Abstract
Among cereal crops, rye is one of the most tolerant species to aluminum. A candidate gene approach was used to determine the likely molecular identity of an Al tolerance locus (Alt4). Using PCR primers designed from a wheat aluminum tolerance gene encoding an aluminum-activated malate transporter (TaALMT1), a rye gene (ScALMT1) was amplified, cloned and sequenced. Subsequently, the ScALMT1 gene of rye was found to be located on 7RS by PCR amplification using the wheat–rye addition lines. SNP polymorphisms for this gene were detected among the parents of three F2 populations that segregate for the Alt4 locus. A map of the rye chromosome 7R, including the Alt4 locus ScALMT1 and several molecular markers, was constructed showing a complete co-segregation between Alt4 and ScALMT1. Furthermore, expression experiments were carried out to clarify the function of this candidate gene. Briefly, the ScALMT1 gene was found to be primarily expressed in the root apex and upregulated when aluminum was present in the medium. Five-fold differences in the expression were found between the Al tolerant and the Al non-tolerant genotypes. Additionally, much higher expression was detected in the rye genotypes than the moderately tolerant “Chinese Spring” wheat cultivar. These results suggest that the Alt4 locus encodes an aluminum-activated organic acid transporter gene that could be utilized to increase Al tolerance in Al sensitive plant species. Finally, TaALMT1 homologous sequences were identified in different grasses and in the dicotyledonous plant Phaseolus vulgaris. Our data support the hypothesis of the existence of a common mechanism of Al tolerance encoded by a gene located in the homoeologous group four of cereals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminum (mainly existing as Al3+, the trivalent cationic form) is very toxic to many crops (Foy et al. 1978; Rao et al. 1993; Kochian 1995). This light metal is the main factor that limits productivity in acidic soils that comprise approximately 30–40% of the world’s arable soils and up to 70% of potentially arable land (Aniol et al. 1980; Haug 1984). The major symptom of Al toxicity is the inhibition of root growth (Delhaize and Ryan 1995; Ma and Furukawa 2003) by destroying the root apex (Ryan et al. 1993).
While liming of soil can ameliorate some Al toxicity, it is expensive and ineffective in the subsoil. For this reason, the development of new crop varieties tolerant to Al seems to be the best solution to that problem. To date, several plant mechanisms to tolerate Al stress have been proposed (Kochian et al. 2002; Samac and Tesfaye 2003; Kochian et al. 2004). One of the mechanisms explaining Al tolerance is the exudation of organic acids (malate, citrate and oxalate) that chelate Al in the rhizosphere (Miyasaka et al. 1991; Delhaize et al. 1993; Ma et al. 2000, 2001; Li et al. 2000).
Molecular strategies have been used to detect genes implicated in the plant response to Al. Several genes have been found to be activated by Al in tolerant and susceptible genotypes in species like wheat (Snowden and Gardner 1993; Richards and Gardner 1994; Cruz-Ortega et al. 1997; Hamel et al. 1998), rice (Yu et al. 1998), tobacco (Ezaki et al. 1995, 1996), Arabidopsis (Richards et al. 1998), pea (Brosche and Strid 1999; Savenstrand et al. 2000) and rye (Rodriguez-Milla et al. 2002). There are also some examples of genes that are expressed in Al tolerant genotypes but not in the susceptible ones in different species: soybean (Ermolayev et al. 2003), wheat (Hamilton et al. 2001; Sasaki et al. 2004) and Arabidopsis (Sivaguru et al. 2003; Larsen et al. 2005; Hoekenga et al. 2006). Among them, particular attention should be placed on the wheat TaALMT1 gene because it encodes an aluminum-activated malate transporter and its Al tolerance function has been demonstrated (Delhaize et al. 2004; Sasaki et al. 2004). Recently, this gene was completely sequenced and located on the 4DL chromosome arm (Raman et al. 2005). In any case, all these genes are good candidates to be studied in other crops to find mechanisms of Al tolerance in plants.
Analysis of populations segregating for Al tolerance has generated a worthy amount of information to identify loci controlling tolerance in plants. In wheat Al tolerance segregates as a single dominant locus (Delhaize et al. 1993; Somers and Gustafson 1995; Riede and Anderson 1996; Basu et al. 1997; Raman et al. 2005). More recently, the AltBH tolerance locus was linked to some molecular markers and located on 4DL (Riede and Anderson 1996; Luo and Dvorak 1996; Rodriguez-Milla and Gustafson 2001). In barley a dominant gene located on the 4HL chromosome arm has been found (Stolen and Andersen 1978; Tang et al. 2000; Raman et al. 2002). In maize tolerance genes located on chromosomes 2, 6 and 10 have been described (Brondani and Paiva 1996; Sibov et al. 1999). In Arabidopsis semidominant Mendelian loci have been found on chromosomes 1 and 4 (Larsen et al. 1996), 1998), and different QTLs that explain 40% of total variation have been described in Arabidopsis and sorghum (Kobayashi and Koyama 2002; Hoekenga et al. 2003; Magalhaes et al. 2004). In rice, 11 QTLs of Al tolerance have been located (Nguyen et al. 2002, 2003). Finally, a single locus, Alt SB, was found to control Al tolerance in two highly Al tolerant sorghum cultivars (Magalhaes et al. 2004).
Cereal crops exhibit variation in Al tolerance, and rye (Secale cereale L.) is the most tolerant cereal (Mugwira et al. 1978; Little 1988; Aniol and Madej 1996), whereas barley is the most Al-sensitive (Foy 1983). Rye can be used as a source of resistance genes for wheat (Triticum ssp.) through wheat–rye introgression and as a component of triticale (xTriticosecale Wittmack). Therefore, the knowledge of the mechanism and the genes that control Al tolerance in rye will provide fundamental information than can be used to increase Al tolerance in other cereals. Using wheat–rye addition lines, at least three different Al tolerance genes have been discovered in rye: Alt1, Alt2 and Alt3 located on 6RS, 3R and 4RL, respectively (Aniol and Gustafson 1984; Ma et al. 2000; Aniol 2004). The analysis of segregating populations has detected three independent and dominant loci that confer Al resistance in rye located on 4RL (Alt3), 6RS (Alt1) and 7RS (Alt4) (Gallego and Benito 1997; Gallego et al. 1998a, b; Miftahudin et al. 2002; Matos et al. 2005).
The secretion of organic acids to the rhizosphere seems to be the main mechanism of Al tolerance in rye (Li et al. 2000). We hypothesized that at least a part of the Al tolerance behavior in rye is conditioned by a gene that is part of an organic acid exudation system. The main objective of the present work was to investigate whether a homolog of the Al tolerance TaALMT1 gene of wheat is a good candidate to be the Alt4 locus located on the 7RS chromosome arm of rye.
Materials and methods
Plant material
To map the Alt4 rye locus three F2 families from crosses between Al tolerant rye plants (cv. “Ailés”) and Al non-tolerant rye plants (“Riodeva” inbred line), segregating 3 Al tolerant:1 Al non-tolerant, were used: AR1-6⊗, AR1-25⊗ and AR6-5⊗. These F2 populations were obtained by selfing three different F1 plants originated from different crosses between a tolerant plant from the “Ailés” (A) rye cultivar and an individual from the non-tolerant inbred line “Riodeva” (R).
To assign loci to rye or barley chromosomes, hexaploid wheat, cv “Chinese Spring” (CS), rye Secale cereale L., cv. “Imperial” (I), barley Hordeum vulgare L., cv. “Betzes” (B), amphiploids wheat–rye and wheat–barley and the wheat–rye and wheat–barley disomic addition lines and the two ditelosomic wheat–rye addition lines 7RS and 7RL were used.
The TaALMT1 wheat marker was located using the ‘Chinese Spring’ nullitetrasomics and ditelocentrics available for homoeologous group four.
Aluminum tolerance screening test
The Al tolerance test was carried out using the nutrient-culture, modified-pulse method (Aniol 1984). Seeds were sterilized for 10 min with HgCl2 (0.1%), well rinsed with water and germinated overnight on filter paper in Petri dishes. Sprouted seeds were transferred the next day onto polyethylene nets fixed in Lucite frames. Styrofoam blocks were attached to the frames with rubber bands and floated on the surface of the vigorously aerated nutrient solution. Containers with the nutrient solution were placed in a water bath at 25°C under 16 h per day illumination. The nutrient solution consisted of: 0.4 mM CaCl2, 0.65 mM KNO3, 0.25 mM MgCl2·6H2O, 0.01 mM (NH4)2SO4 and 0.04 mM NH4NO3. Four days after sowing, seedlings were incubated for 24 h in nutrient solution with aluminum in the form of AlKSO4·12 H2O at the concentration of 150 μM. After each exposure to Al, seedlings were removed from Al-containing solution, thoroughly washed for 2–3 min in running tap water. Then, roots were stained with 0.1% aqueous solution of Eriochrome cyanine R for 10 min. After staining the excess dye was removed by washing under tap water. Subsequently, seedlings were transferred to Al-free nutrient solution for 48 h. Additional root growth after these 2 days was easily assessed. In seedlings where the aluminum treatment did not destroy the root apical meristem, the root segment growing after Al-treatment was white (unstained) and contrasted to the heavily stained root part exposed to aluminum. When the apical meristem was damaged, root tips did not show any regrowth after 48 h in Al-free medium, remaining intensely stained. During all stages of growth, and particularly during Al-treatment the pH of the nutrient solution was adjusted at 4.0 with 1 N HCl. The nutrient solution was changed daily.
For mapping purposes, aluminum tolerance was determined as a dominant trait in the F2 generation.
DNA extraction
Young leaves from plants were frozen in liquid nitrogen and stored at −80°C. The extraction was carried out using a small-scale DNA isolation method (Dneasy Plant Mini Kit, Qiagen).
PCR
Amplifications from genomic DNA were done in reactions of 10 μl containing 60 ng template DNA, 100 μM of each dNTP, 5 pmoles of each primer, MgCl2 2 mM, 0.25 μl of Taq DNA polymerase, and 1× reaction buffer (Tris–HCl 100 mM pH 8.3, KCl 100 mM). Amplifications were carried out in a PTC-100 thermal cycler (MJ Research) with the following program: a preliminary step of 5 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at 59–69°C (depending on the pair of primers, Table 1) and 2 min at 72°C and a final step of 10 min at 72°C. PCR reactions were stored at 4°C until electrophoresis on 2% agarose gels stained with ethidium bromide. Several pairs of primers were designed based on the sequences of the TaALMT1 wheat gene (Sasaki et al. 2004).
Cloning and sequencing of products
After PCR, bands of the approximate expected sizes were excised from 2% agarose TAE gels, purified with a Bioclean kit (Biotools), and cloned using a TA cloning kit (Invitrogen, Inc.). Cloned PCR products were sequenced at the Universidad Complutense facility using an automatic sequencer model 3730 (Applied Biosystems).
Sequence analyses
DNA sequences were analyzed using the Chromas Lite 1.0 program (Technelysium Pty Ltd). To confirm product identity, the GenBank BLAST application was used to compare DNA sequences, and the amino acid sequences predicted from them, with corresponding NCBI sequences (http://www.ncbi.nlm.nih.gov/).
The program Genscan (http://www.genes.mit.edu/GENSCAN.html) was initially used for predicting the locations and exon–intron boundaries of the ScALMT1 gene in genomic sequences of rye.
MegAlign Software (DNAStar Inc.) was utilized to align sequences using the CLUSTAL V method (Higgins and Sharp 1988). In some cases, after the alignment was completed, a neighbor-joining method was employed to reconstruct phylogeny for the putative alignment (MEGA version 2.1, Kumar et al. 2001). Bootstrap resampling of 1,000 replicates was performed to test the robustness of the dendrograms.
In order to predict the secondary structure of the putative protein and to interpret the effect of the amino acid differences among the different wheat and rye ALMT1 variants, the computer programs shown in Table 2 were utilized.
Genetic mapping
Linkage analyses were performed on F2 segregation data using the JoinMap 3.0 computer program (Van Ooijen and Voorrips 2001). Two loci were considered linked if the LOD score was greater than 3 and if the two-point distance was less than 45 cM. Genetic distances were calculated using the Kosambi function (Kosambi 1944).
RNA extraction and RT-PCR
Al tolerant (cv. “Ailés”), non-tolerant (“Riodeva” inbred line) rye plants and the moderately Al tolerant wheat cultivar “Chinese Spring” were grown in the described Al tolerance test conditions with 150 μM Al for five different times (not exposed, 30, 60, 360 and 1,200 min). Root apices, non-apical parts of the roots and leaves were removed and immediately frozen in liquid nitrogen and stored at −80°C.
Total RNA was extracted from 20 different samples per genotype and per exposure time. RNA quality was checked by gel electrophoresis and then quantified with a NanoDrop® ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). Total RNA (1 μg) was reverse transcribed using random hexamer primers and 200 units of MMLV reverse transcriptase (Advantage™ RT-for-PCR Kit, Clontech Laboratories, Inc.) in the presence of RNase-free DNase, under conditions specified by the supplier.
Quantitative PCR
Briefly, based on the complete sequence of the ScALMT1 rye gene one pair of PCR primers (FWD 5′-GCGGCTTTGTTGCAAGTGA-3′ and REV 5′-TCAACCAAGTCCGCGAGAAG-3′) and one TaqMan® probe (5′-6-FAM-ATGGCAGTCACCGCAA-MGB-3′) were designed using the Primer Express® Software Version 2.0 (Applied Biosystems, USA). The PCR reactions contained TaqMan® universal PCR master mix (Applied Biosystems), 6.0 pmol of each primer, 5.0 pmol of FAM-tagged probe (Applied Biosystems), and cDNA made from ∼100 ng of RNA. The PCR was carried out using the Applied Biosystems 7900 HT Fast Real-Time PCR System, under the following conditions: one step at 95°C for 10 min and 40 cycles with steps at 95°C for 15 s and 60°C for 1 min.
A completely conserved region among ribosomal 18S gene sequences from rye, wheat, barley and rice were used to design the following primers and probe: FWD 5′-TCAACGAGGAATGCCTAGTAAGC-3′, REV 5′-ACAAAGGGCAGGGACGTAGTC-3′, probe 5′-6-FAM- AGTCATCAGCTCGCG-MGB-3′. This was to provide an internal constitutively expressed control that would be useful not only for this experiment but also in future expression studies on such species.
All PCR samples and controls were prepared in duplicates using 0.2 ml MicroAmp Optical reaction tubes and MicroAmp Optical tube caps (Applied Biosystems, USA), and all 96-well plates included two standard curves (target gene and endogenous control) in order to apply the “relative standard curve method” supported by Applied Biosystems, to analyze the data. In this approach, unknown sample values are interpolated from the standard curves. Duplicated control reactions for every sample without reverse transcription were included to ensure that PCR products were not due to amplification of contaminant genomic DNA. For comparison purposes, expression level at each time point is expressed as the fold-increase over the level obtained in the non-tolerant genotype without exposure to aluminum.
Results
Identification in rye of sequences related to TaALMT1
The pair of primers previously reported by Sasaki et al. (2004) to amplify the Al tolerance wheat TaALMT1 gene (AB081803 and AB081804) were used in rye. After failing to amplify the complete gene using one set of primers, several primer pairs were designed, and positive results were achieved by amplifying the gene in three different parts with the ScALMT1-1, ScALMT1-2 and ScALMT1-3 primer pairs (Table 1). Then, the amplified fragments were sequenced from the “Riodeva” inbred line (Al-non-tolerant) to confirm the identity. The three sequences were assembled into one that matched with the TaALMT1 gene sequences of wheat (91% similarity at the nucleotide level). The ends of the transcript were not determined experimentally. This new gene in rye was named ScALMT1 and its sequence was sent to the NCBI data bank (DQ158087). Afterwards, the same strategy was conducted to obtain the ScALMT1 sequence in the Al-tolerant cultivar “Ailés” (DQ158086). The genes detected in “Ailés” and “Riodeva” were 3,898 and 3,855 bp long, respectively, and were 98.0% identical.
The program Genscan (http://www.genes.mit.edu/GENSCAN.html) for predicting the locations and exon–intron structures was used on the ScALMT1 genomic sequences of “Ailés” and “Riodeva” and showed a total of five introns with sizes ranging from 109 to 1,423 bp (Fig. 1). The predicted transcription unit was 1,359 bp long and the deduced protein consisted of 253 residues with a predicted molecular mass of 49.4 and 49.6 kDa and a theoretical pI of 6.64 and 7.14, respectively. The cDNAs obtained to carry out the subsequent expression experiments were sequenced confirming the length of exons, introns and transcription units in the Al tolerant and Al non-tolerant genotypes. The predicted proteins each have six hydrophobic putative transmembrane regions (TMHMM software). A search of the NCBI protein-database (http://www.ncbi.nlm.nih.gov/) with the ScALMT1 genomic sequence revealed 86% amino acid sequence identity to the TaALMT1 wheat gene and 63.3% to a rice gene (CAD40928) that encodes a putative protein of unknown function. At least 17 additional putative proteins encoded by expressed sequence tags (ESTs) from rice and Arabidopsis thaliana showed from 38.1 to 35.2% identity to ScALMT1.
Schematic diagram of the structure of ScALMT1 gene. Exons are represented by solid boxes and the triangles indicate the position of the five intercalated introns. Numbers indicate the length in base pairs of each sequence. The approximate position of the primers used for amplifying the ALMT1 gene (Table 1) and for quantitative PCR are indicated
The deduced protein sequences of the ScALMT1 genes from the Al tolerant cultivar “Ailés”, the Al non-tolerant inbred line “Riodeva”, and the sequences previously deduced from wheat were compared (Fig. 2). The rye proteins showed forty residue substitutions and seven amino acid deletions with respect to the wheat proteins. The different programs did not detect any structural changes in the proteins due to these amino acid substitutions and deletions with possible functional implications. On the other hand, the deduced rye proteins differed among themselves at six amino acid residues. Five out of the six changes are substitutions of amino acids with the same or very similar chemical properties. However, the substitution of a glycine in the Al tolerant genotype with an arginine in the Al non-tolerant one in position 73 could have important structural implications. Only one program (PSIPRED) out of the seven programs predicted a secondary structure change from a strand (with the Gly) to a helix (with the Arg).
CLUSTAL W alignment of the ALMT1 proteins deduced from the nucleotide sequences of the rye and wheat Al tolerant (Alt) and Al sensitive (Als) alleles. The wheat TaALMT1 sequences were obtained from the NCBI database, AB081803 and AB081804 (Sasaki et al. 2004). Gaps (represented by hyphens) were introduced into the sequences to maximize the alignment. *Conserved amino acids
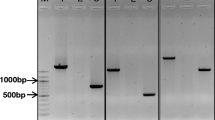
To assign the ScALMT1 gene to a rye chromosome arm, amplifications with primers ScALMT1-4F and ScALMT1-4R in the disomic and ditelosomic wheat Chinese Spring and rye Imperial addition lines were carried out (Fig. 3). The amplification product was present on the 7R and 7RS lines and absent in the rest of them, including 7RL.
Chromosomal location of the rye ScALMT1 gene. Amplification of rye genomic DNA using ScALMT1-4F and ScALMT1-4R primers. M Molecular-weight marker (100 bp ladder), 1R to 7R: wheat “Chinese Spring”–rye “Imperial” disomic addition lines 1R to 7R, 7RS: ditelosomic wheat “CS”–rye “I” addition line 7RS, 7RL: ditelosomic wheat “CS”–rye “I” addition line 7RL, CS: wheat “CS”, CSI: wheat “CS”–rye “I” amphiploid, I: rye “I”
Segregation and mapping of ScALMT1 locus
In order to detect variability that can be mapped, sequencing experiments were carried out in the parents of the F2 progenies that were segregating for the Alt4 locus. From the different polymorphisms detected, a SNP that was distinguishable with the restriction enzyme BspHI was selected.
To establish whether ScALMT1 is associated with the Alt4 locus in rye, we analyzed three F2 populations segregating for this locus. Genomic DNA isolated from F2 plants was analyzed using a cleaved amplified polymorphic sequence (CAPS) marker assay based on the restriction with BspHI enzyme. The ScALMT1 polymorphism showed complete cosegregation with the Al tolerant phenotypes. No recombination between the Al tolerance locus Alt4 and the ScALMT1 locus was found in the 381 F2 plants analyzed in these populations (Fig. 4).
Segregation of the ScALMT1 gene in ten plants of the F2 population AR1-6. Genomic DNA isolated from F2 plants was analyzed using the CAPS marker assay based on the amplification using ScALMT1-3F and ScALMT1-3R primers and ulterior restriction with BspHI enzyme. M Molecular-weight marker (100 bp ladder), A: “Ailés” Al tolerant F2 parent, R: “Riodeva” Al non-tolerant F2 parent, T: F2 Al tolerant plant, NT: F2 Al non-tolerant plant. Arrows show the segregating alleles (1 and 2). 1 Undigested PCR product. 2 Fragments that appear after digestion with BspHI. The different homozygotes and heterozygotes are indicated at the bottom of the figure
A set of markers previously mapped in rye was also included to refine the map position of Alt4 and ScALMT1. Figure 5 shows the map constructed including intermicrosatellites (SCIMs), RAPDs, SCARs, microsatellites (SCMs), the Alt4 locus and the ScALMT1 gene. In total, the map spans 84 cM and consists of 26 loci. The Alt4 locus and the ScALMT1 gene mapped at the same position and they were flanked by the markers SCIM812626 and OPQ4578.
Linkage map of chromosome 7R showing the Alt4 locus, the ScALMT1 gene and several markers previously mapped (Matos et al. 2005). The centromere is located close to SCM40 (Korzun et al. 2001). The markers SCM86 and SCIM8111376 had been located to 7RS in a previous study (Matos et al. 2005). Map distances are in Morgans (cM)
Expression of the ScALMT1 gene
We investigated the level of transcripts of the ScALMT1 gene in rye when exposed to aluminum in acidic conditions. Total RNA was extracted from root apices, non-apical parts of the roots and leaves of Al tolerant cv. “Ailés” and Al non-tolerant “Riodeva” inbred line and then analyzed by real-time PCR. A gene-specific probe and primers were designed from a conserved exonic region between “Ailés” and “Riodeva”.
Firstly, we observed that the transcripts of the ScALMT1 gene were induced by the presence of Al in the hydroponic medium, were more highly expressed in the root apices than in the non-apical parts of the roots and in the leaves (data not shown), and were more abundant in the Al tolerant genotype than the Al sensitive one (Fig. 6). The expression of the gene is not confined to the root apices but is more abundant there than in the non-apical parts of the roots (data not shown). Gene induction in Al tolerant rye seems to start during the first hour of Al exposure and keeps increasing until at least 20 h after exposure. However, induction in the Al non-tolerant inbred line does not seem to start until at least 6 h after the Al treatment. Very little expression of this gene was detected without Al even in the Al tolerant rye cultivar. The ScALMT1 gene transcripts in root apices of Al tolerant rye (“Ailés”) increases ∼20 times between time zero and 20 h after Al exposure. On the other hand, gene expression at the same moment (20 h) was found to be increased just ∼5 times in the root apex of the Al non-tolerant line (“Riodeva”). Five-fold differences in the expression were found between the Al tolerant and the Al non-tolerant genotypes after 20 h of exposure.
Real-time RT-PCR results showing temporal expression patterns of root tip cDNA transcripts of ScALMT1 gene from tolerant (T) and non-tolerant (NT) genotypes over a 20-h period. Change (fold difference) at each time point is expressed as the relative difference in expression compared to the non-tolerant genotype without exposure to aluminum. The columns represent the mean of two replicates per time point and the error bars show the range of values
Transcripts from leaves of rye genotypes show a very weak expression (data not shown). The highest level of expression detected in leaves (“Ailés” at 20 h of Al exposure) was less than the expression observed in the non-tolerant roots without Al treatment. The maximum level of leaf expression (“Ailés” at 20 h of Al exposure) is 125 times less than that detected in its own root apex at the same time. In summary, at all time-points, expression in leaves was many fold lower than in roots.
Chinese Spring is a moderately Al tolerant wheat cultivar. Quantitative PCR experiments were carried out to compare the levels of expression in this wheat cultivar and the rye genotypes utilized in the present work (data not shown). We found in Chinese Spring that the TaALMT1 wheat gene is constitutively expressed, and it is not induced by Al. That is, no significant differences in the expression were found at different times after exposure to Al. On the other hand, 8-fold and 35-fold differences in the expression was found in the Al sensitive and Al tolerant genotypes, respectively, compared to Chinese Spring.
PCR detection of the ALMT1 gene in different cereal species
The ALMT1 primers (Table 1) were utilized to carry out amplifications from different cereal genomes. PCR amplifications from the available ditelocentric and nulli-tetrasomic lines for group four of wheat were performed. The ScALMT1-3 primer pair produced different amplification patterns, assigning the TaALMT1 locus to the 4DL chromosome arm of wheat. Additionally, the fragments amplified in the Nulli 4D Tetra 4A and Nulli 4D Tetra 4B lines were cloned and sequenced and two different sequences homologous to TaALMT1 were detected (DQ158090 and DQ158091, respectively). Using the ALMT1 primers, another two homologous fragments with 94.3–96% identity to the rye one were detected in barley (DQ158088 and DQ158089). Chromosomal location experiments were attempted by amplification from the corresponding wheat–barley addition lines but no size differences between the barley and the wheat genomes were found. Then, after using eight different restriction enzymes to digest the PCR products (EcoRI, HapII, HhaI, HaeIII, HpaI, MseI, PstI, AfaI) a HhaI restriction site was detected in wheat that was absent in barley, allowing the detection of a DNA fragment on the 4H chromosome (Fig. 7). This fragment was sequenced to confirm that it was homologous to ScALMT1. Finally, the ScALMT1-3 pair of primers was used to amplify and sequence DNA fragments from Triticum urartu (DQ158093), Aegilops speltoides (DQ158092), Avena sativa (DQ322703), Saccharum officinarum (DQ322704), Zea mays (DQ322705) and Phaseolus vulgaris (DQ322702). These each showed at least 72.3% similarity to the wheat sequences.
Chromosomal location of the barley hvALMT1 gene. Amplification of genomic DNA using ScALMT1-3F and ScALMT1-3R primers and ulterior restriction with HhaI enzyme. 1H to 7H: wheat “Chinese Spring”–barley “Betzes” disomic addition lines 1H to 7H, B: barley “B”, CS: wheat “CS”, M molecular-weight marker (100 bp ladder)
Discussion
In the research described here, a candidate-gene approach was used to begin to determine the molecular identity of the Alt4 locus (previously located on 7RS). The segregation pattern of the ScALMT1 candidate gene, detected on the basis of a SNP polymorphism, was determined in three mapping populations and completely cosegregated with the Al tolerance phenotype. We thought that the Al tolerance gene TaALMT1 described in wheat (Sasaki et al. 2004) could be the Alt4 gene in rye because the 4DL arm in wheat containing the TaALMT1 gene (Raman et al. 2003, 2005) is partly homoeologous to the 7RS chromosome arm in rye (Naranjo and Fernandez-Rueda 1991; Devos et al. 1993). Subsequently, we found that this gene is primarily expressed in the root tip of rye just like in the wheat TaALMT1 gene (Sasaki et al. 2004), and that there is an increased expression in tolerant versus sensitive genotypes.
The structure of the ScALMT1 genomic region reveals that two of the introns are larger than expected in a plant gene, that is something around 150–350 bp on average (Wendel et al. 2002). Probably, this feature or some secondary structure prevented the amplification of the ScALMT1 gene in just one piece. Lack of large differences in the size of the introns (less than 37 bp) between tolerant and non-tolerant genotypes does not point to an obvious involvement in the introns in determining functional allelic variation in ScALMT1. ScALMT1 has a similar structure to the wheat TaALMT1 gene (Raman et al. 2005). At least two alleles of ScALMT1 are present in rye (ScALMT1-1 and ScALMT1-2). Although six amino acid substitutions between the two alleles were found, most of the protein analysis programs found no structural transformations of statistical significance. Only the PSIPRED software predicted a conformational change due to a transition from a Gly to an Arg. Glycine can influence the conformation of polypeptides since this amino acid can allow the polypeptide to adopt conformations which are sterically forbidden in other amino acids. However, more data are needed to investigate a potential functional significance of this difference. Also, both the wheat and rye proteins had six putative transmembrane regions. The same plasma membrane localization recently demonstrated for the wheat malate channel (Yamaguchi et al. 2005) should be expected for the rye gene. The Arabidopsis AtALMT1 gene, which encodes a malate transporter as well, has been identified as one of the genes critical for A1 tolerance in Arabidopsis (Hoekenga et al. 2006). In our opinion, the minimal sequence differences detected indicate that both wheat and rye alleles are functionally capable of organic acid transport to the rhizosphere. However, a few amino acid changes can modify the function/substrate of a transport protein. Rye releases citrate from its roots as well as malate (Li et al. 2000) so the possibility that the rye ScALMT1 gene facilitates citrate release instead of malate release has to be considered. Experimental works are being done in order to measure the malate and citrate efflux from the roots of the two rye lines (Al tolerant cv. “Ailés” and Al non-tolerant “Riodeva” inbred line) to address this question.
Gene expression experiments have demonstrated that the ScALMT1 transcripts are primarily expressed in the root apex and are induced by aluminum. Numerous authors have presented data suggesting that the secretion of organic acids to the rhizosphere is a common mechanism of Al tolerance in plants (Ryan et al. 1995; Delhaize et al. 1993; Zheng et al. 1998). Two patterns of organic acid secretion have been identified. In pattern I, no discernible delay is observed between the addition of Al and the onset of organic acid release. In wheat the secretion of malate was detectable within 15–30 min after exposure to Al (Delhaize et al. 1993). In pattern II, organic acid secretion is delayed for several hours after exposure to Al. In rye, citrate and malate efflux increases steadily during a 10-h period (Li et al. 2000). These observations could be linked to a different pattern of mRNA expression between these species. Constitutive expression of the TaALMT1 gene, similar to malate secretion behavior, has been reported in wheat (Sasaki et al. 2004). We also found in Chinese Spring that the TaALMT1 homologous wheat gene is constitutively expressed, and it is not induced by Al (data not shown). In our work, time-course experiments suggest that this different organic acid secretion behavior between wheat and rye may be due to a different pattern of gene expression of homologous organic acid transporter genes. Further work will be needed to elucidate the reason for this different behavior between wheat and rye.
Our sequencing, mapping and expression results support the hypothesis that the first isolation of a gene of agronomic interest in rye has probably been reached. Cereal crops exhibit variation in Al tolerance, and rye (Secale cereale L.) is the most tolerant cereal (Aniol and Madej 1996; Kim et al. 2001), whereas barley is the most sensitive (Foy 1983). Therefore, the understanding of the mechanism and the genes that control Al tolerance in rye is important and will provide fundamental information than can be used to increase Al tolerance in other cereals. The introduction of the TaALMT1 gene of wheat into barley cultivars increases their tolerance to aluminum (Delhaize et al. 2004). The cv. Chinese Spring has previously been shown to exhibit a TaALMT1 transcript expression level which is approximately half that of the Al tolerant genotypes (ET8 line and Atlas 66 cultivar) (Raman et al. 2005). We have shown that the Al tolerant “Ailés” rye cultivar is expressing this gene at higher levels than the Chinese Spring wheat cultivar. These results would indicate that the ScALMT1 rye gene might be a source of greater Al tolerance. This fact should be taken with great care because equal expression of the reference gene in wheat and rye has been assumed, and the comparison of the expression of the 18S reference gene has not be done yet. Taking into account this very high level of Al tolerance and gene expression in rye, we will investigate whether the use of the ScALMT1 gene can further increase plant production in acidic soils.
There are previous data showing that the wheat 4DL arm is homoeologous to the barley 4HL and to the rye 7RS and 5RL chromosome arms, and is not homoeologous to the rye 4RL arm (Naranjo and Fernandez-Rueda 1991; Devos et al. 1993; Naranjo et al. 1997). Moreover, Gale and Devos (1998) have obtained a consensus grass comparative map and have pointed out that rice chromosome 3 is homoeologous to wheat chromosome 4. A conserved aluminum tolerance locus in the homoeology group four of triticeae species has been proposed (Rodriguez-Milla and Gustafson 2001; Raman et al. 2002; Nguyen et al. 2003). To date, genes for aluminum tolerance on 4DL (Rodriguez-Milla and Gustafson 2001), 4HL (Raman et al. 2002), chromosome 3 of rice (Nguyen et al. 2002, 2003) and chromosome 7RS (Matos et al. 2005) have been mapped.
Our results support the hypothesis of the existence of a common mechanism of Al tolerance encoded by a gene located in the homoeologous group four of cereals. We have isolated the second member of the family of the Al-activated organic acid transporters in cereals (ScALMT1) and found complete co-segregation between the ScALMT1 genotype and the phenotype of tolerance to aluminum. We have shown that the ALMT1 genes of wheat, rye and barley are located on the 4DL, 7RS, and 4H chromosomes, respectively. The wheat chromosomal location result confirmed those obtained by different authors (Ma et al. 2005; Raman et al. 2005). Our chromosomal location of the putative HvALMT1 gene on 4H chromosome suggests that the barley Al tolerance locus (Alp) previously located on 4HL arm (Raman et al. 2002) could be such gene. Additionally, we have also detected the putative ALMT1 sequence in eight grass genomes and even in one dicotyledoneous species. In conclusion, it is very likely that in the homeology group four of grasses, there is a mechanism of aluminum tolerance of different magnitudes depending on the species based on organic acid efflux from the soil via the ALMT1 gene.
A map of the 7R chromosome including the ScALMT1 gene and the Alt4 locus has been constructed. In rye there are several examples of the use of molecular markers to detect traits of interest (Borner et al. 1999; Miedaner et al. 2000; Korzun et al. 2001; Miftahudin et al. 2002, 2004, 2005). Although there are also some genetic maps, the rye genome has not been mapped to a saturation level (Korzun et al. 2001; Ma et al. 2001; Hackauf and Wehling 2002; Bednarek et al. 2003; Khlestkina et al. 2005). Also, with the genome size of rye reaching approximately 8,000 Mb (Bennett and Smith 1976), a positional cloning is very difficult to achieve in this cereal species. In fact, to date there is no map-based cloned gene in this species. Therefore, as we have shown here, a candidate gene approach is a logical choice in some cases.
References
Aniol A (1984) Introduction of aluminum tolerance into aluminum sensitive wheat cultivars. Zeitschrift Fur Pflanzenzuchtung-J Plant Breed 93:331–339
Aniol A (2004) Chromosomal location of aluminum tolerance genes in rye. Plant Breed 123:132–136
Aniol A, Gustafson JP (1984) Chromosome location of genes-controlling aluminum tolerance in wheat, rye, triticale. Can J Genet Cytol 26:701–705
Aniol A, Madej L (1996) Genetic variation for aluminum tolerance in rye. Vortr Pflanzenz chtg 35:201–211
Aniol A, Hill RD, Larter EN (1980) Aluminum tolerance of spring rye inbred lines. Crop Sci 20:205–208
Basu U, Mcdonaldstephens JL, Archambault DJ, Good AG, Briggs KG et al (1997) Genetic and physiological analysis of doubled-haploid, aluminum-resistant lines of wheat provide evidence for the involvement of a 23 kD, root exudate polypeptide in mediating resistance. Plant Soil 196:283–288
Bednarek PT Piotr M, Renata L, Beata Mylknw EE (2003) Saturating rye genetic map with amplified fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) markers. J Appl Genet 44(1):21–33
Bennett MD, Smith JB (1976) Nuclear-DNA amounts in angiosperms. Philos Trans R Soc Lond Ser B Biol Sci 274:227–274
Borner A, Korzun V, Voylokov AV, Weber WE (1999) Detection of quantitative trait loci on chromosome 5R of rye (Secale cereale L.). Theor Appl Genet 98:1087–1090
Brondani C, Paiva E (1996) ‘‘RFLP’’ analysis of aluminum tolerance in chromosome 2 in maize. Pesquisa Agropecuaria Brasileira 31:575–579
Brosche M, Strid A (1999) Cloning, expression, molecular characterization of a small pea gene family regulated by low levels of ultraviolet B radiation and other stresses. Plant Physiol 121:479–487
Claros MG, Vonheijne G (1994) Toppred-Ii—an improved software for membrane-protein structure predictions. Comput Appl Biosci 10:685–686
Cruz-Ortega R, Cushman JC, Ownby JD (1997) cDNA clones encoding 1,3-beta-glucanase and a fimbrin-like cytoskeletal protein are induced by Al toxicity in wheat roots. Plant Physiol 114:1453–1460
Cserzo M, Eisenhaber F, Eisenhaber B, Simon I (2002) On filtering false positive transmembrane protein predictions. Protein Eng 15:745–752
Delhaize E, Ryan PR (1995) Aluminum toxicity and tolerance in plants. Plant Physiol 107:315–321
Delhaize E, Ryan PR, Randall PJ (1993) Aluminum tolerance in wheat (Triticum-Aestivum L). 2. Aluminum-stimulated excretion of malic-acid from root apices. Plant Physiol 103:695–702
Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T et al (2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA 101:15249–15254
Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL et al (1993) Chromosomal rearrangements in the Rye genome relative to that of wheat. Theor Appl Genet 85:673–680
Ermolayev V, Weschke W, Manteuffel R (2003) Comparison of Al-induced gene expression in sensitive and tolerant soybean cultivars. J Exp Bot 54:2745–2756
Ezaki B, Yamamoto Y, Matsumoto H (1995) Cloning and sequencing of the Cdnas induced by aluminum treatment and P-I starvation in cultured tobacco cells. Physiologia Plantarum 93:11–18
Ezaki B, Tsugita S, Matsumoto H (1996) Expression of a moderately anionic peroxidase is induced by aluminum treatment in tobacco cells: Possible involvement of peroxidase isozymes in aluminum ion stress. Physiologia Plantarum 96:21–28
Foy C (1983) The physiology of plant adaptation to mineral stress. Iowa State J Res 57:355–392
Foy CD, Chaney RL, White MC (1978) Physiology of metal toxicity in plants. Annu Rev Plant Physiol Plant Mol Biol 29:511–566
Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95(5):1971–1974
Gallego FJ, Benito C (1997) Genetic control of aluminum tolerance in rye (Secale cereale L.). Theor Appl Genet 95:393–399
Gallego FJ, Calles B, Benito C (1998a) Molecular markers linked to the aluminum tolerance gene Alt1 in rye (Secale cereale L). Theor Appl Genet 97:1104–1109
Gallego FJ, Lopez-Solanilla E, Figueiras AM, Benito C (1998b) Chromosomal location of PCR fragments as a source of DNA markers linked to aluminum tolerance genes in rye. Theor Appl Genet 96:426–434
Hackauf B, Wehling P (2002) Identification of microsatellite polymorphisms in an expressed portion of the rye genome. Plant Breed 121:17–25
Hamel F, Breton C, Houde M (1998) Isolation and characterization of wheat aluminum-regulated genes: possible involvement of aluminum as a pathogenesis response elicitor. Planta 205:531–538
Hamilton CA, Good AG, Taylor GJ (2001) Induction of vacuolar ATPase and mitochondrial ATP synthase by aluminum in an aluminum-resistant cultivar of wheat. Plant Physiol 125:2068–2077
Haug A (1984) Molecular aspects of aluminum toxicity. Crc Crit Rev Plant Sci 1:345–373
Higgins DG, Sharp PM (1988) Clustal—a package for performing multiple sequence alignment on a microcomputer. Gene 73:237–244
Hoekenga OA, Vision TJ, Shaff JE, Monforte AJ, Lee GP et al (2003) Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta × Columbia) by quantitative trait locus mapping. A physiologically simple but genetically complex trait. Plant Physiol 132:936–948
Hoekenga OA, Maron LG, Piñeros MA, Cançado GMA, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, Matsumoto H, Yamamoto Y, Koyama H, Kochian LV (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA 103:9738–9743
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202
Khlestkina EK, Than MHM, Pestsova EG, Roder MS, Malyshev SV et al (2005) Mapping of 99 new microsatellite-derived loci in rye (Secale cereale L.) including 39 expressed sequence tags (vol 110, pg 990, 2005). Theor Appl Genet 110:990–991
Kim BY, Baier AC, Somers DJ, Gustafson JP (2001) Aluminum tolerance in triticale, wheat, and rye. Euphytica 120:329–337
Kobayashi Y, Koyama H (2002) QTL analysis of Al tolerance in recombinant inbred lines of Arabidopsis thaliana. Plant Cell Physiol 43:1526–1533
Kochian LVPC (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46:237–260
Kochian LV, Pence NS, Letham DLD, Pineros MA, Magalhaes JV et al (2002) Mechanisms of metal resistance in plants: aluminum and heavy metals. Plant Soil 247:109–119
Kochian LV, Hoekenga OA, Pineros MAPC (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55:459–493
Korzun V, Malyshev S, Voylokov AV, Borner A (2001) A genetic map of rye (Secale cereale L.) combining RFLP, isozyme, protein, microsatellite and gene loci. Theor Appl Genet 102:709–717
Kosambi D (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Krogh A, Larsson B, Vonheijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Larsen PB, Tai CY, Kochian LV, Howell SH (1996) Arabidopsis mutants with increased sensitivity to aluminum. Plant Physiol 110:743–751
Larsen PB, Degenhardt J, Tai CY, Stenzler LM, Howell SH et al (1998) Aluminum-resistant Arabidopsis mutants that exhibit altered patterns of aluminum accumulation and organic acid release from roots. Plant Physiol 117:9–18
Larsen PB, Geisler MJB, Jones CA, Williams KM, Cancel JD (2005) ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J 41:353–363
Li XF, Ma JF, Matsumoto HPC (2000) Pattern of aluminum-induced secretion of organic acids differs between rye and wheat. Plant Physiol 123:1537–1543
Little R (1988) Plant–soil interactions at low Ph problem-solving—the genetic approach. Commun Soil Sci Plant Anal 19:1239–1257
Luo MC, Dvorak J (1996) Molecular mapping of an aluminum tolerance locus on chromosome 4D of Chinese Spring wheat. Euphytica 91:31–35
Ma JF, Furukawa J (2003) Recent progress in the research of external Al detoxification in higher plants: a minireview. J Inorg Biochem 97:46–51
Ma JF, Taketa S, Yang ZM (2000) Aluminum tolerance genes on the short arm of chromosome 3R are linked to organic acid release in triticale. Plant Physiol 122:687–694
Ma XF, Wanous MK, Houchins K, Milla MAR, Goicoechea PG et al (2001) Molecular linkage mapping in rye (Secale cereale L.). Theor Appl Genet 102:517–523
Ma HX, Bai GH, Carver BF, Zhou LL (2005) Molecular mapping of a quantitative trait locus for aluminum tolerance in wheat cultivar Atlas 66. Theor Appl Genet 112:51–57
Magalhaes JV, Garvin DF, Wang YH, Sorrells ME, Klein PE et al (2004) Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the Poaceae. Genetics 167:1905–1914
Matos M, Camacho MV, Pérez-Flores V, Pernaute B, Pinto-Carnide O et al (2005) A new aluminum tolerance gene located on rye chromosome arm 7RS. Theor Appl Genet 111:360–369
Miedaner T, Glass C, Dreyer F, Wilde P, Wortmann H et al (2000) Mapping of genes for male-fertility restoration in ‘Pampa’ CMS winter rye (Secale cereale L.). Theor Appl Genet 101:1226–1233
Miftahudin T, Scoles GJ, Gustafson JP (2002) AFLP markers tightly linked to the aluminum-tolerance gene Alt3 in rye (Secale cereale L.). Theor Appl Genet 104:626–631
Miftahudin T, Scoles GJ, Gustafson JP (2004) Development of PCR-based codominant markers flanking the Alt3 gene in rye. Genome 47:231–238
Miftahudin T, Chikmawati T, Ross K, Scoles GJ, Gustafson JP (2005) Targeting the aluminum tolerance gene Alt3 region in rye, using rice/rye micro-colinearity. Theor Appl Genet 110:906–913
Miyasaka SC, Buta JG, Howell RK, Foy CD (1991) Mechanism of aluminum tolerance in snapbeans—root exudation of citric-acid. Plant Physiol 96:737–743
Mugwira LM, Elgawhary SM, Patel SU (1978) Aluminum tolerance in triticale, wheat and rye as measured by root-growth characteristics and aluminum concentration. Plant Soil 50:681–690
Naranjo T, Fernandez-Rueda P (1991) Homeology of rye chromosome arms to wheat. Theor Appl Genet 82:577–586
Naranjo T, Fernandez-Rueda P, Maestra B (1997) Chromosome rearrangements and homoeologous pairing: implications for the introgression of alien genes into wheat. In: Lelley T (ed) Current topics in plant cytogenetics related to plant improvement. WUV-Universitätsverlag, Austria, pp 198–205
Nguyen VT, Nguyen BD, Sarkarung S, Martinez C, Paterson AH et al (2002) Mapping of genes controlling aluminum tolerance in rice: comparison of different genetic backgrounds. Mol Genet Genomics 267:772–780
Nguyen BD, Brar DS, Bui BC, Nguyen TV, Pham LN et al (2003) Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., into indica rice (Oryza sativa L.). Theor Appl Genet 106:583–593
Persson B, Argos P (1997) Prediction of membrane protein topology utilizing multiple sequence alignments. J Protein Chem 16:453–457
Raman H, Moroni JS, Sato K, Read BJ, Scott BJ (2002) Identification of AFLP and microsatellite markers linked with an aluminum tolerance gene in barley (Hordeum vulgare L.). Theor Appl Genet 105:458–464
Raman H, Zhang K, Appels R, Moroni S, Cakir M et al (2003) Molecular mapping and mechanism of aluminum tolerance in five doubled haploid populations of Triticum aestivum L (Bread wheat). In: Proceedings of 10th international wheat genetics symposium, Italy, pp 404–406
Raman H, Zhang K, Cakir M, Appels R, Garvin DF et al (2005) Molecular characterization and mapping of ALMT1, the aluminum-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48:781–791
Rao IM, Zeigler RS, Vera R, Sarkarung S (1993) Selection and breeding for acid-soil tolerance in crops. Bioscience 43:454–465
Richards KD, Gardner RC (1994) The effect of aluminum treatment on wheat roots—expression of heat-shock, histone and Shh genes. Plant Sci 98:37–45
Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC (1998) Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol 116:409–418
Riede CR, Anderson JA (1996) Linkage of RFLP markers to an aluminum tolerance gene in wheat. Crop Sci 36:905–909
Rodriguez-Milla MA, Gustafson JP (2001) Genetic and physical characterization of chromosome 4DL in wheat. Genome 44:883–892
Rodriguez-Milla MA, Butler E, Huete AR, Wilson CF, Anderson O et al (2002) Expressed sequence tag-based gene expression analysis under aluminum stress in Rye. Plant Physiol 130:1706–1716
Rost B (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. In: Computer Methods for Macromolecular Sequence Analysis, pp 525–539
Ryan PR, Ditomaso JM, Kochian LV (1993) Aluminum toxicity in root —an investigation of spatial sensitivity and the role of the root cap. J Exp Bot 44:437–446
Ryan PR, Delhaize E, Randall PJ (1995) Characterization of Al-stimulated efflux of malate from the apices of Al-tolerant wheat roots. Planta 196:103–110
Samac DA, Tesfaye M (2003) Plant improvement for tolerance to aluminum in acid soils—a review. Plant Cell Tissue Organ Cult 75:189–207
Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ et al (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37:645–653
Savenstrand H, Brosche M, Angehagen M, Strid A (2000) Molecular markers for ozone stress isolated by suppression subtractive hybridization: specificity of gene expression and identification of a novel stress-regulated gene. Plant Cell Environ 23:689–700
Sibov ST, Gaspar M, Silva MJ, Ottoboni LMM, Arruda P et al (1999) Two genes control aluminum tolerance in maize: genetic and molecular mapping analyses. Genome 42:475–482
Sivaguru M, Ezaki B, He ZH, Tong HY, Osawa H et al (2003) Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol 132:2256–2266
Snowden KC, Gardner RC (1993) 5 Genes induced by aluminum in wheat (Triticum aestivum L) roots. Plant Physiol 103:855–861
Somers DJ, Gustafson JP (1995) The expression of aluminum stress induced polypeptides in a population segregating for aluminum tolerance in wheat (Triticum aestivum L). Genome 38:1213–1220
Stolen O, Andersen S (1978) Inheritance of tolerance to low soil-Ph in barley. Hereditas 88:101–105
Tang Y, Sorrells ME, Kochian LV, Garvin DF (2000) Identification of RFLP markers linked to the barley aluminum tolerance gene Alp. Crop Sci 40:778–782
Tusnady GE, Simon I (1998) Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 283:489–506
Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0, software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Wendel JF, Cronn RC, Alvarez I, Liu B, Small RL et al (2002) Intron size and genome size in plants. Mol Biol Evol 19:2346–2352
Yamaguchi M, Sasaki T, Sivaguru M, Yamamoto Y, Osawa H et al (2005) Evidence for the plasma membrane localization of Al-activated malate transporter (ALMT1). Plant Cell Physiol 46:812–816
Yu LH, Umeda M, Liu JY, Zhao NM, Uchimiya H (1998) A novel MT gene of rice plants is strongly expressed in the node portion of the stem. Gene 206:29–35
Zheng SJ, Ma JJ, Matsumoto H (1998) High aluminum resistance in buckwheat – I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol 117:745–751
Acknowledgments
We appreciate the comments of A. Ramos on the manuscript. We wish to thank A.J. Lukaszewski for kindly providing the wheat–rye disomic addition lines. This work was supported by the research grants AGL 2003-06470 from the Ministerio de Educación y Ciencia de España, PR1/05 from the Universidad Complutense and PR27/05-13599 from the Santander/Complutense. G. Fontecha is a recipient of Programa Alban predoctoral fellowship (UE).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Langridge.
G. Fontecha and J. Silva-Navas contributed equally to this work.
Rights and permissions
About this article
Cite this article
Fontecha, G., Silva-Navas, J., Benito, C. et al. Candidate gene identification of an aluminum-activated organic acid transporter gene at the Alt4 locus for aluminum tolerance in rye (Secale cereale L.). Theor Appl Genet 114, 249–260 (2007). https://doi.org/10.1007/s00122-006-0427-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0427-7