Abstract
This work was aimed at compiling the world’s first linkage map of redcurrant with the use of microsatellite and SNP DNA markers. As a result of mapping, eight linkage groups with an LOD level of 10 were constructed, which included 299 DNA markers, including 12 microsatellite and 287 SNP markers; the total length of all linkage groups was 741 cM; the average was 93 cM. In seven of the eight linkage groups obtained, at least one microsatellite DNA marker was mapped. Most of the microsatellite loci localized on the redcurrant map correspond to their position on the blackcurrant linkage groups (loci e3-B02, g1-A01, and g1-L12 are located on the same linkage group no. 5; loci g2-J08, g2-B20, e1-O21, and e1-O01 on the resulting map are each in a separate linkage group 2, 3, 4, and 6, respectively), with the exception of the loci mapped at blackcurrant on linkage group 1, which in our work were unmapped. The g2-J11 locus in our studies amplifies fragments in two ranges, which suggests its duplication in the genome of the studied samples. This locus was mapped in two linkage groups of redcurrant. Two new, not previously mapped and unpublished microsatellite loci were also localized on linkage map of redcurrant. In the future, it is planned to localize economically useful traits in the redcurrant genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The genus currant Ribes L. includes more than 150 species, growing mainly in the temperate zone of the Northern Hemisphere. Blackcurrant (Ribes nigrum L.), redcurrant (Ribes rubrum L.), and gooseberry (Ribes uva-crispa L.) are economically important [1].
The most extensively studied using DNA markers culture of the genus Ribes L. is blackcurrant. For it, several genetic maps (GM) of linkage groups were compiled and a number of techniques for marker-assisted selection were developed [2–4].
Redcurrant is today widespread in the industrial berry industry of the countries of the European Union [5]. In addition to the color and biochemical composition of berries, redcurrant has a number of significant differences from blackcurrant, including the fact that redcurrant that does not have an aroma inherent to blackcurrant, secreted by special glands on the leaves. Redcurrants are characterized by a more rapid entry into a dormant state, more durable fruit-bearing shoots, a later onset of vegetation, etc. Hybrids between blackcurrant and redcurrant are barren [6].
The purpose of this work is to compile the world’s first genetic map of redcurrant linkage groups using microsatellite and SNP DNA markers.
In this work, we used a hybrid population of 139 hybrids (six of them were excluded during the mapping process because of a large number of dying plants) obtained from crossing the Belaya Potapenko variety (Krasnyi Krest × Krasnaya sibiryachka) and hybrid forms 1426-21-80 (82-4-11 (Rote Spatlese × Chulkovskaya) × 78-2-118 (Rote Spatlese × Maarsis Prominent)). The mapped family has a complex genetic origin in which species such as redcurrant (R. rubrum L.), common currant (R. sativum L.), large-fruited currant (R. sativum var. macrocarpum), rocky currant (R. petreum Wulf.), and multiflorous currant (R. multiflorum Kit.) are involved. More than 60 pairs of primers for microsatellite loci were taken for analysis on a small sample of redcurrant samples. Of these, 38 pairs of primers were previously used to study representatives of the currant genus [7–11]. The nucleotide sequences of 30 pairs of primers (hereinafter referred to as new) were provided by Nahla Bassil (unpublished data, personal communications) and were created on the basis of the nucleotide sequences of currants placed in the NCBI database. Polymorphism of microsatellite loci was detected in 8% PAAG stained with silver nitrate. To identify SNP markers, a DNA library sequencing approach with reduced genome complexity (genotyping by sequencing, GBS) was used. The library ready for sequencing was sequenced on one track of the Illumina HiSeq 4000. The genetic map was compiled using the Join Map3/0 program, regression mapping was prepared using the Kosambi function.
Of the tested pairs of primers for microsatellite loci, the majority were unsuitable for mapping for the following reasons: they amplified fragments of more than 500 bp, they amplified non-polymorphic PCR products, they did not amplify on the DNA of the parents of the hybrid population at a minimum annealing temperature of 50°C, they showed an atypical profile of amplification for microsatellite loci (a large number of fragments in a wide range that do not disappear with increasing annealing temperature), etc. A typical microsatellite profile and the presence of polymorphic fragments were observed during amplification with 13 pairs of primers (previously published gr2-J05, g1-M07, g1-K04, g2-J08, g2-B20, e1-O21, e3-B02, g1-A01, g1-L12, e1-O01, g2-G12 and the new Cra-531 and Cra-489). Fragments in two ranges (250–260 and 315–320 bp) are amplified at the g2-J11 locus in the mapped population, which suggests the duplication of this locus in the genome of the studied samples. Thus, data on polymorphism of 14 microsatellite loci were used for mapping (Table 1).
Sequencing of DNA libraries with reduced genome complexity created on the DNA of a hybrid population and parents resulted in 382 373 581 readings of good quality. After removing adapter sequences and detecting SNP using Tassel 5.0, 5741 SNPs were detected. After analysis of cleavage in the population, 667 SNP markers were used for mapping.
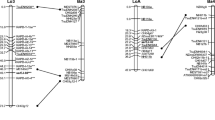
As a result of mapping, eight cohesion groups were constructed with a LOD level of 10 (Fig. 1), which included 299 loci, among them 12 microsatellite and 287 SNP markers; the total length of all cohesion groups was 741 cM; the average length was 93 cM (Table 2). Three microsatellite loci were not included in any of the linkage groups (ungrouped)—these are gr2-J05, g1-M07, and g1-K04. It is interesting to note that these loci are located on the same linkage group no. 1 of the genetic map of blackcurrant. More than half of the SNP markers were also outside the linkage groups; several markers were excluded because of the high contribution to the parameter Mean chisquare contributions. In seven of the eight linkage groups, at least one microsatellite DNA marker was mapped.
Genetic map of redcurrant (the numbering of linkage groups corresponds to the map of blackcurrant taking into account the location of microsatellite loci; the presented linkage groups combine markers at LOD 10 and are maps of the first round of mapping, except for linkage group 2, which is represented by a map of the second round of mapping).
On the resulting map of the redcurrant, the linkage groups were numbered according to the linkage groups of the previously published genetic map of the blackcurrant [2], based on the localization of identical microsatellite loci. In this case, the loci e3-B02, g1-A01, and g1-L12 mapped together on the fifth linkage group of blackcurrant and on the map of redcurrant we obtained also are located on one linkage group. Loci g2-J08, g2-B20, e1-O21, and e1-O01 on the map of blackcurrant, located respectively in linkage groups 2, 3, 4, and 6, and on our map are also each in a separate linkage group (Table 2). The position of the locus g2-J11 contradicts the localization on the blackcurrant map. Mapped to the blackcurrant genome on the seventh linkage group together with the g2-g12 locus, on our map, the g2-J11 locus did not join the g2-g12 locus but entered a separate linkage group (tentatively designated as 7.2?). In addition, in connection with the g2-J11 duplication assumed in the redcurrant genome, this locus was also mapped to linkage group 2 together with the g2-J08 locus. One of the obtained linkage groups is made up entirely of SNP markers and, possibly, represents the eighth linkage group, since the microsatellite loci mapped to the linkage group 8 of the blackcurrant genetic map did not pass the initial test for one of the reasons described above and were not used in the mapping of the redcurrant genome. Thus, the majority of microsatellite loci located by us on the map of redcurrant correspond to the position of loci on the linking groups of blackcurrant. Two new, not previously mapped and unpublished microsatellite loci are also localized. Design of primers for them was carried out on the basis of sequences of Ribes americanum. These are the Cra-489 locus (F: 5'-TCTATTATCACACCCTCAACAA-3', R: 5'-GTTTATACGACACATCAACTTTCCA-3') to pentanucleotide repeats in the genome and the Cra-531 locus (F: 5'-AGAAGTGAAAGTGGAAGAACC-3', R: 5'-GTTTGTTTGAAGGAAGACAGAGA-3') to trinucleotide repeats, localized in this work on redcurrant linkage groups no. 3 and no. 10, respectively.
In the future, it is planned to increase the number of mapped microsatellite loci and to search for the localization of economically useful traits in the redcurrant genome.
REFERENCES
Pomologiya (Pomology), vol. IV: Smorodina, kryzhovnik (Currant, Gooseberry), Sedov, E.N., Ed., Orel: Vserossiiskii Nauchno-Issledovatel’skii Institut Selektsii Plodovykh Kul’tur, 2009.
Brennan, R., Jorgensen, L., Hackett, C., et al., The development of a genetic linkage map of blackcurrant (Ribes nigrum L.) and the identification of regions associated with key fruit quality and agronomic traits, Euphytica, 2008, vol. 161, pp. 19—34. https://doi.org/10.1007/s10681-007-9412-8
Russell, J.R., Bayer, M., Booth, C., et al., Identification, utilisation and mapping of novel transcriptome-based markers from blackcurrant (Ribes nigrum), BMC Plant Biol., 2011, no. 11(1), p. 147. https://doi.org/10.1186/1471-2229-11-147
Russell, J., Hackett, C., Hedley, P., et al., The use of genotyping by sequencing in blackcurrant (Ribes nigrum): developing high-resolution linkage maps in species without reference genome sequences, Mol. Breed., 2014, vol. 33, no. 44, pp. 835—849. https://doi.org/10.1007/s11032-013-9996-8
Malinovskii, B., 2017. https://propozitsiya.com/krasnaya-smorodina-priznana-samoy-rentabelnoy-nishevoy-yagodoy. https://doi.org/10.1051/fruits/2011049.
Buchenkov, I.E., Chernetskaya, A.G., Ryshkel’, I.V., and Ryshkel’, O.S., Interspecific distant hybridization of black currant (Ribes nigrum L.) and red currant (Ribes rubrum L.), Vestn. Poless. Gos. Univ., Ser. Prirodoved. Nauk, 2014, no. 2, pp. 48—55.
Brennan, R., Jorgensen, L., Woodhead, M., et al., Development and characterization of SSR markers in Ribes species, Mol. Ecol., 2002, no. 2(3), pp. 327—330. https://doi.org/10.1046/j.1471-8286.2002.00233.x
Cavanna, M., Marinoni, D.T., Beccaro, G.L., et al., Microsatellite-based evaluation of Ribes spp. germplasm, Genome, 2009, vol. 52, no. 10, pp. 839—848. https://doi.org/10.1139/G09-057
Antonius, K., Karhu, S., Kaldm, H., et al., Development of the Northern European Ribes core collection based on a microsatellite (SSR) marker diversity analysis, Plant Genet. Resour.: Charact. Util., 2012, pp. 70—73. https://doi.org/10.1017/S1479262111000980
Palmieri, L., Grando, M.S., Sordo, M., et al., Establishment of molecular markers for germplasm management in a worldwide provenance Ribes spp. collection, Plant Omics, 2013, vol. 6, no. 3, pp. 165—174.
Pikunova, A.V., Knyazev, S.D., Bakhotskaya, A.Yu., et al., Microsatellite loci polymorphism in Russian black currant (Ribes nigrum L.) varieties from collection of All-Russian Research Institute for Fruit Crops Breeding, S.-kh.Biol., 2015, vol. 50, no. 1, pp. 46—54. https://doi.org/10.15389/agrobiology.2015.1.46rus
ACKNOWLEDGMENTS
We are grateful to Nahla Bassil for the design of the primers for new previously unpublished microsatellite loci.
Funding
This work was supported by a grant from the Russian Science Foundation “Study of the Currant Genome (Ribes L.) Using DNA Markers,” no. 18-76-0032.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest.
This article does not contain any research using animals as an object. This article does not contain any research involving people as an object.
Rights and permissions
About this article
Cite this article
Pikunova, A.V., Goryunova, S.V., Goryunov, D.V. et al. The Development of a Genetic Linkage Map of Redcurrant (Ribes rubrum L.) by Means of SSR and SNP DNA Markers. Russ J Genet 56, 1387–1390 (2020). https://doi.org/10.1134/S1022795420100105
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795420100105