSTI1, its positive additive effect was from MY23. Four, two and one QTLs were detected for E1, E2 and E3, five and three QTLs were identified in aSSM and CSTI, respectively.
qSST1, qSSM1 and qCSTI1 were located within a 20 cM interval and were considered the same QTL (named qCtb1). qCtb1 was located in the physical region between markers RM446 and RM488 and explained 4.23%-6.05% of PVE. Similarly, qSST5, qSSM5 and qCSTI5 were considered the same QTL (named qCtb5). qCtb5 was located between markers CMB0526.3 and ID5014265 and explained 3.34%-8.67% of the phenotypic variation. qSST12, qSSM12 and qCSTI12 were considered the same QTL (named qCtb12). qCtb12 was located between markers S12011B and RM277 with PVE ranging from 4.53%- 12.02. Thus, qCtb1, qCtb5 and qCtb12 were repeatedly identified under different conditions and were considered stable expression QTLs. The major QTL qCtb1, in which the positive additive effect was from MY23, was detected in E1, aSSM and CSTI. Two major QTLs, qCtb5 and qCtb12, in which the positive additive effect was from JL1. qCtb5 was repeatedly identified in E2, aSSM and CSTI. qCtb12 was repeatedly detected in E3, aSSM and CSTI. Among these QTLs, qCtb12 had the highest LOD value and PVE, which were 4.76 and 12.02%, respectively.
QTL for SST by environmental interactions
To some extent, genotype-environment interactions may play an important role in determining cold resistance of rice. Thus, the effects and contributions of QTL-by-environment for cold tolerance in three years (E1, E2 and E3) were examined in this study. Thirteen environmental interaction QTLs were detected on chromosomes 1, 2, 3, 4, 5, 7, 8, 10, 11 and 12 under three natural low-temperature conditions (Table 3). The QTLs, qeSST1, qeSST2.1, qeSST2.2, qeSST2.3, qeSST3, qeSST4, qeSST5, qeSST7, qeSST8.1, qeSST8.2, qeSST10, qeSST11 and qeSST12 were detected with environmental effects. Among these QTLs, including qeSST1, qeSST2.1, qeSST2.2, qeSST2.3, qeSST3, qeSST5, qeSST7, qeSST11 and qeSST12, co-localized with QTL mapping in single environment, as shown in Table 2. Among these QTLs, qeSST2.1, qeSST3, qeSST8.1 and qeSST10 have large environmental effect values, and the other QTLs, including qeSST2.2, qeSST4 and qeSST12, have relatively smaller environmental effects. These results indicate that environmental interaction QTLs played an important role in explaining SST phenotypic variation under natural low-temperature conditions.
Discussion
Phenotypic variation under natural low-temperature conditions
Low temperature at the booting and heading stages is a serious abiotic stress in rice, and cold tolerance is a complex trait controlled by many quantitative trait loci and environmental factors. Most cold-tolerance QTLs at the booting and heading stages have been detected by deep cold-water irrigation and artificial chamber environments (Saito et al. 2001; Endo et al. 2016; Sun et al. 2018). Compared to the cold-water irrigation and growth chamber method, the natural low-temperature treatment method for temperature control is a relatively simple and more effective and is suitable for mass verification (Xu et al. 2008a, b). In addition, phenotypic identification and identified QTL through the natural low-temperature treatment can be directly applied to production. Yunnan Province is favorable environment to carry out natural low-temperature stress treatment due to its high altitude and climatic conditions for each growth and development period, especially the booting and flowering stages in rice. Therefore, many previous studies on cold tolerance have been carried out in Yunnan (Jiang et al. 2010, 2011; Xu et al. 2008a, b; Zhou et al. 2010). Zhu et al. (2015) identified six cold-tolerance QTLs on the chromosomes 3, 4 and 12 at the booting stage under natural low-temperature conditions by association analysis. The major QTL qCTB7 associated with cold tolerance in the Yunnan natural low-temperature environment was detected on chromosome 7, is approximately 92 kb in length and contains 12 putative candidate genes (Zhou et al. 2010). Under natural low temperature in Yunnan, 129 loci associated with cold tolerance at the reproductive stage were identified, and 24 loci were co-localized with reported QTLs indicating the reliability (Guo et al. 2020). In a previous study, we selected the strong cold-resistance japonica rice variety JL1, and the cold-sensitive indica rice variety MY23 as parents and developed F2, F2:3 and RIL populations, in which the values of plant height, panicle length, panicle extraction and SST were largely reduced in natural low-temperature environments (data not shown), indicating that cold stress has a major impact on the phenotype of rice (Han et al. 2005c). In this study, RIL population from JL1 and MY23 was used to evaluate cold tolerance and QTLs under natural low-temperature conditions in muti-environments. Lower temperature had a significant adverse effect on SST. The distribution of SST and CSTI of the RIL population exhibited an continuous distribution. Moreover, transgressive segregation that fell beyond the parents was observed. Twenty-seven cold-tolerance QTLs were detected under different environments, including fifteen addictive QTLs and thirteen environment interaction QTLs. These results suggested that natural low-temperature conditions can help to identify cold resistance and novel QTLs.
Comparison with previous studies for cold-tolerance QTLs
As a crop originated from tropical and subtropical areas, rice is cold sensitive. Low temperature influences multiple stages of growth and development in rice, such as germination, and seedlings exposed to low temperatures exhibit slow development, reduced tillers, yellowish leaves and rot (Andaya and Mackill 2003a). The booting and flowering stages of rice is the most sensitive period for cold stress, which affects pollen development, the setting rate, and ultimately the yield. In this study, fifteen cold-tolerance QTLs were detected on chromosomes 1, 2, 3, 5, 7, 11 and 12, compared with the F2:3 population derived from the same parents, qSST2.2 adjacent to qCTB2 (Han et al. 2005b).
Among nine QTLs, qCtb1, qCtb5 and qCtb12 repeatedly detected were considered stable expression QTL. The positive additive effect of qCtb1 was from MY23, and qCtb5 and qCtb12, in which the positive additive effect was from JL1. The major QTL, qCtb12, was identified on chromosome 12, which was expressed in different environments (E3, aSSM and CSTI) and had the highest LOD and PVE. Comparison with previous studies revealed that many cold-resistance QTLs have been mapped in this region. Among them, qLTG-12, qLTG12a, qLTG12b, qLTG12c and qLTG12a were determined for low-temperature germinability (Li et al. 2013; Fujino et al. 2015; Jiang et al. 2020)). qCTS12a and qCTS-12 are related to cold tolerance at the seedling stage (Andaya and Mackill 2003a; Zhang et al. 2013). qCTB12 is related to cold tolerance and was detected in RM292-RM260 on chromosome 12 at the booting stage by Andaya and Mackill (Andaya and Mackill 2003b).
The major QTL qCtb5 detected on chromosome 5 between CMB0526.3 and ID5014265 was repeatedly identified in E2, aSSM and CSTI. Comparison with previous studies revealed that this region has been mapped to many cold-tolercance QTLs (Fig. 4). Among these QTLs, qSV-5c is related to seed vigor at the germination stage and has been mapped to a 400-kb genomic region on chromosome 5 (Xie et al. 2014). Association mapping based on 5 K rice array of 249 indica rice varieties widely distributed in China determined the QTL qCTSR5-2 using the severity of damage and seed survival rate as cold-tolerant indices (Zhang et al. 2018). qLTG(I)5, qLTG(II)5 and qLTGS(I)5 were detected on chromosomes 5 and were associated with low-temperature germination (LTG) and low-temperature stress index (Najeeb et al. 2020). qCST5 affecting cold tolerance at the seedling stage was identified from the cold-tolerant variety IL112 (Liu et al. 2013). qCTSS-5 was mapped on chromosomes 5 by NGS-assisted BSA QTL method at the seedling stage (Yang et al. 2013). OsRAN2 is associated with cold tolerance of the seedling stage (Zang et al. 2010). qCTB5, qCTB-5–1 and qCTB-5–2 are related to cold tolerance and have been detected on chromosome 5 at the booting stage (Andaya and Mackill 2003b; Xu et al. 2008a, b). These cold-tolerance QTLs were detected at different stages or environments, indicating that the QTL qCtb5 is a major QTL and expressed at all growth stages. Thus, qCtb5 really exists and plays a key role in enhancing cold tolerance in rice and can be as a candidate QTL for further fine mapping and breeding application.
Meta-analysis of cold-resistance QTLs on the chromosome 5. Gray circles: QTLs for low-temperature germination reported by Xie et al. 2014, Zhang et al. 2018 and Najeeb et al. 2020. Red circles: QTLs for cold tolerance at the seedling stage reported by Liu et al. 2013 and Yang et al. 2013. Violet circles: the QTLs for cold tolerance at the booting stage reported by Andaya and Mackill 2003b and Xu et al. 2007. (Color figure online)
Conclusion
To identify QTLs related to cold tolerance at the booting and flowering stages in rice, an RIL population was constructed using the cold-tolerant japonica cultivated variety JL1 and the cold-sensitive indica cultivated variety MY23. The RIL population and the parental lines were planted under natural low-temperature conditions, and theirs SST were investigated. Fifteen QTLs were detected on chromosomes 1, 2, 3, 5, 7, 11 and 12, with LOD values ranging from 2.64 to 4.76, and with PVE ranging from 3.34 to 12.02%. Three QTLs, including qCtb1, qCtb5 and qCtb12, were repeatedly detected under different conditions and were considered stable major QTL. qCtb5 was identified on chromosome 5 between CMB0526.3 and ID5014265. In this interval or nearby, many cold-resistance QTLs have been identified in previous studies. qCtb5 is considered as a candidate region for fine mapping and cold-tolerance gene resources for breeding utilization.
Data availability
All data included in this study are available upon request by contact with the corresponding author.
References
Andaya VC, Mackill DJ (2003a) Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. J Exp Bot 54(392):2579–2585. https://doi.org/10.1093/jxb/erg243
Andaya VC, Mackill DJ (2003b) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica x indica cross. Theor Appl Genet 106(6):1084–1090. https://doi.org/10.1007/s00122-002-1126-7
Cruz RPd, Sperotto RA, Cargnelutti D, Adamski JM, FreitasTerra TD, Fett JP (2013) Avoiding damage and achieving cold tolerance in rice plants. Food Energy Secur 2(2):96–119. https://doi.org/10.1002/fes3.25
Endo T, Chiba B, Wagatsuma K, Saeki K, Ando T, Shomura A, Mizubayashi T, Ueda T, Yamamoto T, Nishio T (2016) Detection of QTLs for cold tolerance of rice cultivar ‘Kuchum’ and effect of QTL pyramiding. Theor Appl Genet 129(3):631–640. https://doi.org/10.1007/S00122-015-2654-2
Fujino K, Obara M, Shimizu T, Koyanagi KO, Ikegaya T (2015) Genome-wide association mapping focusing on a rice population derived from rice breeding programs in a region. Breed Sci 65(5):403–410. https://doi.org/10.1270/jsbbs.65.403
Guo HF, Zeng YW, Li JL, Ma XQ, Zhang ZY, Qj L, Li J, Gu YS, Zhang HL, Li JJ, Li ZC (2020) Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnol J 18(12):2491–2503. https://doi.org/10.1111/PBI.13424
Han LZ, Zhang SY, Qiao YL, Ruan RC, Zhang JG, Cao GL, Koh HJ (2005a) QTL analysis of root traits at the seedling stage in rice under cold water irrigation. Acta Agron Sin 31(11):1415–1421 (in Chinese)
Han LZ, Qiao YL, Zhang YY, Cao GL, Yea JD, Koh HJ (2005b) Identification of QTLs for cold tolerance at the booting stage in rice. Acta Agron Sin 31(5):653–657 (in Chinese)
Han LZ, Qiao YL, Zhang SY, Cao GL, Ye CR, Xu FR, Dai LY, Ye JD, Koh HJ (2005c) QTL analysis of some agronomic traits in rice under different growing environments. Scientia Agricultura Sinica 38(6):1080–1087 (in Chinese)
Jiang S, Yang C, Xu Q, Wang L, Yang X, Song X, Wang J, Zhang X, Li B, Li H, Li Z, Li W (2020) Genetic dissection of germinability under low temperature by building a resequencing linkage map in japonica rice. Int J Mol Sci 21(4):1284. https://doi.org/10.3390/ijms21041284
Jiang WZ, Lee JH, Chu SH, Ham TH, Woo MO, Cho YI, Chin JY, Han LZ, Xuan YS, Yuan DL, Xu FR, Dai LY, Yea JD, Koh HJ (2010) Genotype×environment interactions for chilling tolerance of rice recombinant inbred lines under different low temperature environments. Field Crops Res 117(2):226–236. https://doi.org/10.1016/J.FCR.2010.03.007
Jiang WZ, Jin YM, Lee JH, Lee KL, Piao RH, Han LZ, Shin JC, Jin RD, Cao TH, Pan HY, Du XL, Koh HJ (2011) Quantitative trait loci for cold tolerance of rice recombinant inbred lines in low temperature environments. Mol Cells 32(6):579–587. https://doi.org/10.1007/S10059-011-0186-4
Kuroki M, Saito K, Matsuba S, Yokogami N, Shimizu H, Ando I, Sato Y (2007) A quantitative trait locus for cold tolerance at the booting stage on rice chromosome 8. Theor Appl Genet 115(5):593–600. https://doi.org/10.1007/S00122-007-0589-Y
Li HH, Ye GY, Wang JK (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374. https://doi.org/10.1534/genetics.106.066811
Li J, Pan Y, Guo H, Zhou L, Yang S, Zhang Z, Yang J, Zhang H, Li J, Zeng Y, Li Z (2017) Fine mapping of QTL qCTB10-2 that confers cold tolerance at the booting stage in rice. Theor Appl Genet 131(1):157–166. https://doi.org/10.1007/s00122-017-2992-3
Li L, Liu X, Xie K, Wang Y, Liu F, Lin Q, Wang W, Yang C, Lu B, Liu S, Chen L, Jiang L, Wan J (2013) qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.). Theor Appl Genet 126(9):2313–2322. https://doi.org/10.1007/s00122-013-2137-2
Li SS, Wang JK, Zhang LY (2015) Inclusive composite interval mapping of QTL by environment interactions in biparental populations. PLoS ONE 10(7):e0132414. https://doi.org/10.1371/journal.pone.0132414
Liu FX, Xu WY, Song Q, Tan LB, Liu JY, Zhu ZF, Fu YC, Su Z, Sun CQ (2013) Microarray-assisted fine-mapping of quantitative trait loci for cold tolerance in rice. Mol Plant 6(3):757–767. https://doi.org/10.1093/MP/SSS161
Meng L, Li HH, Zhang LY, Wang JK (2015) QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283. https://doi.org/10.1016/j.cj.2015.01.001
McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newsl 14:11–13
Najeeb S, Ali J, Mahender A, Pang YL, Zilhas J, Murugaiyan V, Vemireddy LR, Li Z (2020) Identification of main-effect quantitative trait loci (QTLs) for low-temperature stress tolerance germination- and early seedling vigor-related traits in rice (Oryza sativa L.). Mol Breed 40(1):10. https://doi.org/10.1007/S11032-019-1090-4
Saito K, Miura K, Nagano K, Hayano-Saito Y, Araki H, Kato A (2001) Identification of two closely linked quantitative trait loci for cold tolerance on chromosome 4 of rice and their association with anther length. Theor Appl Genet 103:862–868. https://doi.org/10.1007/s001220100661
Saito K, Hayano-Saito Y, Maruyama-Funatsuki W, Sato Y, Kato A (2004) Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor Appl Genet 109(3):515–522. https://doi.org/10.1007/s00122-004-1667-z
Saito K, Hayano-Saito Y, Kuroki M, Sato Y (2010) Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci 179(1–2):97–102. https://doi.org/10.1016/j.plantsci.2010.04.004
Sasaki T, Burr B (2000) International rice genome sequencing project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3:138–141. https://doi.org/10.1016/S1369-5266(99)00047-3
Sun J, Yang L, Wang J, Liu H, Zheng H, Xie D, Zhang M, Feng M, Jia Y, Zhao H, Zou D (2018) Identification of a cold-tolerant locus in rice (Oryza sativa L.) using bulked segregant analysis with a next-generation sequencing strategy. Rice (N Y) 11(1):24. https://doi.org/10.1186/s12284-018-0218-1
Ooijen JV, Ooijen JV, Verlaat JV, Ooijen J, Tol JV, Dalen J, Buren J, Meer JVD, Krieken JV, Ooijen J, Kessel JV, Van O, Voorrips R, Heuvel LVD (2006) JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen, Netherlands
Tang JH, Ma XD, Cui D, Han B, Geng LY, Zhao ZW, Li YF, Han LZ (2019) QTL analysis of main agronomic traits in rice under low temperature stress. Euphytica. https://doi.org/10.1007/s10681-019-2507-1
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and qtls. J Hered 93(1):77–78
Xie LX, Tan ZW, Zhou YY, Xu RB, Feng LB, Xing YZ, Qi XQ (2014) Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice. J Integr Plant Biol 56(8):749–759. https://doi.org/10.1111/JIPB.12190
Xu FR, Yu TQ, Tang CF, Xin-Xiang A, Fan CZ, Hu YL, Zhang DY, Dong C, Dai LY (2008) Low-temperature response to major agronomic traits by using recombinant inbred line (RIL) populations derived from Towada/Kunmingxiaobaigu. Sci Agric Sin 41(11):3437–3447 (in Chinese)
Xu LM, Zhou L, Zeng YW, Wang FM, Zhang HL, Shen SQ, Li ZC (2008b) Identification and mapping of quantitative trait loci for cold tolerance at the booting stage in a japonica rice near-isogenic line. Plant Sci 174(3):340–347. https://doi.org/10.1016/j.plantsci.2007.12.003
Yang ZM, Huang DQ, Tang WQ, Zheng Y, Liang KJ, Cutler AJ, Wu WR (2013) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS One. https://doi.org/10.1371/JOURNAL.PONE.0068433
Zang AP, Xu XJ, Neill S, Cai WM (2010) Overexpression of OsRAN2 in rice and Arabidopsis renders transgenic plants hypersensitive to salinity and osmotic stress. J Exp Bot 61(3):777–789. https://doi.org/10.1093/JXB/ERP341
Zeng YW, Yang SM, Cui H, Yang XJ, Xu LM, Du J, Pu XY, Li ZC, Cheng ZQ, Huang XQ (2009) QTLs of cold tolerance-related traits at the booting Stage for NIL-RILs in rice revealed by SSR. Genes Genom 31(2):143–154. https://doi.org/10.1007/Bf03191147
Zhang S, Zheng J, Liu B, Peng S, Leung H, Zhao J, Wang X, Yang T, Huang Z (2013) Identification of QTLs for cold tolerance at seedling stage in rice (Oryza sativa L.) using two distinct methods of cold treatment. Euphytica 195(1):95–104. https://doi.org/10.1007/s10681-013-0977-0
Zhou L, Zeng YW, Zheng WW, Tang B, Yang SM, Zhang HL, Li JJ, Li ZC (2010) Fine mapping a QTL qCTB7 for cold tolerance at the booting stage on rice chromosome 7 using a near-isogenic line. Theor Appl Gene 121(5):895–905. https://doi.org/10.1007/s00122-010-1358-x
Zhang MC, Ye J, Xu Q, Feng Y, Yuan XP, Yu HY, Wang YP, Wei XH, Yang YL (2018) Genome-wide association study of cold tolerance of Chinese indica rice varieties at the bud burst stage. Plant Cell Rep 37(3):529–539. https://doi.org/10.1007/S00299-017-2247-4
Zhang Z, Li J, Pan Y, Li J, Zhou L, Shi H, Zeng Y, Guo H, Yang S, Zheng W, Yu J, Sun X, Li G, Ding Y, Ma L, Shen S, Dai L, Zhang H, Yang S, Guo Y, Li Z (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat Commun 8:14788. https://doi.org/10.1038/ncomms14788
Zhu YJ, Chen K, Mi XF, Chen TX, Ali J, Ye GY, Xu JL, Li ZK (2015) Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS One 10(12):e0145704
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2016YFD0100101,2016YFD0100301), the National Natural Science Foundation of China (31671664), the National Natural Sciences Foundation (31670326), Technology Innovation and Application Development Program in Chongqing (cstc2019jscx-msxmX0353), CAAS Science and Technology Innovation Program, National Infrastructure for Crop Germplasm Resources (NICGR2018-01), Protective Program of Crop Germplasm of China (2018NWB036-01, 2018NWB036-122).
Author information
Authors and Affiliations
Contributions
LZ conducted field work, generated phenotypic data, preformed data analysis and wrote the manuscript; JT generated phenotypic data and genotypic data; DC preformed the genotyping of the mapping population; CT helped for field work; XM helped for field work; XA helped for field work; BH helped for field work; GC helped for field work; ZZ designed the research and manuscript revision; H-JK designed the research and manuscript revision; LH conceived the experiment, guided experiments and manuscript revision. All authors read and approved the final version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, L., Tang, J., Cui, D. et al. Identification of QTLs for cold tolerance at the booting and flowering stages in rice (Oryza sativa L.). Euphytica 217, 214 (2021). https://doi.org/10.1007/s10681-021-02898-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-021-02898-6