Abstract
A higher understanding of genetic and genomic bases of partial resistance in plants and their diversity regarding pathogen variability is required for a more durable management of resistance genetic factors in sustainable cropping systems. In this study, we investigated the diversity of genetic factors involved in partial resistance to Aphanomyces euteiches, a very damaging pathogen on pea and alfalfa, in Medicago truncatula. A mapping population of 178 recombinant inbred lines, from the cross F83005.5 (susceptible) and DZA045.5 (resistant), was used to identify quantitative trait loci for resistance to four A. euteiches reference strains belonging to the four main pathotypes currently known on pea and alfalfa. A major broad-spectrum genomic region, previously named AER1, was localized to a reduced 440 kb interval on chromosome 3 and was involved in complete or partial resistance, depending on the A. euteiches strain. We also identified 21 additive and/or epistatic genomic regions specific to one or two strains, several of them being anchored to the M. truncatula physical map. These results show that, in M. truncatula, a complex network of genetic loci controls partial resistance to different pea and alfalfa pathotypes of A. euteiches, suggesting a diversity of molecular mechanisms underlying partial resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are exposed to numerous invading pathogenic organisms and have developed a wide array of active or passive defense mechanisms to defeat them, leading to complete or partial resistance. While the molecular bases of complete resistance conferred by resistance genes with qualitative effects are increasingly well-understood (Glazebrook 2005; Jones and Dangl 2006; Bent and Mackey 2007), knowledge concerning the molecular mechanisms underlying partial resistance, often controlled by multiple genes with small effects, remains limited. Complex genetic networks of additive and epistatic-effect loci controlling partial resistance have been identified in cereals and Brassicaceae (Li et al. 2006; Ma et al. 2006; Rowe and Kliebenstein 2008). To date, different hypotheses about the genes underlying partial resistance loci have been proposed. Poland et al. (2009) hypothesized that partial resistance loci could correspond to (a) “defeated” genes controlling complete resistance, (b) genes involved in basal defense, defense signal transduction, plant architecture or development, (c) components of chemical warfare or (d) genes previously unidentified. The spectrum of action of partial resistance loci toward pathogen races also remains unclear. Partial resistance was often thought to be non-race-specific but several studies have provided evidence that these loci can be race-specific or effective against a subset of pathogen races, especially in the case of resistance to biotrophic pathogens (Calenge et al. 2004; Rocherieux et al. 2004; Wisser et al. 2005).
Model species, such as Arabidopsis thaliana, rice and tomato, are of important interest for deciphering the genetic and molecular bases of partial resistance. They provide useful tools, including large genomic resources generated from the complete sequencing of their genome, for identifying and cloning plant disease resistance genes. Natural genetic variation toward pathogen resistance was observed within model species germplasm collections, allowing genetic studies of resistance to various pathogens to be carried out (Rose et al. 2007; Shindo et al. 2007). Studies to elucidate the genetic mechanisms controlling partial resistance in A. thaliana (Perchepied et al. 2006; Jubault et al. 2008; Rowe and Kliebenstein 2008), rice (Ramalingam et al. 2003; Carrillo et al. 2005; Yu et al. 2006; Hu et al. 2008) and tomato (Smart et al. 2003; Brouwer et al. 2004; Finkers et al. 2007) have led to progress in unraveling genes underlying partial resistance loci (Poland et al. 2009).
In 1990, Medicago truncatula was proposed as a model legume for the analysis of rhizobia–legume symbiosis (Barker et al. 1990), and then emerged as a model species for studying plant–pathogen interactions (Rose 2008). In particular, the model legume offers an original opportunity for gaining knowledge about the molecular bases of partial resistance, by comparison with increasingly well-understood symbiosis mechanisms. In particular, the molecular pathways leading to plant symbiosis were shown to share components in common with those involved in defense responses to pathogens (Samac and Graham 2007). Various levels of resistance toward legume pathogens were identified within M. truncatula germplasm collections, facilitating genetic studies to identify and isolate resistance genes (Tivoli et al. 2006). Genes or QTL have been identified in M. truncatula for resistance to root rot nematode (Dhandaydham et al. 2008), aphids (Klingler et al. 2005, 2007), Phoma medicaginis (Kamphuis et al. 2008), Colletotrichum trifolii and Erysiphe pisi (Ameline-Torregrosa et al. 2008a; Yang et al. 2008), Ralstonia solanacearum (Vailleau et al. 2007) and recently Aphanomyces euteiches (Djebali et al. 2009; Pilet-Nayel et al. 2009). In these studies, specific or broad-spectrum resistance to the pathogens studied mainly involved major genes and few quantitative trait loci.
A. euteiches Drechsler is a major soil borne oomycete pathogen, considered as biotrophic, which causes economically important damage to various legume species. A. euteiches was isolated from a number of legume species, including pea (Wicker and Rouxel 2001; Levenfors et al. 2003) and alfalfa (Malvick and Grau 2001). Studies on the pathogenic variability of A. euteiches were conducted on pea and alfalfa. On pea, Wicker and Rouxel (2001) identified two main pathotypes of A. euteiches, based on reactions of six pea genotypes: a predominant pathotype, present in France, other European countries and the USA, and a more specific pathotype, including only strains from the USA. On alfalfa, Malvick and Grau (2001) identified two races of A. euteiches, races 1 and 2, based on the reactions of three alfalfa populations (Saranac, WAPH-1 and WAPH-5). M. truncatula is a host for A.euteiches and shows genetic variability for resistance to the pea and alfalfa strains of A. euteiches (Vandemark and Grunwald 2004; Moussart et al. 2007). To date, no data are available regarding the pathogenic variability of A. euteiches on M. truncatula differential genotypes. Complete and partial levels of resistance have been identified in M. truncatula, which depend on host genotypes and A. euteiches pathotypes (Vandemark and Grunwald 2004; Moussart et al. 2007). The M. truncatula/A. euteiches pathosystem is therefore a good model to investigate the genetic bases of partial resistance and their relation with resistance genes, regarding natural variation in the host and pathogen.
Previously, genetic analysis of resistance to A. euteiches strains from the predominant pea pathotype was carried out from two different RIL populations of M. truncatula, derived from crosses involving the susceptible line F83005.5 and two different sources of resistance, DZA045.5 and A17, expressing high and partial levels of resistance, respectively. A monogenic control of the resistance was identified in both studies and major loci, named AER1 and prAe1, expressing high dominant and partial recessive genetic effects on resistance to the main pea pathotype of A. euteiches, respectively, were mapped to the distal part of chromosome 3 (Djebali et al. 2009; Pilet-Nayel et al. 2009). Partial levels of resistance to the specific American pea pathotype and the two alfalfa races described were identified in DZA045.5 (unpublished data). Till date, no loci were identified in M. truncatula for resistance to those pathotypes. The aim of this study was to analyze the diversity of genetic factors involved in resistance of M. truncatula DZA045.5 to the main pea and alfalfa pathotypes of A. euteiches. By doing this, a more extensive knowledge of the genetic architecture of resistance to A. euteiches in M. truncatula could be obtained, which is potentially transferable to leguminous crops, especially pea and alfalfa, because of the high levels of synteny between the model and cultivated legumes (Choi et al. 2004). In this study, we mapped, throughout the whole genome, additive and epistatic loci associated with resistance to four reference strains of A. euteiches belonging to the main pathotypes currently known on pea and alfalfa. We used a F5:6 recombinant inbred lines (RIL) population derived from a cross between the two lines mentioned above, F83005.5 and DZA045.5. We analyzed the specificity of the identified loci toward the strains tested and compared them to those previously identified (Djebali et al. 2009; Pilet-Nayel et al. 2009). We anchored several of the identified regions to the physical map of M. truncatula for future prospects of identifying candidate genes underlying the resistance.
Materials and methods
Pathogen material
Two pea-infecting strains of A. euteiches were used in disease resistance tests, namely RB84, isolated from an infested field at Riec-sur-Belon (Finistère, France) (Moussart et al. 2007) and Ae109 (Wicker and Rouxel 2001), isolated from an infested field in Winconsin (USA) and referred to as strain Ae467 in Malvick et al. (1998) and Malvick and Percich (1999). The RB84 and Ae109 strains belong to the main French pea pathotype of A. euteiches and the minor pea pathotype from the USA, respectively, identified by Wicker and Rouxel (2001). RB84 strain has a broad host-spectrum within legumes since it has been reported pathogenic to pea, alfalfa and M. truncatula, bean, lentil and vetch (Moussart et al. 2008). Ae109 is pathogenic to pea and alfalfa (Malvick et al. 1998). The RB84 strain was previously used in Pilet-Nayel et al. (2009). Two alfalfa-infecting strains of A. euteiches were also used in this study, namely MF-1 and NC-1, isolated in the USA (Wisconsin and North Carolina, respectively). MF-1 and NC-1 belong to race 1 (R1) and race 2 (R2) of A. euteiches, respectively, identified by Malvick and Grau (2001). These two strains have a narrow host-range, since they were reported only pathogenic to alfalfa, but not to pea, bean or red-clover (Malvick et al. 1998).
Plant material
A population of 178 F5:6-derived RILs, produced by single seed descent from a cross between two M. truncatula inbred line accessions, F83005.5 (female parent, originating from southern France) and DZA045.5 (male parent, originating from Algeria), was used in this study. Previously, the F5 RIL generation was used for genetic analysis of the resistance (Pilet-Nayel et al. 2009) and for construction of a genetic map (Avia et al., submitted). In this study, the F6 RIL generation was used for resistance evaluation in disease screening tests. The two parental lines, F83005.5 and DZA045.5, are included in the smallest core-collection set (eight lines) built by Ronfort et al. (2006) (M. truncatula stock center, INRA, France; http://www.montpellier.inra.fr/BRC-MTR/). F83005.5 and DZA045.5 were shown to be susceptible and resistant to the RB84 French strain, respectively (Moussart et al. 2007). Next, these lines were also observed to express contrasted responses when inoculated with Ae109, MF-1 and NC-1 (unpublished data). Two Pisum sativum genotypes, Baccara (F. Desprez, France) and PI180693 (USDA Plant Introduction Station, USA), susceptible and partially resistant to A. euteiches, respectively (Wicker et al. 2003), were used as controls in all disease-resistance experiments. One additional pea genotype, MN313 (Davis et al. 1995), resistant to Ae109 and susceptible to RB84 was included in the disease-resistance tests using the Ae109 strain. Three M. sativa cultivars, Saranac (susceptible to R1 and R2 strains of A. euteiches), WAPH-1 (resistant to R1 strains and susceptible to R2 strains) and WAPH-5 (resistant to R1 and R2 strains), were added as controls in the disease-resistance tests using the two alfalfa MF-1 and NC-1 strains (Malvick and Grau 2001).
Inoculation procedures and resistance evaluation
All tests to screen for resistance to A. euteiches were performed as described by Pilet-Nayel et al. (2009). Each RIL test was conducted using a randomized complete block design with two blocks. In each block, five plants per recombinant line, parent and control line were grown in a pot, in a moistened vermiculite substrate and were then inoculated with one strain and evaluated. Each RIL test was repeated twice for each of the four strains. The screening tests were conducted in a growth chamber at 25°C for 16 h day and 23°C for 8 h night. Twelve days after sowing, seedlings were inoculated with a suspension of zoospores at a concentration adjusted to 2 × 103 zoospores/ml for the pea strains and 102 zoospores/ml for the alfalfa strains. The method for producing the inoculum was described by Moussart et al. (2001). Seedlings were inoculated by applying 25 ml of inoculum suspension per pot. Vermiculite was saturated with water after inoculation to favor disease development. Fourteen days after inoculation, the plants were uprooted and disease severity was scored on each individual plant using the 0-5 disease scoring scale described in Moussart et al. (2007) and modified in Pilet-Nayel et al. (2009). A root rot index (RRI) ranging from 0 to 5 was then calculated as the mean disease score on all plants in a pot.
Statistical analyses of the disease resistance data
The phenotypic data obtained from each pair of resistance tests per A. euteiches strain were statistically analyzed using a generalized linear model [(PROC GLM of Statistical Analysis System (SAS) software (SAS Institute Inc., 2000)], as followed: Pijk = μ + Li + Tj + Bk/j + (Li × Tj) + eijk, where Pijk is the mean disease score of the ith RIL located in the kth block of the jth test, μ is the mean of all the data, Li is the RIL i effect, Tj is the test j effect, Bk/j is the block k effect in the jth test, Li × Tj is the RIL i × test j interaction effect, and eijk is the residual. Normality of residues and homogeneity of variances were checked using Skewness, Kurtosis and Shapiro–Wilk (P ≥ 0.05) statistics and Bartlett’s test (P > 0.05), respectively. Broad sense heritability (h 2) was estimated from ANOVA using the formula h 2 = σ 2 g/[σ 2 g + (σ 2 e/n)], where σ 2 g is the genetic variance, σ 2 e the residual variance and n is the number of replicates per line. As missing data were observed from our experiments, RIL least-square means were estimated from ANOVA and used for linkage analysis. Spearman correlation coefficients (r 2) were calculated from RRI adjusted means obtained between the four strains and between the two RIL generations tested for RB84 strain (F5 in Pilet-Nayel et al. 2009 and F6 in this study), using the PROC CORR procedure of the SAS software.
Molecular markers and genetic linkage analysis
A framework genetic map, mainly based on SSR markers, was developed from the F83005.5 × DZA045.5 F5-RIL population to identify QTL for freezing tolerance (Avia et al., submitted). This genetic map included 178 markers, distributed over eight linkage groups (LG) and covered 1,197 cM Kosambi (≈1,317 cM Haldane). In this study, we genotyped the RIL population with additional SSR markers designed from M. truncatula BAC accessions, anchored to the integrated genetic-physical map (http://www.medicago.org/genome/map.php, Univ. Minn. 2006). Information on SSR primer sequences and BAC anchoring is described in Mun et al. (2006), on the Medicago website (http://www.medicago.org/genome/genetic_map_table.html) and in Djebali et al. (2009), except for the SSR MTIC1179 (marker information: BAC accession CR940308; forward primer: 5′-AGTGTGATTTTTACACCAAAGA-3′; reverse primer: 5′-TGTCAAGCTTCAGTTTTTCC-3′; motif: (TA)6; C. Jacquet, personal communication). SSR markers from Mun et al. (2006) are named “MtB” followed by the SSR number and SSR markers from Djebali et al. (2009) are designated “MTIC” followed by the SSR number. SSR amplification reactions were carried out using the procedure described for pea by Loridon et al. (2005). Mendelian segregation of parental alleles in the RIL population (1:1 ratio) was tested for each marker by a Chi-square test (α = 0.01). Additional SSR markers were positioned within the existing framework map (Avia et al., submitted), using the “assign” and “try” commands of MAPMAKER/EXP version 3.0b (Lincoln et al. 1992). All genetic distances were expressed in centimorgan values, calculated with the Haldane mapping function.
Additive-effect QTL analysis
For QTL analysis, composite interval mapping (CIM, Zeng 1994) was performed using Windows QTL Cartographer 2.5 software (Wang et al. 2005). We used the standard model 6 of the program with ten cofactors selected by forward–backward regression (P < 0.05) and a window size of 10 cM. Walk speed was set at 2.0 cM to scan the entire genome. This procedure estimated the log-likelihood (LOD) score, additive effect and percentage of phenotypic variance (R 2) every 2.0 cM along each chromosome. A LOD threshold of 2.9 was defined for QTL detection (α = 0.05) with Ae109, MF-1 and NC-1 and a LOD threshold of 3.0 was defined for QTL detection (α = 0.05) with the RB84 strain, using permutation tests (n = 1,000) according to the method of Churchill and Doerge (1994). Two-LOD support intervals were established as >95% QTL confidence intervals (Van Ooijen 1992) from the CIM module. QTL were considered to co-localize if their 2-LOD confidence intervals overlap. Additive-effect QTL were named “Ae-Mt” (for A phanomyces e uteiches M edicago t runcatula) followed by the LG number and the QTL number within the LG.
Epistatic-effect QTL analysis
We searched for pairwise epistatic interactions between all possible marker couples of the genetic map, using a two-way ANOVA model with an interaction component, implemented in SAS software (SAS/IML, SAS 1989), as followed : Pijk = μ + Xi + Yj + (Xi × Yj) + eijk where Pijk is the least-square mean disease score of the kth RIL within the population, μ the mean of all the data, Xi the ith genotype effect at the X locus, Yj the jth genotype effect at the Y locus, Xi × Yj the interaction effect between the ith genotype at the X locus and the jth genotype at the Y locus, and eijk is the residual. The search for pairwise epistatic interactions was conducted using a SAS program adapted from the EPISTACY program (Holland 1998). To survey the whole genome for pairwise epistatic effects, we evaluated 20,100 [n(n − 1)/2] possible interactions for a map of n = 201 markers. The threshold to claim a statistically significant interaction was set at P < 2.5 × 10−4 and R 2 < 5%. The threshold (P < 2.5 × 10−4) corresponded to the number of false positive interactions expected, set at five in this study, divided by the total number of marker pairs tested (Holland 1998). This threshold calculation method has been used in different studies (Jubault et al. 2008; Liu et al. 2006; Manzanares-Dauleux et al. 2000). We preferred this liberal but reasonable threshold rather than the Bonferroni correction that is considered to be overly conservative when a large number of tests is performed (Ungerer and Rieseberg 2003). The percentage of phenotypic variation explained by all the QTL detected for each strain (R 2) was estimated with a full ANOVA model including all additive and pairwise epistatic regions detected. Epistatic-effect QTL were named “Ae-MtE” (for A phanomyces e uteiches M edicago t runcatula Epistasis) followed by the epistatic interaction number.
Identification of physical genomic regions associated with QTL
The list of BAC accessions included within the support interval of the A. euteiches resistance QTL identified was established from the position of the upper and lower bound markers, using the v2.0 of the draft genome assembly generated by the MGSC (http://www.medicago.org/genome/), with gene prediction from the International Medicago Genome Annotation Group (IMGAG). Genes predicted on these BAC accessions were examined to identify those known to be involved in disease resistance in plants.
Results
Phenotyping of the RIL population
The mean adjusted RRI scores obtained on pea controls inoculated with RB84, Ae109, MF-1, and NC-1 were 4.93, 4.51, 0.10 and 0.29, respectively, for Baccara and 3.57, 3.95, 0.12, and 0.36, respectively, for PI180693. These results confirmed that the alfalfa strains, MF-1 and NC-1, are not pathogenic to pea. In the Ae109 disease-resistance test, MN313 was more resistant than PI180693 (RRI = 3.04 and 3.95, respectively) confirming that Ae109 belongs to the US-specific pathotype, as described by Wicker and Rouxel (2001). The mean adjusted RRI scores obtained on alfalfa controls inoculated with MF-1 and NC-1 were 2.63 and 2.61, respectively, for Saranac, 1.78 and 2.86, respectively, for WAPH-1 and 1.36 and 2.05, respectively, for WAPH-5. These results confirmed that the MF-1 and NC-1 strains belong to race 1 (R1) and race 2 (R2) of A. euteiches, respectively, as previously described (Malvick et al. 2009).
The main results of the variance analysis (ANOVA) obtained from RRI scores with the four strains are summarized in Table 1. For the four strains, ANOVA showed highly significant genotypic and disease test effects (P < 0.005) on the phenotypic variation observed. No significant genotype × test interaction effects were identified (P > 0.05), except with the NC-1 strain (P = 0.02).
Mean-based heritabilities were very high in the RB84 and Ae109 strain tests (h 2 = 0.93 and 0.73, respectively). They were lower in the MF-1 and NC-1 strain tests (h 2 = 0.45 and 0.39, respectively) in accordance with smaller phenotypic ranges obtained with both these strains. Error variances obtained in the variance analysis for the MF-1 and NC-1 strains were higher than those obtained for RB84 and Ae109 (0.39, 0.33, 0.27, 0.19 for MF-1, NC-1, RB84, and Ae109, respectively) suggesting that the results obtained with the alfalfa strains were less reproducible than with the pea strains.
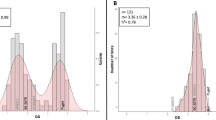
Distribution of RRI adjusted means in the F6-RIL with RB84 strain showed a bimodal curve (Fig. 1a), as previously observed in the F5-RIL (Pilet-Nayel et al. 2009) \( (r_{{{\text{F5}} - {\text{F6 RILs}}}}^{2} = 0.89). \) The RRI adjusted means obtained with Ae109, MF-1, and NC-1 showed normal distributions, with disease severity median values of 2.83, 2.59 and 2.53, respectively (Fig. 1b–d). The mean adjusted RRI scores obtained for parental lines inoculated with the RB84, Ae109, MF-1, and NC-1 strains were 3.75, 3.4, 2.82, and 3.26, respectively, for F83005.5 and 1.23, 2.03, 2.2, and 2.14, respectively, for DZA045.5. In all experiments, transgressive segregations were observed, with lines that were more resistant and/or susceptible than the parents (Fig. 1).
Spearman correlation coefficients were highly significant between RRI adjusted means from resistance tests using strains RB84, Ae109, and MF-1 (Table 2). MF-1 RRI adjusted means were moderately correlated to NC-1 RRI scores. The RRI scores obtained for NC-1 were not significantly correlated with those observed for RB84 and Ae109.
Genetic mapping
QTL analysis was conducted using a genetic map comprising 201 markers, distributed over eight LG, and covering 1,434 cM Haldane (≈1,308 cM Kosambi) (Fig. 2). Compared to the framework genetic map described by Avia et al. (submitted), this updated genetic map included 23 additional markers, including (a) 21 SSR markers anchored to the integrated genetic-physical map (Mun et al. 2006) (MtB2, MtB205, MtB45, and MtB304 on LG2; MtB319, MtB141, MtB76, MtB6, MtB122, MtB172, and h2_11A20a on LG3; MtB253, MtB34, MtB66, MtB306, MtB124, and MtB289 on LG7; MtB311, MtB247, MtB225, and MtB3 on LG8) and (b) two SSR markers designed from the M. truncatula BAC accessions AC135103 and CR940308, containing the prAe1 recessive QTL previously associated with resistance of A17 to A. euteiches on LG3 (MTIC742, Djebali et al. 2009), MTIC1179, C. Jacquet, personal communication. Out of the 23 additional markers mapped, 12 markers did not segregate according to expected Mendelian ratios at α = 1%. Among these 12 markers, one marker (MtB122) favored a higher frequency of F83005.5 alleles and 11 markers favored the DZA045.5 parental alleles (MtB2, MtB45, and MtB304 on LG2; MtB253, MtB34, MtB66, MtB306, MtB306, MtB124, and MtB289 on LG7; MtB225 and MtB3 on LG8). All the markers were mapped as expected, according to their position on the integrated genetic-physical map.
a Genomic localization, on the eight linkage groups (LG) of M. truncatula, of QTL detected for resistance to the RB84, Ae109, MF-1 and NC-1 strains of A. euteiches from the F83005.5 × DZA045.5 F5:6-RIL population. Additive and epistatic-effect QTL are referenced to the Tables 3 and 4, respectively, and are colored in blue, purple, red and green for RB84, Ae109, MF-1 and NC-1, respectively. Lengths of additive-effect QTL boxes correspond to the LOD-1 support interval (from the peak marker) and QTL lines are extended to the LOD-2 support interval. Markers associated with additive and epistatic-effect QTL are indicated. The 23 additional markers, compared to the framework genetic map described by Avia et al. (submitted) are indicated by one asterisk. SSR markers are coded “MtB” (Mun et al. 2006), “MTIC” (Djebali et al. 2009; C. Jacquet, personal communication) or h2 11A20a (http://www.medicago.org/genome/map.php. Univ. Minn. 2006). AFLP markers are coded “E12M”, “E13M” or “E14M” and RAPD markers are “L07.600” and “M10.950” (Avia et al., submitted). The size of each LG is given in cM Haldane. b The AER1 genomic region. The length of the additive-effect QTL box for resistance to Ae109 strain correspond to the LOD-1 support interval (from the peak marker) and QTL lines are extended to the LOD-2 support interval. Markers involved in epistatic interactions in the AER1 genomic region involved in resistance to MF-1 and NC-1 strains are indicated by red and green arrows, respectively. Genetic distances between markers are indicated on the left of the linkage group, in centimorgans Haldane (cM). The highest probable position of AER1 obtained for resistance to RB84 strain using qualitative disease scores is indicated in blue. The name of BAC accessions included within the AER1 genomic region are indicated by black arrows
Additive-effect QTL mapping
From the F83005.5 × DZA045.5 F5:6-RIL population, a total of 11 additive-effect QTL and one major-effect locus associated with resistance to the four strains of A. euteiches were detected over seven LG of the M. truncatula genetic map (Fig. 2a). QTL results are detailed in Table 3.
As expected, the major dominant AER1 locus, previously identified from the F83005.5 × DZA045.5 F5-RIL population for resistance to RB84 (Pilet-Nayel et al. 2009), was detected again from the F6-RIL population with the same strain using quantitative disease scores. We mapped additional markers to gain more precise information about its position at the top of LG3 and estimated the confidence interval at 2.6 cM Haldane (≈2.1 cM Kosambi). The closest SSR marker to AER1 was MtB199, which was anchored to the same M. truncatula BAC accession as the MTIC742 SSR marker. MTIC742 was closely linked to the prAe1 locus detected from a A17 × F85005.5 RIL population when evaluated for resistance in in vitro infection assays (Djebali et al. 2009) using the ATCC201684 strain, belonging to the same pathotype as RB84 (data not shown). At the LOD score peak, AER1 explained 78.9% of the total phenotypic variation observed for resistance to RB84. Genomic localization of AER1 was also identified using qualitative disease scores (resistant and susceptible) obtained in the RIL population for resistance to RB84. Using qualitative scores, the AER1 locus was localized on LG3 between the MTIC1179 and MTIC742 markers, at estimated genetic distances of 0.9 and 0.7 cM from the closest markers MTIC1179 and MTIC742, respectively (Fig. 2b). A QTL for resistance to Ae109 was also identified in the same genomic region, close to the MtB319 marker. At the LOD score peak, it explained 25.5% of the total phenotypic variation observed for resistance to Ae109. In this region, the DZA045.5 alleles contributed to resistance to both strains. The AER1 genomic region was anchored by four markers (MtB199, MTIC742, MTIC1179, and MtB319) to contig 953 of the M. truncatula physical map.
On LG1 and LG8, two genomic regions, Ae-Mt1.1 and Ae-Mt8.1, were identified with both the Ae109 and MF-1 strains, which individually explained 4–10.5% of the phenotypic variation, depending on the QTL and the strain. At these two regions, the DZA045.5 alleles contributed to resistance to the two strains. The Ae-Mt8.1 genomic region was anchored by the MtB311 marker to contig 214 of the M. truncatula physical map.
Nine QTL were identified with a single A. euteiches strain, individually accounting for 4.5–14.5% of the phenotypic variation: on LG3 (Ae-Mt3.2) with Ae109; on LG2 (Ae-Mt2.1), LG3 (Ae-Mt3.1), LG4 (Ae-Mt4.1) and LG7 (Ae-Mt7.1 and Ae-Mt7.2) with MF-1; on LG2 (Ae-Mt2.2), LG5 (Ae-Mt5.1) and LG8 (Ae-Mt8.2) with NC-1. For three of these QTL (Ae-Mt2.1, Ae-Mt2.2, and Ae-Mt4.1), resistant alleles were contributed by the F83005.5 susceptible parent line. Six of these QTL (Ae-Mt2.1, Ae-Mt3.1, Ae-Mt3.2, Ae-Mt5.1, Ae-Mt7.2, and Ae-Mt8.2) were anchored to the M. truncatula physical map by the closest markers indicated in Table 3.
Epistatic-effect QTL mapping
A total of 12 significant pairwise epistatic interactions was identified among all marker pairs of the genetic map, for resistance to the four strains studied, that individually accounted for up to 11.5% of the phenotypic variation (Fig. 2a; Table 4). Most of the significant digenic interactions (11 out of 12) were identified with the alfalfa strains, especially MF-1. One significant interaction, contributing to resistance to the NC-1 strain (Ae-MtE11), was found between the major additive effect genomic region AER1 (detected with RB84 and Ae109) and a genomic region that shows a significant additive effect on resistance to the MF-1 strain (QTL Ae-Mt7.2). The two genomic regions implicated in the Ae-MtE11 epistatic interaction were anchored to the M. truncatula physical map (Fig. 2b). Seven significant interactions (Ae-MtE1, Ae-MtE3, Ae-MtE4, Ae-MtE5, Ae-MtE8, Ae-MtE10, and Ae-MtE12) were found between a genomic region with a significant additive effect on resistance (including the AER1 genomic region, QTL Ae-Mt1.1, Ae-Mt3.1, and Ae-Mt7.2) and a genomic region that did not have an additive effect on resistance. These epistatic interactions could contribute to resistance to different strains from those for which additive-effect loci involved in the interactions were detected. Particularly, the AER1 genomic region was involved in the Ae-MtE8 interaction contributing to resistance to MF-1. Four significant interactions (Ae-MtE2, Ae-MtE6, Ae-MtE7, and Ae-MtE9) were identified between genomic regions that did not have an additive effect on resistance to A. euteiches. For five of the total significant epistatic interactions identified (Ae-MtE2, Ae-MtE6, Ae-MtE, Ae-MtE10, and Ae-MtE12), F83005.5 alleles contributed to increase levels of resistance to the alfalfa strains, at loci that did not have an additive effect on resistance to those particular strains. Seven markers out of the 24 markers implicated in the significant pairwise epistatic interactions were shown to have individual significant effect on the variable tested.
For each of the four strains, the part of phenotypic variation explained by all the additive-effect QTL detected was higher than that explained by all the epistatic-effect QTL, especially for the pea strains. In the full ANOVA model including the additive and epistatic QTL detected, the overall additive and epistatic R 2 values were 78.2 and 2.9%, respectively, for resistance to RB84, 31.2 and 4.0%, respectively, for resistance to Ae109, 13.8 and 10.9%, respectively, for resistance to MF-1 and 25.7 and 11.0%, respectively, for resistance to NC-1.
Physical genomic regions underlying Aphanomyces resistance loci
The AER1 genomic region, identified for the four strains with an additive (RB84 and Ae109) or epistatic effect (MF-1 and NC-1), was anchored to three M. truncatula BAC accessions, including the same BAC accessions, AC135103 and CR940308, as the region prAe1 (Djebali et al. 2009) and the BAC accession AC144723 (Fig. 2b). It covered a region of 440 kb, including 89 annotated genes (BAC AC135103, CR940308, CT030192, CT963080, and AC144723), among which: (a) the 13 candidate genes identified by Djebali et al. (2009) in a 135 kb region [nine genes encoding putative cyclin-like F-box proteins, a ubiquitin-associated enzyme, a cinnamyl alcohol gene (CAD), one vegetative cell wall protein gp1 precursor and a gene with homology to ethylene insensitive 3], (b) three genes encoding disease-resistance proteins, including a RPM1-like protein (BAC AC135103), (c) nine other genes encoding putative cyclin-like F-box proteins (BAC CT030192 and CT963080), and (d) two genes encoding late nodulins (BAC CT030192).
One additive-effect genomic region identified with both the Ae109 and MF-1 strains (Ae-Mt8.1) and six additive and/or epistatic-effect QTL detected with one strain (Ae-Mt2.1, Ae-Mt3.1, Ae-Mt3.2, Ae-Mt5.1, Ae-Mt7.2, and Ae-Mt8.2) were also anchored to the M. truncatula physical map (Table 3). A total of 315 annotated genes were identified on 18 BAC accessions covering the seven genomic regions associated with resistance to one or two strains of A. euteiches. Among these genes, 36% were identified as of hypothetical proteins. For each genomic region, BAC identification and prediction of a selection of genes known to be involved in plant disease resistance are summarized in Table 5.
Discussion
A complex network of loci controls resistance to different pathotypes of A. euteiches in M. truncatula
This study is the first report evaluating the diversity of genetic factors involved in resistance of M. truncatula, toward the pathogenic variability of A. euteiches described on pea and alfalfa. A total of 12 additive and 12 epistatic genetic factors, distributed over the eight M. truncatula chromosomes, were identified for resistance to A. euteiches, using two pea and two alfalfa-infecting strains from France and the USA (Fig. 2).
AER1, a major broad-spectrum genomic region
The AER1/prAe1 genomic region was detected on LG3 for different sources of resistance (DZA045.5 and A17, respectively) expressing different genetic effects (dominant and recessive, respectively) leading to high or partial levels of resistance in the DZA045.5 and A17 lines, respectively (Djebali et al. 2009; Pilet-Nayel et al. 2009). In this region, the AER1 locus confidence interval, detected for resistance to RB84, spans the same two BAC accessions as the prAe1 locus confidence interval. The AER1 and prAe1 loci may therefore correspond to different alleles of a same locus or to different closely linked loci. In this study, the AER1 region had a high or reduced additive effect on resistance to the RB84 and Ae109 strains, respectively, and an epistatic effect on resistance to MF-1 and NC-1. The AER1 region therefore has a broad-spectrum of action on pea and alfalfa-infecting A. euteiches strains, acting either alone or by interacting with other genes for conferring expression of resistance. These results suggest that the AER1 genomic region contains a resistance gene or multiple closely linked genes contributing to high levels of resistance in M. truncatula against RB84-pathotype strains occurring in Europe and the USA. In the case of the existence of a resistance gene underlying the AER1 genomic region, the resistance gene may have evolved under selection pressure from other pathotypes of A. euteiches occurring in the USA, resulting in its residual additive or epistatic expression against those pathotypes. However, it is also possible that the AER1 genomic region includes different closely linked genes, contributing to the resistance to different A. euteiches pathotypes, as observed in other pathosystems (Huang et al. 2004).
Ae-Mt8.1 and Ae-Mt1.1, two genomic regions associated with resistance to two A. euteiches strains isolated from different hosts
The Ae-Mt1.1 and Ae-Mt8.1 genomic regions, with additive and/or epistatic effects on resistance to A. euteiches, were common to the pea-infecting Ae109 and alfalfa-infecting MF-1 strains. A third genomic region, Ae-Mt7.1, detected for significant resistance to MF-1, was also associated with resistance to Ae109 with a LOD score close but lower than the LOD threshold defined (LOD = 2.7 and R 2 = 3%, data not shown). These results suggest that some common genes in M. truncatula could control resistance to both the Ae109 and MF1 strains, which may interact with common pathogenicity factors to the two A. euteiches strains, despite their different host-specificities (MF-1 is pathogenic on alfalfa only, while Ae109 is able to infect both pea and alfalfa; Malvick et al. 1998). Ae109 and MF-1 were both isolated in Wisconsin (USA) from pea and alfalfa, respectively (Malvick and Percich 1999; Malvick and Grau 2001), where A. euteiches was first described as an important pathogen of pea in 1925, and was associated with root rot in alfalfa in 1927 (Jones and Drechsler 1925; Linford 1927). Peas have been grown intensively in Wisconsin for the last century and the central USA also has fields with a history of alfalfa production (Malvick et al. 2009). Holub et al. (1991) showed that 97% of isolates from pea fields in Wisconsin were able to infect alfalfa. Recently, reactions of the three reference alfalfa populations (Saranac, WAPH-1 and WAPH-5) to infection by Ae109 were found to be typical of responses observed with race 1 strains (Malvick et al. 2009) (data not shown). Genotypic relationships, based on RAPD, between 62 strains of A. euteiches, showed that Ae109 and MF-1 are close genetically and are closer than MF-1 and NC-1 (Malvick et al. 1998). These findings suggest that Ae109 and MF-1 may be derived from each other or from a common ancestral isolate, and evolved through their adaptation to different hosts (pea and/or alfalfa). Consequently, these strains could share common pathogenicity genes which may interact with common genes involved in resistance in the host M. truncatula plant.
Ae-Mt3.1 and Ae-Mt7.2, two genomic regions associated with resistance to two strains isolated from a same host
Two other genomic regions (Ae-Mt3.1 and Ae-Mt7.2) were identified with additive or epistatic effects on resistance to the two alfalfa strains. In addition, two minor additive-effect QTL (data not shown) were identified on LG2 (near the marker MtB2, LOD = 2.7 and R 2 = 5%) and LG4 (near the marker MTIC249, LOD = 2.6 and R 2 = 5%) for resistance to NC-1, with a LOD score close to but lower than the defined LOD threshold, close to markers having a significant epistatic effect on resistance to MF-1. These results suggest that common or linked genes may control resistance to strains from different pathotypes isolated from a same host that would share genetic similarities. However, seven additive-effect QTL and 12 loci involved in epistatic interactions were also significantly detected for a single strain, mainly MF-1, which indicates that there are also specific genes controlling resistance to the different pathotypes described on pea and alfalfa.
At present, knowledge of A. euteiches genetic diversity is limited (Malvick et al. 1998; Grunwald and Hoheisel 2006; Akamatsu et al. 2007). Further studies and tools would be valuable to investigate genetic relationships among strains and the diversity of genetic factors controlling pathogenicity in A. euteiches, leading to a better understanding of the diversity of genetic loci involved in resistance to A. euteiches pathotypes in M. truncatula. Recently, two cDNA libraries were constructed (Gaulin et al. 2008), to generate a unigene set of about 8,000 sequences that was organized in a database named AphanoDB (Madoui et al. 2007). This genomic resource would be useful for developing new molecular markers to elucidate A. euteiches genetic diversity and study pathogen gene expression during the M. truncatula/A. euteiches interaction.
Genomic bases of partial resistance to A. euteiches in M. truncatula
Molecular mechanisms and genes involved in partial resistance are currently poorly understood. Hypothesis about function of genes underlying partial resistance loci was recently reviewed in Poland et al. (2009). Based on our results, we hypothesize that partial resistance is controlled by a diversity of mechanisms which depend on pathogen and plant genotypes, have some overlap with complete resistance and involve a network of genes expressing different genetic effects. A few hypotheses about function of genes underlying genetic factors identified for resistance to A. euteiches in M. truncatula can be discussed.
The AER1/prAe1 genomic region, conferring resistance against a broad-spectrum of A. euteiches pathotypes, is involved in complete or partial resistance, depending on the strain and the plant genotype. Similar results were previously observed in other pathosystems, leading to the hypothesis that resistance QTL may be weak forms of R-genes, such as in apple (Soufflet-Freslon et al. 2008), Brassicaceae (Manzanares-Dauleux et al. 2000; Rocherieux et al. 2004) or pepper (Caranta et al. 1997). Li et al. (1999) also suggested that “defeated” major resistance genes can have residual effects on different races of the same pathogen or different pathogens. However, the first gene cloned associated with partial resistance, namely pi21 for blast resistance in rice, is not a classical RGA (Fukuoka et al. 2009). In our study, the AER1/prAe1 genomic region, anchored to M. truncatula BAC contig 953 at one end of LG3, was localized to the same region as a supercluster of nucleotide binding site leucine-rich repeat (NBS-LRR) genes, encoding approximately 40% of all coiled-coil-NBS-LRRs, including three coiled-coil-NBS-LRRs from the BAC AC135103 (Ameline-Torregrosa et al. 2008b). However, Djebali et al. (2009) fine mapped the prAe1 locus to a 135 kb sequence interval, within which no RGA was identified. In this study, we showed that the 440 kb broad-spectrum AER1 genomic region overlaps with this same prAe1 135 kb interval and particularly includes (a) genes encoding proteins associated with the proteasome (cyclin-like F-box proteins, ubiquitin-associated enzyme) and defense responses (hydroxyproline-rich glycoprotein family, cinnamyl alcohol dehydrogenase) (Djebali et al. 2009), (b) three genes encoding disease-resistance proteins, including a RPM1-like protein, that was shown, in A. thaliana, to confer dual specificity to pathogens expressing either of two unrelated Pseudomonas syringae avr genes (Grant et al. 1995), and (c) two genes encoding late nodulins. In legumes, Graham et al. (2004) showed that nodule-specific genes encoded cystein-cluster proteins (CCPs), such as late nodulins, have strong similarities to plant defensins and proposed that nodule-specific CCPs are induced as a secondary defense to protect the nodule from pathogenic organism, while allowing the symbiosis to continue.
Among the genes known to be involved in disease-resistance plant underlying the four genomic regions associated with resistance to two strains anchored to the physical map, the most striking features were (a) the presence of a cluster of nine lipoxygenase encoding genes at Ae-Mt8.1, (b) genes involved in defense signal transduction, such as a WRKY transcription factor, serine/threonine protein kinases and cyclin-like F-box proteins at Ae-Mt3.1 and (c) a cluster of three heat shock proteins at Ae-Mt7.2. Lipoxygenases play a role in signal transduction and are toxic to microbes (Rosahl 1996). In A. thaliana, Hubert et al. (2003) showed that mutations in a heat shock protein can modulate RPM1 function. Interestingly, the Ae-Mt7.2 QTL, containing a cluster of heat shock proteins, was shown to interact with the AER1 genomic region, including a RPM1-like protein, for contributing to resistance.
Among the three other QTL anchored to the M. truncatula physical map, associated with resistance to one strain, two QTL (Ae-Mt5.1 and Ae-Mt8.2) particularly contain NBS-LRR genes, which may be in accordance with the narrow-specificity of these resistance QTL (Poland et al. 2009). We also identified genes involved in defense responses, such as a hydroxyproline-rich glycoprotein family, at Ae-Mt2.1.
Among the total eight genomic regions anchored to the physical map, one striking feature is the presence of genes encoding cyclin-like F-box proteins underlying four genomic regions (AER1, Ae-Mt2.1, Ae-Mt3.1, and Ae-Mt8.1), suggesting another hypothesis that these genes may correspond to paralogs playing a role in resistance to A. euteiches. In A. thaliana, Bikard et al. (2009) suggested that epistatic interactions between loci controlling a recessive embryo lethality could be explained by divergent evolution occurring among paralogs of an essential duplicate gene. In the M. truncatula genome, duplication events remain poorly known.
Many other genes, not or less known to be involved in disease resistance in plants, have been identified underlying confidence intervals of the identified QTL. Further fine mapping and functional validation studies will be necessary to conclude on the function(s) of genes underlying A. euteiches resistance in M. truncatula.
Conclusion
This study provides insight into the complex architecture of genetic factors involved in the resistance of M. truncatula, toward the pathogenic variability of A. euteiches described on pea and alfalfa. We identified a major broad-spectrum genomic region, namely AER1, four additive and/or epistatic genomic regions associated with resistance to two strains and several additive or epistatic-effect loci associated with resistance to a specific strain.
It will be of significant interest to isolate the gene(s) underlying the AER1 region associated with broad-spectrum resistance to A. euteiches in order to study its (their) regulation and regulated molecular pathways leading to complete or partial resistance. Consequently, the gene functions underlying a part of the genetic network controlling resistance to A. euteiches may be elucidated.
Furthermore, transferring knowledge on the genetics and genomics of resistance to A. euteiches from the model legume to cultivated leguminous crops infected by A. euteiches (especially pea and alfalfa), in which few (Pilet-Nayel et al. 2005) or no genetic mapping studies of the resistance have been developed, would also be of great interest for optimizing breeding strategies for resistance to A. euteiches in legumes.
References
Akamatsu HO, Grunwald NJ, Chilvers MI, Porter LD, Peever TL (2007) Development of codominant simple sequence repeat, single nucleotide polymorphism and sequence characterized amplified region markers for the pea root rot pathogen, Aphanomyces euteiches. J Microbiol Methods 71:82–86
Ameline-Torregrosa C, Cazaux M, Danesh D, Chardon F, Cannon SB, Esquerre-Tugaye MT, Dumas B, Young ND, Samac DA, Huguet T, Jacquet C (2008a) Genetic dissection of resistance to anthracnose and powdery mildew in Medicago truncatula. Mol Plant Microbe Interact 21:61–69
Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB (2008b) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146:5–21
Barker DG, Bianchi S, Blondon F, Dattee Y, Duc G, Essad S, Flament P, Gallusci P, Genier G, Guy P, Muel X, Tourneur J, Denarie J, Huguet T (1990) Medicago truncatula, a model plant for studying the molecular genetics of the rhizobium–legume symbiosis. Plant Mol Biol Report 8:40–49
Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45:399–436
Bikard D, Patel D, Le Mette C, Giorgi V, Camilleri C, Bennett MJ, Loudet O (2009) Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323:623–626
Brouwer DJ, Jones ES, St Clair DA (2004) QTL analysis of quantitative resistance to Phytophthora infestans (late blight) in tomato and comparisons with potato. Genome 47:475–492
Calenge F, Faure A, Goerre M, Gebhardt C, Van de Weg WE, Parisi L, Durel CE (2004) Quantitative trait loci (QTL) analysis reveals both broad-spectrum and isolate-specific QTL for scab resistance in an apple progeny challenged with eight isolates of Venturia inaequalis. Phytopathol 94:370–379
Caranta C, Lefebvre V, Palloix A (1997) Polygenic resistance of pepper to potyviruses consists of a combination of isolate-specific and broad-spectrum quantitative trait loci. Mol Plant Microbe Interact 10:872–878
Carrillo G, Wu J, Liu B, Sugiyama N, Ona I, Variar M, Courtois B, Leach JE, Goodwin PH, Leung H, Cruz CMV (2005) Association of candidate defense genes with quantitative resistance to rice blast and in silico analysis of their characteristics. Rice is life: scientific perspectives for the 21st century. In: Proceedings of the World Rice Research Conference held in Tsukuba, Japan, 4–7 November 2004, pp 479–482
Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, Young ND, Cook DR (2004) Estimating genome conservation between crop and model legume species. PNAS 101(43):15289–15294
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Colditz F, Nyamsuren O, Niehaus K, Eubel H, Braun HP, Krajinski F (2004) Proteomic approach: Identification of Medicago truncatula proteins induced in roots after infection with the pathogenic oomycete Aphanomyces euteiches. Plant Mol Biol 55:109–120
Colditz F, Niehaus K, Krajinski F (2007) Silencing of PR-10-like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches. Planta 226:57–71
Davis DW, Fritz VA, Pfleger FL, Percich JA, Malvick DK (1995) MN 144, MN 313, and MN 314: garden pea lines resistant to root rot caused by Aphanomyces euteiches Drechs. HortScience 30(3):639–640
Dhandaydham M, Charles L, Zhu H, Starr JL, Huguet T, Cook DR, Prosperi JM, Opperman C (2008) Characterization of root-knot nematode resistance in Medicago truncatula. J Nematol 40:46–54
Djebali N, Jauneau A, Ameline-Torregrosa C, Chardon F, Jaulneau V, Mathé C, Bottin A, Cazaux M, Pilet-Nayel M-L, Baranger A, Aouani ME, Esquerré-Tugayé M-T, Dumas B, Huguet T, Jacquet C (2009) Partial resistance of Medicago truncatula to Aphanomyces euteiches is associated with protection of the root stele and is Controlled by a major QTL rich in proteasome-related genes. Mol Plant Microbe Interact 22:1043–1055
Esquerré-Tugayé M-T, Campargue C, Mazau D (1999) The response of plant cell wall hydroxyprolin-rich glycoproteins to microbial pathogens and their elicitors. In: Datta SK, Muthukrishnan S (eds) Pathogenesis-related proteins in plants. CRC Press, Boca Raton, pp 157–170
Finkers R, van den Berg P, van Berloo R, ten Have A, van Heusden AW, van Kan JAL, Lindhout P (2007) Three QTLs for Botrytis cinerea resistance in tomato. Theor Appl Genet 114:585–593
Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325:998–1001
Gaulin E, Madoui MA, Bottin A, Jacquet C, Mathe C, Couloux A, Wincker P, Dumas B (2008) Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS ONE 3:e1723
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Graham MA, Silverstein KAT, Cannon SB, VandenBosch KA (2004) Computational identification and characterization of novel genes from legumes. Plant Physiol 135:1179–1197
Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL (1995) Structure of the Arabidopsis Rpm1 gene enabling dual-specificity disease resistance. Science 269:843–846
Grunwald NJ, Hoheisel GA (2006) Hierarchical analysis of diversity, selfing, and genetic differentiation in populations of the oomycete Aphanomyces euteiches. Phytopathol 96:1134–1141
Hardie DG (1999) Plant protein serine threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol 50:97–131
Holland JB (1998) EPISTACY: A SAS program for detecting two-locus epistatic interactions using genetic marker information. J Heredity 89:374–375
Holub EB, Grau CR, Parke JL (1991) Evaluation of the forma-specialis concept in Aphanomyces euteiches. Mycol Res 95:147–157
Hu KM, Qiu DY, Shen XL, Li XH, Wang SP (2008) Isolation and manipulation of quantitative trait loci for disease resistance in rice using a candidate gene approach. Mol Plant 1:786–793
Huang S, Vleeshouwers VGAA, Werij JS, Hutten RCB, van Eck HJ, Visser RGF, Jacobsen E (2004) The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. Mol Plant Microbe Interact 17:428–435
Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22:5679–5689
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Jones FR, Drechsler C (1925) Root rot of peas in the United States caused by Aphanomyces euteiches. J Agric Res 30:293–325
Jubault M, Lariagon C, Simon M, Delourme R, Manzanares-Dauleux MJ (2008) Identification of quantitative trait loci controlling partial clubroot resistance in new mapping populations of Arabidopsis thaliana. Theor Appl Genet 117:191–202
Kamphuis LG, Lichtenzveig J, Oliver RP, Ellwood SR (2008) Two alternative recessive quantitative trait loci influence resistance to spring black stem and leaf spot in Medicago truncatula. BMC Plant Biol 8:30
Klingler J, Creasy R, Gao L, Nair RM, Calix AS, Jacob HS, Edwards OR, Singh KB (2005) Aphid resistance in Medicago truncatula involves antixenosis and phloem-specific, inducible antibiosis, and maps to a single locus flanked by NBS-LRR resistance gene analogs. Plant Physiol 137:1445–1455
Klingler JP, Edwards OR, Singh KB (2007) Independent action and contrasting phenotypes of resistance genes against spotted alfalfa aphid and bluegreen aphid in Medicago truncatula. New Phytol 173:630–640
Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P (2006) F-box proteins everywhere. Curr Opin Plant Biol 9:631–638
Levenfors JP, Wikström M, Persson L, Gerhardson B (2003) Pathogenicity of Aphanomyces spp. from different leguminous crops in Sweden. Eur J Plant Pathol 109:535–543
Li ZK, Luo LJ, Mei HW, Paterson AH, Zhao XZ, Zhong DB, Wang YP, Yu XQ, Zhu L, Tabien R, Stansel JW, Ying CS (1999) A "defeated" rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol Gen Genet 261:58–63
Li ZK, Arif M, Zhong DB, Fu BY, Xu JL, Domingo-Rey J, Ali J, Vijayakumar CHM, Yu SB, Khush GS (2006) Complex genetic networks underlying the defensive system of rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. PNAS 103:7994–7999
Lincoln S, Daly M, Lander ES (1992) Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute technical report, 3rd edn. Whitehouse Technical Institute, Cambridge
Linford MB (1927) Additional hosts of Aphanomyces euteiches, the pea root rot fungus. Phytopathol 17:133–134
Liu ZQ, Adamczyk K, Manzanares-Dauleux M, Eber F, Lucas MO, Delourme R, Chevre AM, Jenczewski E (2006) Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 174:1583–1596
Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, Rameau C, Baranger A, Coyne C, Lejeune-Hénaut I, Burstin J (2005) Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor Appl Genet 111:1022–1031
Ma HX, Bai GH, Zhang X, Lu WZ (2006) Main effects, epistasis, and environmental interactions of quantitative trait loci for fusarium head blight resistance in a recombinant inbred population. Phytopathol 96:534–541
Madoui MA, Gaulin E, Mathe C, San Clemente H, Couloux A, Wincker P, Dumas B (2007) AphanoDB: a genomic resource for Aphanomyces pathogens. BMC Genomics 8:471
Malvick DK, Grau CR (2001) Characteristics and frequency of Aphanomyces euteiches races 1 and 2 associated with Alfalfa in the Midwestern United States. Plant Dis 85:740–744
Malvick DK, Percich JA (1999) Identification of Pisum sativum germ plasm with resistance to root rot caused by multiple strains of Aphanomyces euteiches. Plant Dis 83:51–54
Malvick DK, Grau CR, Percich JA (1998) Characterization of Aphanomyces euteiches strains based on pathogenicity tests and random amplified polymorphic DNA analyses. Mycol Res 102:465–475
Malvick D, Grunwald N, Dyer A (2009) Population structure, races, and host range of Aphanomyces euteiches from alfalfa production fields in the central USA. Eur J Plant Pathol 123:171–182
Manzanares-Dauleux MJ, Delourme R, Baron F, Thomas G (2000) Mapping of one major gene and of QTLs involved in resistance to clubroot in Brassica napus. Theor Appl Genet 101:885–891
Moussart A, Wicker E, Duparque M, Rouxel F (2001) Development of an efficient screening test for pea resistance to Aphanomyces euteiches. In: 4th European conference on grain legumes, Cracow, pp 272–273
Moussart A, Onfroy C, Lesné A, Esquibet M, Grenier E, Tivoli B (2007) Host status and reaction of Medicago truncatula accessions to infection by three major pathogens of pea (Pisum sativum) and alfalfa (Medicago sativa). Eur J Plant Pathol 117:57–69
Moussart A, Even MN, Tivoli B (2008) Reaction of genotypes from several species of grain and forage legumes to infection with a French pea isolate of the oomycete Aphanomyces euteiches. Eur J Plant Pathol 122:321–333
Mun JH, Kim DJ, Choi HK, Gish J, Debellé F, Mudge J, Denny R, Endré G, Saurat O, Dudez AM, Kiss GB, Roe B, Young ND, Cook DR (2006) Distribution of microsatellites in the genome of Medicago truncatula: a ressource of genetic markers that integrate genetic and physical maps. Genetics 172:2541–2555
Perchepied L, Kroj T, Tronchet M, Loudet O, Roby D (2006) Natural variation in partial resistance to Pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana. PLoS ONE 1:e123
Pilet-Nayel ML, Muehlbauer FJ, McGee RJ, Kraft JM, Baranger A, Coyne CJ (2005) Consistent quantitative trait loci in pea for partial resistance to Aphanomyces euteiches isolates from the United States and France. Phytopathol 95:1287–1293
Pilet-Nayel ML, Prosperi JM, Hamon C, Lesne A, Lecointe R, Le Goff I, Herve M, Deniot G, Delalande M, Huguet T, Jacquet C, Baranger A (2009) AER1, a major gene conferring resistance to Aphanomyces euteiches in Medicago truncatula. Phytopathol 99:203–208
Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 14:21–29
Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, Carbonne F, Griffiths G, Esquerre-Tugaye MT, Rosahl S, Castresana C, Hamberg M, Fournier J (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 139:1902–1913
Ramalingam J, Cruz CMV, Kukreja K, Chittoor JM, Wu JL, Lee SW, Baraoidan M, George ML, Cohen MB, Hulbert SH, Leach JE, Leung H (2003) Candidate defense genes from rice, barley, and maize and their association with qualitative and quantitative resistance in rice. Mol Plant Microbe Interact 16:14–24
Rocherieux J, Glory P, Giboulot A, Boury S, Barbeyron G, Thomas G, Manzanares-Dauleux MJ (2004) Isolate-specific and broad-spectrum QTLs are involved in the control of clubroot in Brassica oleracea. Theor Appl Genet 108:1555–1563
Ronfort J, Bataillon T, Santoni S, Delalande M, David JL, Prosperi JM (2006) Microsatellite diversity and broad scale geographic structure in a model legume: building a set of nested core collection for studying naturally occurring variation in Medicago truncatula. BMC Plant Biol 6:28
Rosahl S (1996) Lipoxygenases in plants—their role in development and stress response. Zeitschrift für Naturforschung. (C) 51:123–138
Rose RJ (2008) Medicago truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: past, present and future. Funct Plant Biol 35:253–264
Rose LE, Michelmore RW, Langley CH (2007) Natural variation in the Pto disease resistance gene within species of wild tomato (Lycopersicon). II. Population genetics of Pto. Genetics 175:1307–1319
Rowe HC, Kliebenstein DJ (2008) Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180:2237–2250
Samac DA, Graham MA (2007) Recent advances in legume-microbe interactions: recognition, defense response, and symbiosis from a genomic perspective. Plant Physiol 144:582–587
SAS (1989) SAS/STAT users guide, version 6, 4th edn. SAS Institute, Cary
Shindo C, Bernasconi G, Hardtke CS (2007) Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Ann Bot 99:1043–1054
Smart CD, Myers KL, Restrepo S, Martin GB, Fry WE (2003) Partial resistance of tomato to Phytophthora infestans is not dependent upon ethylene, jasmonic acid, or salicylic acid signaling pathways. Mol Plant Microbe Interact 16:141–148
Soufflet-Freslon V, Gianfranceschi L, Patocchi A, Durel CE (2008) Inheritance studies of apple scab resistance and identification of Rvi14, a new major gene that acts together with other broad-spectrum QTL. Genome 51:657–667
Tivoli B, Baranger A, Sivasithamparam K, Barbetti MJ (2006) Annual Medicago: from a model crop challenged by a spectrum of necrotrophic pathogens to a model plant to explore the nature of disease resistance. Ann Bot 98:1117–1128
Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Ungerer MC, Rieseberg LH (2003) Genetic architecture of a selection response in Arabidopsis thaliana. Evolution 57:2531–2539
Vailleau F, Sartorel E, Jardinaud MF, Chardon F, Genin S, Huguet T, Gentzbittel L, Petitprez M (2007) Characterization of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula. Mol Plant Microbe Interact 20:159–167
Van Ooijen JW (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet 84:803–811
Vandemark GJ, Grunwald NJ (2004) Reaction of Medicago truncatula to Aphanomyces euteiches race 2. Arch Phytopathol Plant Prot 37:59–67
Wang S, Basten CJ, Zeng ZB (2005) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
Wicker E, Rouxel F (2001) Specific behaviour of french Aphanomyces euteiches Drechs. populations for virulence and aggressiveness on pea, related to isolates from Europe, America and New Zealand. Eur J Plant Pathol 107:919–929
Wicker E, Moussart A, Duparque M, Rouxel F (2003) Further contributions to the development of a differential set of pea cultivars (Pisum sativum) to investigate the virulence of isolates of Aphanomyces euteiches. Eur J Plant Pathol 109:47–60
Wisser RJ, Sun Q, Hulbert SH, Kresovich S, Nelson RJ (2005) Identification and characterization of regions of the rice genome associated with broad-spectrum, quantitative disease resistance. Genetics 169:2277–2293
Yan JQ, Wang J, Zhang H (2002) An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J 29:193–202
Yang SM, Gao MQ, Xu CW, Gao JC, Deshpande S, Lin SP, Roe BA, Zhu HY (2008) Alfalfa benefits from Medicago truncatula: The RCT1 gene from M. truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proc Natl Acad Sci USA 105:12164–12169
Yu J, Zhou Y, Cang J, Xu J (2006) Mapping of quantitative resistance loci to bacterial leaf blight and their race specificity in rice (Oryza sativa L.). Acta Agronom Sin 32:1611–1617
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Acknowledgments
This work was funded by a pre-doctoral fellowship from INRA, Département de Génétique et Amelioration des Plantes, French Ministry of Agriculture and Fishing and UNIP (Union Nationale Interprofessionnelle des Plantes riches en proteines, Paris, France), that we greatly acknowledge. It was also supported by the FP6 Grain Legume Integrated Project (FOOD-CT-2004-506223) (GLIP). We wish to thank Dr Malvick for kindly providing the alfalfa-infecting strains of A. euteiches.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Varshney.
Rights and permissions
About this article
Cite this article
Hamon, C., Baranger, A., Miteul, H. et al. A complex genetic network involving a broad-spectrum locus and strain-specific loci controls resistance to different pathotypes of Aphanomyces euteiches in Medicago truncatula . Theor Appl Genet 120, 955–970 (2010). https://doi.org/10.1007/s00122-009-1224-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1224-x