Abstract
The domestication of emmer wheat (Triticum turgidum spp. dicoccoides, genomes BBAA) was one of the key events during the emergence of agriculture in southwestern Asia, and was a prerequisite for the evolution of durum and common wheat. Single- and multilocus genotypes based on restriction fragment length polymorphism at 131 loci were analyzed to describe the structure of populations of wild and domesticated emmer and to generate a picture of emmer domestication and its subsequent diffusion across Asia, Europe and Africa. Wild emmer consists of two populations, southern and northern, each further subdivided. Domesticated emmer mirrors the geographic subdivision of wild emmer into the northern and southern populations and also shows an additional structure in both regions. Gene flow between wild and domesticated emmer occurred across the entire area of wild emmer distribution. Emmer was likely domesticated in the Diyarbakir region in southeastern Turkey, which was followed by subsequent hybridization and introgression from wild to domesticated emmer in southern Levant. A less likely scenario is that emmer was domesticated independently in the Diyarbakir region and southern Levant, and the Levantine genepool was absorbed into the genepool of domesticated emmer diffusing from southeastern Turkey. Durum wheat is closely related to domesticated emmer in the eastern Mediterranean and likely originated there.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The emergence of agriculture in the Fertile Crescent, an arc of land stretching from the Levant to southeastern (SE) Turkey, northern Iraq, and western Iran, was founded on the domestication of three cereals: diploid einkorn wheat (Triticum monococcum L., genomes AmAm), tetraploid emmer wheat (T. turgidum L. ssp. dicoccon, genomes BBAA), and diploid barley (Hordeum vulgare L., genomes HH). Domesticated einkorn and emmer began to appear in southern Levant and SE Turkey about 9,600 to 9,000 bp, and there is no evidence that one of the two wheats was domesticated earlier than the other (Nesbitt and Samuel 1996). The domestication of hulled emmer was the first step that ultimately resulted in the evolution of free-threshing tetraploid durum wheat (T. turgidum ssp. durum, genomes BBAA) and hexaploid bread wheat (T. aestivum ssp. aestivum, genomes BBAADD) that are today globally two of the most important field crops.
A long temporal transition from wild forms with brittle spike rachis to cultivated forms with non-brittle spike rachis and almost simultaneous appearance of domesticated forms at archeological sites across the Fertile Crescent has thwarted the effort to identify the site(s) at which the cereals were domesticated on the basis of archeology alone (Willcox 1998). If it could be assumed that the distribution and genetic structure of the present-day populations of wild ancestors of the three cereals approximate the distribution and genetic structure of these populations at the time of origins of agriculture, it should be possible to identify the domestication site of each cereal from genetic relationships between wild and domesticated populations. Several studies based on this rationale have been reported. Heun et al. (1997) used amplified fragment length polymorphism (AFLP) to assess genetic relatedness between wild and domesticated einkorn wheat and concluded that the Karaca Dag mountains in SE Turkey was the site of einkorn domestication. Badr et al. (2000), also using AFLP, concluded that barley was domesticated in southern Levant, which approximately consists of modern Jordan, Israel, Palestine, Lebanon, and southwestern (SW) Syria. The site of emmer domestication has remained inconclusive.
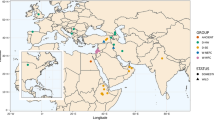
Wild emmer (T. turgidum ssp. dicoccoides) grows in a discontinuous arc from southern Levant to northwestern (NW) Syria, SE Turkey, northern Iraq, and NW Iran (Fig. 1). In SE Turkey, it grows from the Kartal Dagi mountains northwest of Gaziantep, throughout the Urfa plateau, the Karaca Dag region west of Diyarbakir, and the Silvan region NE of Diyarbakir.
Distribution of wild emmer across the Fertile Crescent. The geographic locations of local populations are indicated by dotted lines. They are designated by numbers corresponding to populations identified by multilocus genotype analyses. Modern countries and mountain ranges are in bold. Relevant cities are indicated by solid circles. Archeological sites are indicated by open circles
Morphologically, wild emmer consists of two races. Most emmer is slender and is classified as race horanum. A robust judaicum race grows sporadically in Israel. Blumler (1998) suggested that the judaicum race originated from hybridization between wild emmer and durum wheat.
Ozkan et al. (2002) compared wild and domesticated emmer using AFLP and showed that wild emmer formed two populations: southern and northern. Domesticated emmer was more related to the northern than to the southern wild emmer. Since populations from the Kartal Dagi mountains, Urfa plateau, Iraq, and Iran were sampled inadequately or not at all, the identification of the site at which emmer was domesticated remained inconclusive. To fill this gap, Mori et al. (2003) investigated chloroplast microsatellite variation in a sample that included these populations. They discovered a large number of ctDNA haplotypes that could be allocated into two broad lineages, I and II. Haplotype 10, belonging to lineage I, was present in 39.6% of domesticated emmer accessions and 90% of common wheat (T. aestivum) accessions. This haplotype was present in three accessions of wild emmer from the Kartal Dagi region that represented 4% of the total number of wild emmer accessions investigated. They hence concluded that emmer was domesticated in the Kartal Dagi region. In addition, they found haplotypes 22 and 59 belonging to lineage II in domesticated emmer and T. aestivum but not in wild emmer. Closely related haplotypes were encountered in geographically diverse populations of wild emmer. These findings led Mori et al. (2003) to hypothesize that emmer was domesticated twice, once in the Kartal Dagi region and a second time somewhere else.
Ozkan et al. (2005) acquired accessions investigated by Mori et al. (2003), performed AFLP on them, and concluded that Kartal Dagi was not the site of emmer domestication. Wild emmer accessions from the Karaca Dag region and from the Sulaimanyia region along the Iraq/Iran border appeared equally closely related to domesticated emmer (Ozkan et al. 2005). This finding left the question of emmer domestication open to various interpretations. Emmer could have been domesticated only once. In that case the site could have been either the Karaca Dag region or Sulaimaniya region. Alternatively, emmer could have been domesticated in both regions independently.
The question of emmer domestication is revisited here, employing experimental techniques different from those used previously. Restriction fragment length polymorphism (RFLP) at genetically characterized loci is used, which facilitated the use of multilocus genotype analyses in allocation of emmer accessions into populations (Pritchard et al. 2000; Falush et al. 2003). Several experimental approaches are used to assess the significance of the past gene flow for genetic relationships between the populations of wild and domesticated emmer. Finally, the geographic region where durum wheat may have originated is inferred.
Materials and methods
Plants
A total of 277 accessions of wild emmer, 186 landraces of domesticated emmer, and 55 landraces and varieties of durum wheat were used. Geographic origins, allocation to populations, and admixture are summarized in electronic supplementary material of this paper (S1). The distribution of wild emmer accessions across the Fertile Crescent and the location of the populations are shown in Fig. 1. The location of accessions can be obtained from the US National Germplasm Repository web site. All taxa used in this project are self-pollinating.
Markers and genotypes
DNAs were isolated from a single plant per accession, landrace, or cultivar according to Dvorak et al. (1988). RFLPs at 131 single-copy loci that have previously been mapped in T. monococcum (Dubcovsky et al. 1996), Ae. speltoides (Luo et al. 2005) and Ae. tauschii (J. Dvorak, K. R. Deal and M. C. Luo, unpublished) were detected in Southern blots with 58 cDNA or PstI genomic clones (Dvorak et al. 2004). DNA of T. aestivum cv ‘Chinese Spring’ was inserted into one lane of each Southern blot and used as a standard to which the restriction fragments in the blot were compared. Restriction fragments were assigned to chromosomes with the aid of ‘Chinese Spring’ wheat nulisomic-tetrasomic lines (Sears 1966) and Chinese Spring-Lophopyrum elongatum disomic substitution lines (Dvorak 1980). Homozygosity of the plants was exploited for the determination of allelism among restriction fragments at polymorphic loci. Restriction fragments simultaneously present in a restriction profile of a single plant were assumed not to be allelic. This methodology made it possible to allocate all restriction fragments to loci and genomes.
Multilocus cluster analysis
To assign accessions and landraces to populations, multilocus genotype based clustering implemented in Structure v2.1 was used (Pritchard and Wen 2004). Multilocus genotype analysis assuming linkage as implemented in the program was performed. Because the plants were homozygous, haploid data option was used. On the basis of preliminary trials, population models varying from two to nine populations (K = 1 to 9) were tested with the Structure v2.1 program using 70,000 burnin runs and 30,000 iterations; ten independent runs were performed at each level of K. Symmetrical similarity coefficients (SSC) among the ten independent runs were computed as described by Rosenberg et al. (2002). The statistical significance of differences between mean log likelihoods at the neighboring levels of K (between K = 2 and K = 3, between K = 3 and K = 4, etc.) was tested with Wilcoxon two-sample test. Clustering of accessions and landraces at a K value that showed mean log likelihood that did not significantly differ from the mean log likelihood at the next higher K level was used in analyses. The most frequent clustering at that K was used for the allocation of accessions and landraces into populations. Plants showing admixture constituting more than 10% genotype at K = 8 were classified as admixture and removed from the dataset in some analyses. Admixture indicates that an individual has a mixed ancestry and shares a fraction of its genome with individuals allocated to two or more populations. The population source of admixture was determined for each accession and landrace classified as being an admixture in ten runs at K = 8.
In the follow-up population structure analyses, wild emmer accessions allocated into the northern population and southern population were separately analyzed with Structure v2.1 program, using 70,000 burnin runs and 130,000 iterations. Ten independent runs at K = 2 to 6 were performed. Accessions were allocated into populations on the basis of the most frequent clustering at a selected K level.
Gene diversity analysis
Allele frequencies were used to compute mean gene diversity in each population. Mean gene diversity was expressed as expected heterozygosity (He) implemented in the GDA computer program (Lewis and Zaykin 1997). Allele frequencies were used to estimate variance components with analysis of variance (ANOVA). Four-level sampling hierarchy design (Weir 1996) was employed: Level 1 was the domesticated and wild status, level 2 was northern and southern geographic region within the domesticated or wild status, level 3 was populations within geographic region, and level 4 was accessions, landraces or varieties within populations. F-statistic was computed for each level from the variance components. The option not assuming Hardy–Weinberg equilibrium was used. Confidence intervals (CIs) of Fst estimates based on 1,000 bootstrap replications across loci were constructed.
A two-level sampling hierarchy ANOVA was performed using allele frequencies of wild emmer accessions from the northern geographic region (population 3) and the 95% bootstrap confidence interval of Fst was constructed. A similar two-level sampling hierarchy ANOVA was performed for the wild emmer accessions from the southern geographic region of wild emmer.
Genetic distance analysis
Allele frequencies were used to compute Nei’s genetic distances (Nei 1978) among individual accessions of wild emmer and landraces of domesticated emmer with the GDA program package. An unrooted phylogenetic tree was built using neighbor-joining method (NJM) with the GDA program package and visualized with TreeView (Page 1996). In further analyses, populations were formed on the basis of allocation of accessions and landraces into populations on the basis of multilocus genotype clustering. Allele frequencies were used to compute Nei’s genetic distances between populations. Genetic distances were also computed between populations from which accessions and landraces showing more than 10% admixture at K = 8 were removed. The CIs about each genetic distance were constructed from 1,000 bootstrap replications for P values ranging from 0.70 to 0.95 (Weir 1996). To determine P of a difference between two genetic distance estimates, CIs about means were compared. The lowest P at which CIs did not overlap was used as a bootstrap P value of the difference between the mean genetic distances.
Morphological analysis
Plants were grown in a twice replicated trial in the field in a completely randomized design. The spikelets were harvested and the length without awns and width of 5–10 spikelets per plant were measured and averaged. Average seed weight of 50 seeds was determined. One-way ANOVA was performed with the GLM Procedure and Duncan’s multiple range test (SAS 9.1).
Results
Population structure
The hybridization of 58 cDNA or PstI clones with DNA of 277 accessions of wild emmer, 186 landraces of domesticated emmer, and 55 landraces and varieties of durum resulted in the detection of 131 loci, of which 109 were polymorphic. A total of 529 informative alleles were found, all of which were assigned to genomes and loci. On the basis of preliminary trials, population models varying from two to nine populations (K = 2 to 9) were clustered by multilocus genotype analysis. The mean log likelihoods differed significantly from K = 2 to K = 8 but not between K = 8 and K = 9. Complex data, such as those analyzed here, produce many clustering schemes, which is reflected by low SSCs among the runs. SSCs ranged from 0.35 to 0.69, being the lowest at K = 2 and the highest at K = 4. SSC was 0.38 at K = 8. For the total sample, K = 8 was assumed to be optimal.
The entire species was subdivided into wild and domesticated (Fig. 2a). Wild emmer was subdivided into two clusters coinciding with the southern (Levant) and northern (Turkey, Iraq and Iran) geographic regions (Fig. 2a). At K = 8, wild emmer accessions clustered into three populations. Populations 1 and 2 were located in Levant and population 3 coincided with wild emmer in the northern geographic region. Domesticated emmer was subdivided into four populations. Landraces from Ethiopia, Oman and India were included into population 4, those from Mediterranean and Levant into population 5, those from Balkans, Russia and NW Turkey into population 6, and those from NE Turkey, Georgia, Armenia, Dagestan, and Iran into population 7. Durum wheat formed population 8.
a Clustering based on multilocus genotype analysis for K = 2 to 8 for the entire set of wild and domesticated emmer and durum accessions and landraces. Accessions are arranged according to their geographic locations. Populations suggested by the analyses are numbered as in text. b Clustering for K = 2, 3, and 4 for southern wild emmer accessions in Israel, SW Syria and Lebanon (left) and for K = 2, 3, 4 and 5 for northern wild emmer in Turkey, Iraq, and Iran. c A tree produced by the neighbor joining method from Nei’s genetic distances among individual plants of wild and domesticated emmer. Accessions and land races are designated according to their geographic origins: Israel (I), Lebanon (L), southwestern Syria (S), Kartal Dagi region (K), Urfa plateau (U), Iran (N), Iraq (M), Karaca Dag region (D), and domesticated emmer (C). The correspondence of branches to populations suggested by multilocus analyses is indicated. Accessions and land races showing anomalous positions are boxed and, if being admixture, populations involved are indicated
Multilocus genotype clustering was further performed with wild emmer accessions from southern region only (K = 2 to 6). Mean log likelihoods of runs at the neighboring levels of K significantly differed at all K levels. All runs were identical at K = 2 and 3 and SSC was 0.70 at K = 4. At K = 5, SSC among runs decreased to 0.38. K = 4 was therefore selected for further analyses. At K = 4, population 2 was the same as in the previous analysis but population 1 was subdivided into populations 1a, 1b and 1c (Figs. 1, 2b). Population 1a consisted of accessions from central Israel and SW Syria, population 1b consisted of accessions from northern Israel and population 1c consisted of accessions from Lebanon and Lebanon–Syria border (Fig. 1).
Similar analyses were performed on northern wild emmer (K = 2 to 6). SSC ranged from 0.35 (K = 4) to 1.0 (K = 3). Significant differences were observed between mean log likelihoods for neighboring levels of K until K = 5. No significant difference was observed between mean log likelihoods at K = 5 and K = 6 (P = 0.09). At K = 5, SSC was 0.55, and population 3 was subdivided into 3a (Kartal Dagi region west of Gaziantep), 3b (Urfa plateau west of Karaca Dag mountains), 3c (Al Mawsil region in Iraq and Sulaimaniya region in Iraq/Iran), 3d (Karaca Dag mountains and west of Diyarbakir), and 3e (a population 36.2 km west of Diyarbakir) (Figs. 1, 2b).
To examine differentiation of populations suggested by the preceding analyses, a four-level hierarchical ANOVA was performed examining population differentiation due to domestication status (wild and domesticated), geographic region within a domestication status (north and south), populations within a geographic region, and individual accessions and landraces within populations. The largest Fst value was obtained for populations within geographic regions. Fst values due to geographic region and domestication status did not differ from each other at P = 95% (Table 1).
A two-level ANOVA was performed with wild emmer populations 3a to 3e (northern geographic region) and separately with wild emmer in the southern geographic region. Fst was 0.308 (95% bootstrap CI was 0.265–0.0.349) in the northern region and 0.260 (95% bootstrap CI was 0.222–0.303) in southern region. Overlapping of the two CI intervals suggested that wild emmer populations in the two regions were differentiated to a similar degree.
A tree constructed with NJM from Nei’s genetic distances among individual accessions and landraces largely, although not entirely, corroborated results obtained with the multilocus clustering (Fig. 2c). Individual accessions of wild emmer formed distinct branches corresponding to the northern and southern regions observed in the multilocus analysis. Within the northern region, wild emmer formed two branches in the tree. One was composed of accessions from Kartal Dagi, Urfa plateau, Iraq, and Iran. This branch was equivalent to populations 3a, 3b and 3c in the multilocus analysis. The other branch of northern emmer involved populations 3d and 3e. Southern wild emmer also formed two branches. One consisted of populations 1a, 1b and 2 (Fig. 2c). Population 2 formed a long branch within the 1a + 1b branch. The other branch involved all accessions from Lebanon plus few accessions from the Lebabon/Syria boarder. This branch corresponded to population 1c (Fig. 2c). Domesticated emmer formed a monophyletic branch that was juxtaposed to the branch 3d + 3e of wild emmer (Fig. 2c). Populations 4, 5, 6, and 7 formed four distinct branches within the branch. The branch of population 7 (NE Turkey, Georgia, Armenia, Dagestan, and Iran) was the shortest indicating that individual landraces in population 7 show the shortest distances to wild emmer.
To minimize the potential effects of admixture on the determination of genetic relationships between wild and domesticated emmer populations, accessions showing admixture in the multilocus analysis at K = 8 were removed from data (indicated by asterisks in S1). All accessions from NW Turkey and Dagestan were eliminated because they were an admixture. Nei’s genetic distances between populations were computed (Table 2). The distances of northern wild emmer population 3d were shorter to all populations of domesticated emmer than those of 3a, 3b, 3c, and 3e (bootstrap probabilities ranged from P = 0.30 to 0.22).
Population 3c was divided on the basis of geography to accessions from the Al Mawsil area (3 cM) and Sulaimaniya region (3 cN) and genetic distances were computed. The two populations were similar (D = 0.097) and both showed longer genetic distances to domesticated emmer populations than population 3d (Table 2).
Genetic distances of populations 3 cM and 3 cN to all wild and domesticated emmer were longer than those of the original population 3c, which was caused by reduced gene diversity in the 3 cM and 3 cN populations as compared to the gene diversity in the parental 3c population (Table 3). Genetic distances of population 3 cN to the populations of domesticated emmer were longer than those of population 3 cM, which was likely caused by very low gene diversity of population 3 cN (Table 3).
Genetic distances in Table 2 showed an important fact that was hidden in the neighbor-joining tree in Fig. 2c. Southern populations of domesticated emmer 4 and 5 showed equally short distances to southern wild emmer populations 1a, 1b and 1c as to northern wild emmer population 3d (Table 2). Northern domesticated emmer (populations 6 and 7) did not show this proximity to southern wild emmer.
Genetic distances were also computed between populations from which accessions and landraces showing admixture were not removed. Distances between populations were almost universally shorter than those computed from the dataset without admixture (Table 2). This is undoubtedly caused by the increase in gene diversity in most populations with admixture (Table 3). However, the pattern observed was very similar to that obtained with populations from which admixture was removed, as indicated by correlation r = 0.97 (P < 0.001) between the two datasets.
Both datasets in Table 2 showed an intriguing trend. Most southern wild emmer populations showed shorter distances to southern domesticated emmer populations than to those located in the northern geographic region. Northern populations of wild emmer often showed shorter distance to northern populations of domesticated emmer than to the southern populations of domesticated emmer. We hypothesized that this observation reflected past gene flow between sympatric wild emmer and domesticated emmer populations. Two strategies were used to test statistically this hypothesis.
First, Nei’s genetic distances of each of the nine wild emmer populations to the northern and southern domesticated emmer were computed (Table 4). In seven of nine comparisons, Nei’s genetic distance of a wild emmer population to the domesticated emmer population in the same geographic region (sympatric) was shorter than that to domesticated emmer in the other geographic region (allopatric) (P = 0.05, paired t test).
Second, the source of admixture was determined for all accessions and landraces showing more than 10% admixture from a single population at K = 8. In the southern geographic region, 19 and 3 wild emmer accessions showed admixture from southern and northern domesticated emmer, respectively. In the northern geographic region, 5 and 12 wild emmer accessions showed admixture from southern and northern domesticated emmer, respectively. These data show that domesticated emmer is more closely related to sympatric wild emmer than to allopatric wild emmer (P < 0.001, Fisher exact test) across the wild emmer distribution. For domesticated emmer, 13 and 3 landraces of southern domesticated emmer showed admixture from southern and northern wild emmer accessions, respectively, and 1 and 2 landraces of northern domesticated emmer showed admixture from southern and northern wild emmer, respectively (P = 0.15, Fisher exact test).
Durum showed the shortest genetic distances to southern domesticated emmer populations, population 5 (D = 0.109) and population 4 (D = 0.128) (Table 2). Durum was also more related to southern wild emmer than to northern wild emmer; D ranged from 0.181 to 0.188 between durum and southern wild emmer populations and from 0.200 to 0.326 between durum and northern wild emmer populations. Admixture in durum was detected for southern domesticated emmer populations 4 and 5 but not for northern domesticated emmer populations 6 and 7. Nei’s genetic distance of durum to southern population 2 of wild emmer was as long as to the northern wild emmer populations (Table 2).
Gene diversity
Gene diversity was estimated as expected heterozygosity (He) (Table 3). Wild emmer populations 1a, 1b and 1c were more diverse than population 2 and any of the northern wild emmer populations. In the northern area of wild emmer distribution, gene diversity was similar among the populations (He = 0.12 to 0.17) except for population 3e (He = 0.04). In domesticated emmer, the highest diversity was observed in population 5 (He = 0.18). The removal of accessions showing admixture from the dataset reduced gene diversity in most populations (Table 3).
Morphology
Wild emmer from the Diyarbakir region (populations 3d + 3e) and Lebanon (population 1c) had the smallest spikelets and both populations significantly differed from population 2, which had large spikelets (Table 5). The average spikelet size of population 1a + 1b, although being intermediate, did not significantly differ from populations 1c and 3d + 3e (Table 5). The spikelet sizes indicated that wild emmer in population 2 belonged to the robust race judaicum whereas populations 1 and 3 belonged to the slender race horanum. No significant differences were found among these populations in seed weight.
Discussion
Diversity
The same clones that were used here as probes to detect RFLP had previously been used to investigate RFLP in six diploid Aegilops species closely related to emmer (Dvorak et al. 1998). Gene diversity He ranged from 0.14 to 0.72 among those species and was correlated with observed heterozygosity, largely reflecting the cross-pollination rate. Gene diversity in the highly self-pollinating wild emmer, He = 0.19, was comparable to that in its highly self-pollinating diploid relatives.
Several lines of evidence were obtained here for gene flow between wild and domesticated emmer, which accounts for the relatively high gene diversity in domesticated emmer (He = 0.11). Statistic Fst provides an idea about partitioning of global genetic variation due to population structure. Only about 8% of global variation in emmer is due to the domestication status. In comparison, about 17% of global variation is due to geographic division of emmer into northern and southern regions and 46% is due to subdivision of regions into populations. Genetic distances between wild and domesticated emmer are shorter and admixture is more common between sympatric populations of wild and domesticated emmer than between allopatric populations, which is indicative of gene flow between sympatric populations. These relationships were observed from Levant to Iran, indicating that gene flow between wild and domesticated emmer took place across the entire area of wild emmer distribution.
Population structure of wild emmer
Clustering based on multilocus genotypes showed that wild emmer is composed of two genetically distinct populations: northern and southern. The same observation was reported by Ozkan et al. (2002) on the basis of AFLP. Wild emmer populations in both regions are further subdivided. In the southern region, the subdivision parallels the morphological differentiation into the slender race horanum (population 1) and the robust race judaicum (population 2). The former race is then further subdivided into three populations: population 1a is located in central Israel and SW Syria, population 1b is located further north in Israel and population 1c is located in Lebanon. In contrast to our results, Ozkan et al. (2005) separated southern wild emmer into two populations: IV (Israel) and V (Israel, Jordan, Syria and Lebanon).
The robust judaicum race has an unusual position. Genetically, it is closely related to populations 1a and 1b and forms a distinct branch within the 1a + 1b branch in the NJ tree. The sample analyzed here consisted of 13 accessions located north and northwest of the Sea of Galilee. Blumler (1998) studied the judaicum race in the upper Jordan Valley, and concluded that it originated recently via hybridization of wild emmer with durum wheat. Genetic distances of population 2 to durum or domesticated emmer provided no evidence that either of the two domesticated wheats was its ancestor. Genetic distances were computed individually between the 55 varieties and landraces of durum and individual accessions of wild emmer populations 1b and 2 (data not shown). This exercise also failed to show that the judaicum race originated via hybridization of wild emmer with durum.
The northern wild emmer is differentiated into several populations. Accessions from Diyarbakir region were allocated into two populations: 3d consisting of accessions over a wide area west of Diyarbakir and population 3e located 36.2 km west of Diyarbakir. Population 3e was represented by 20 accessions but its gene diversity was only 0.04. This population was almost certainly derived from the more diverse 3d population.
In the multilocus clustering, wild emmer in Iraq and Iran clustered together forming a single population (3c). In contrast, Ozkan et al. (2005), clustered wild emmer population in the Al Mawsil region in Iraq (population III′) separately from the geographically nearby population III in the Sulaimaniya region. The Sulaimaniya population clustered together with population II located in the Diyarbakir region more than 500 km away. To reinvestigate this geographically incongruous finding, population 3c was divided in to the Al Mawsil region (population 3 cM) and Sulaimaniya region (population 3 cN). In contrast to the conclusion made by Ozkan et al. (2005), genetic distances suggested that population 3 cN was more closely related to population 3 cM than to wild emmer in the Diyarbakir region (population 3d).
Emmer domestication
A previous attempt to identify the site of emmer domestication on the basis of ctDNA microsatellite variation led to the conclusion that emmer was domesticated in the Kartal Dagi region (Mori et al. 2003). Neither Ozkan et al. (2005) nor our findings support this hypothesis.
Because wild emmer in the Al Mawsil region and the Sulaimaniya region formed two branches in the Ozkan et al. (2005) study (III and III′) and because the plants of branch III (Sulaimaniya region) clustered with emmer from the Diyarbakir region, Ozkan et al. (2005) concluded that the founding population of domesticated emmer was the Diyarbakir and the Sulaimaniya populations. Our study did not confirm this conclusion; genetic distances of the Sulaimaniya wild emmer were longer to the domesticated emmer populations than those of the Diyarbakir wild emmer.
Two plants with admixture in the 3 cN population had a strong effect on the relationships of this population. When they were included, the 3 cN population was equidistant to the four domesticated emmer populations. However, when they were removed, the 3 cN population appeared distant to allopatric domesticated emmer populations 4, 5 and 6 (D = 0.223 to 0.268) but closely related to the sympatric population 7 (D = 0.185). The relatedness of the core 3 cN population to only the sympatric population of domesticated emmer suggests the existence of gene flow between wild and domesticated emmer in the Sulaymaniya region in the past.
The identification of the site of emmer domestication is predicated on the assumption that the genetic structure of modern wild and domesticated emmer populations has not been altered by gene migration during the past 10,000 years (Blumler 1998). To minimize the potentially confounding effects of past gene flow, accessions and landraces that showed admixture were removed from the dataset used to compute genetic distances between populations. Even then, genetic distances between the populations of wild and domesticated emmer make sense only if the existence of substantial gene flow between wild and domesticated emmer populations is considered or, potentially, if it is assumed that southern domesticated emmer (populations 4 and 5) was domesticated in southern Levant and northern domesticated emmer (populations 6 and 7) in the Diyarbakir region. If the dual domestication scenario were true, domesticated emmer populations 4 and 5, but not 6 and 7, should be closely related to wild emmer population 1, and only populations 6 and 7, but not populations 4 and 5, should be closely related to northern wild emmer population 3d. While the former requirement is true the latter is not. All four domesticated emmer populations are closely related to population 3d, which is more consistent with single domestication site in SE Turkey and introgression in southern Levant than with two independent domestications. Fixation of the ABCT-A1a haplotype in all domesticated emmer and the ABCT-A1b haplotype in wild emmer (Dvorak et al. 2006) also argues against domestication of emmer at two sites geographically as distant as SE Turkey and southern Levant.
Virtually all domesticated emmer landraces clustered into a single branch, suggesting that domesticated emmer is monophyletic; only 4 of 192 landraces were outside of this branch, and these were admixture. The concern that the topology of a phylogenetic tree may conceal polyphylesis (Allaby and Brown 2003), particularly if confounded by subsequent gene flow between populations, is highlighted by the fact that the dual proximity of southern domesticated emmer to wild emmer population 3d and 1, suggested by genetic distances, was unnoticed by Ozkan et al. (2005) and not apparent in the tree in Fig. 2c.
Gene flow between domesticated wheat and wild wheat shown here and elsewhere (Dvorak et al. 2006) may account for the presence of domesticated wheat ctDNA haplotypes as rare polymorphisms in the Kartal Dagi wild emmer reported by Mori et al. (2003). Although domesticated emmer admixture was not as extensive in the Kartal Dagi wild emmer population as in southern Levant, it was detected in three of the 21 accessions from that region.
An alternative possibility that could account for finding ctDNA haplotype 10 in the Kartal Dagi wild emmer as a rare polymorphism (Mori et al. 2003) is that the Kartal Dagi haplotype 10 is a homoplasy due to microsatellite mutation from a related haplotype. The nuclear microsatellite mutation rate was estimated for wheat to be 2.4 × 10−4 per microsatellite allele per generation (Thuillet et al. 2002). If ctDNA microsatellites mutate with similar rates, a mutation causing a microsatellite homoplasy in wild and domesticated emmer is likely during the 9,000 to 10,000 years that have elapsed since emmer domestication.
While the present study and that of Ozkan et al (2005) disagree on the role of the Sulaimaniya population in emmer domestication, both point to the Dyiarbakir region as the site of emmer domestication. This region is near archeological sites on the upper Euphrates and Tigris, such as Cafer Höyük, Nevali Cori, and Cayönü, with one of oldest records of the transition from hunting–gathering to farming. Remnants interpreted as domesticated emmer were recorded at the Prepottery Neolithic B (PPNB) Cafer Höyük (Willcox 1991; de Moulins 1993) and Cayönü (van Zeist and de Roller 1991/1992). However, they were also recorded much further to the south, at the PPNB Tell Aswad near Damascus (van Zeist and Bakker-Heeres 1975) and at Jericho (Hopf 1983). Whether the contemporary appearance of domesticated emmer in Turkey and Levant signals a rapid spread of agriculture across the Fertile Crescent or independent domestication of emmer that was later overshadowed by the spread of domesticated emmer from the north is unclear and both scenarios are possible. Nevertheless, genetic distances between wild and domesticated populations discussed earlier and the presence of domesticated einkorn, which was very likely domesticated in SE Turkey (Heun et al. 1997), along with domesticated emmer at PPNB Jericho (Hopf 1983) seem to be more consistent with the former possibility.
Population structure of domesticated emmer
Domesticated emmer differentiated into two principal populations: northern and southern. The former is subdivided into NW population 6 and NE population 7 (Fig. 3). Population 7 consists of landraces located in NE Turkey, Transcaucasia and Iran. This group probably evolved by diffusion of domesticated emmer from the place of its origin in SE Turkey. Population 6 is located in Greece, northern Balkans (Serbia, Bosnia and Croatia) but includes also Yaroslav region in northern Russia and emmer in NW Turkey.
Origin and diffusion of domesticated emmer. Populations are numbered as in text. The presumed directions of diffusion are indicated by open or closed arrows. Emmer diffusion to India could have occurred by either of the two indicated routes (dotted arrows). The solid star indicates primary origin and the open star indicates the putative second site of domestication or gene flow from wild emmer
Crops may spread from the site of origin via cultural diffusion (spread of crops without movement of people) or demic diffusion (movement of people) (Ammerman and Cavalli-Sforza 1984). Genetic similarity of emmer in Russia and the Balkans parallels Slavic migration. The presence of landraces of population 6 in the Balkans, Greece and NW Turkey may have its roots in cultural diffusion before or during the Byzantine and Ottoman empires. An alternative possibility is that the distribution of population 6 reflects demic diffusion north, west and south as a consequence of the biblical flood of settlements along the shores of a fresh-water lake that is hypothesized to have been located at the site of today’s Black Sea (Ryan and Pitman 2000).
The southern domesticated emmer is subdivided into populations 4 and 5. Population 4 consists of landraces from Ethiopia, Oman, southern India, and Levant, but also several landraces scattered elsewhere. Emmer was the dominant cereal in ancient Egypt until the third millennium BP when it was replaced by free-threshing durum (Nesbitt and Samuel 1996), and has been an important cereal in Ethiopia. It is highly probable, although not shown, that Ethiopian emmer is a remnant of Egyptian emmer. Population 4 has affinity to population 5 of domesticated emmer and population 1 of wild emmer and likely originated in southeastern Mediterranean. The close proximity of Ethiopian emmer to landraces from Oman and southern India suggests that emmer reached those regions from NE Africa via maritime trade (Fig. 3). Emmer cultivation has been limited in India and largely restricted to Andhra Pradesh. India has been the destination of ships that sailed from Red Sea across the Arabian Sea or directly across the Indian Ocean since the Greco-Roman times (Hourani 1963). It is possible that maritime trade was also responsible for the location of several landraces of population 4 in the Balkans and the eastern area of the Black Sea.
Population 5 is almost entirely comprised of landraces from the Levant and Mediterranean and two accessions from SE Turkey and two from Armenia. Population 5 is sympatric with southern emmer and presumably originated by geographic isolation from the northern domesticated emmer accompanied by gene migration from southern wild emmer into the sympatric domesticated emmer, which could have happened by introgressive hybridization or via dual domestication and absorption of the southern domesticate into the genepool of domesticated emmer arriving from the north. Population 5 shows the greatest gene diversity of the four domesticated emmer populations paralleling the greatest diversity of Levantine wild emmer populations.
Durum first appears in the archeological record in Egypt during the Greco-Roman times (reviewed in Nesbitt and Samuel 1996). Genetic relationships are consistent with archeology and suggest that durum evolved in the eastern Mediterranean.
Purity of gene pools
Evidence for gene flow between wild and domesticated emmer ranging from southern Levant to Iran and significant impact on the genetic structure of the populations shows that if sufficient time is available, sympatric wild and domesticated genepools will suffer considerable loss of their original identity, even in a highly self-pollinating species such as emmer. Significant levels of gene flow may occur not only between conspecific wild and domesticated populations but also between those of related species, even if they differ by ploidy (Dvorak et al 2006). This fact must be considered in the design of in situ germplasm conservation programs, studies addressing the geography of plant domestication, and similar genetic projects.
References
Allaby RG, Brown TA (2003) AFLP data and the origins of domesticated crops. Genome 46:448–453
Ammerman AJ, Cavalli-Sforza LL (1984) The neolithic transition and the genetics of populations in Europe. Princeton University Press, Princeton
Badr A, Muller K, Schafer-Pregl R, El Rabey H, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F (2000) On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17:499–510
Blumler MA (1998) Introgression of durum in to wild emmer and the agricultural origin question. In: Damania AB, Valkoun, J, Willcox G, Qualset CO (eds) The origins of agriculture and crop domestication, ICARDA, Aleppo, Syria, ICARDA, IPGRI, FAO and UC/GRCP, pp 252–268
de Moulins D (1993) Les restes de plantes carbonisees de Cafer Hoyuk. Cahiers de l’Euphrate 7:191–234
Dubcovsky J, Luo MC, Zhong GY, Bransteitter R, Desai A, Kilian A, Kleinhofs A, Dvorak J (1996) Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics 143:983–999
Dvorak J (1980) Homoeology between Agropyron elongatum chromosomes and Triticum aestivum chromosomes. Can J Genet Cytol 22:237–259
Dvorak J, McGuire PE, Cassidy B (1988) Apparent sources of the A genomes of wheats inferred from the polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome 30:680–689
Dvorak J, Luo M-C, Yang Z-L (1998) Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics 148:423–434
Dvorak J, Yang Z-L, You FM, Luo MC (2004) Deletion polymorphism in wheat chromosome regions with contrasting recombination rates. Genetics 168:1665–1675
Dvorak J, Akhunov ED, Akhunov AR, Deal KR, Luo MC (2006) Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol Biol Evol 23:1386–1396
Falush D, Pritchard J, Stephens M (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Heun M, Schafer-Pregel R, Klawan D, Castagna R, Accerbi M, Borghi B, Salamini F (1997) Site of einkorn wheat domestication identified by DNA fingerprinting. Science 278:1312–1314
Hopf M (1983) The plants found at Jericho. In: Kenyon KM, Holland TA (eds) Excavations at Jericho V. British School of Archaeology in Jerusalem, London, pp 580–621
Hourani GF (1963) Arab seafaring in the Indian Ocean in ancient and early medieval times. Princeton Oriental Studies, vol 13. Princeton University Press, Princeton
Lewis PO, Zaykin D (1997) Genetic data analysis: Computer program for the analysis of allelic data. Version 1.0. A free program distributed by the authors over the Internet from the GDA Home Page at http://www.chee.unm.edu/gda
Luo MC, Deal KR, Young ZL, Dvorak J (2005) Comparative genetic maps reveal extreme crossover localization in the Aegilops speltoides chromosomes. Theor Appl Genet 111:1098–1106
Mori N, Ishi T, Ishido T, Hirosawa S, Watatani H, Kawahara T, Nesbitt M, Belay G, Takumi S, Ogihara Y, Nakamura C (2003) Origins of domesticated emmer and common wheat inferred from chloroplast DNA fingerprinting. In: Pogna NE, Romano M, Pogna EA, Galterio G (eds) Proceedings of the 10th international wheat genet symposium, Paestum, Italy. Instituto Sperimentale per la Cerealicoltura, Rome, Italy, pp 25–28
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nesbitt M, Samuel D (1996) From staple crop to extinction? The archaeology and history of hulled wheats. In: Padulosi S, Hammer K, Heller J (eds) Hulled wheats. Promoting the conservation and use of underutilized and neglected crops 4 Proc 1st Internatl Workshop on hulled wheats, Castelvecchio Pacoli, Tuscany, Italy. International Plant Genetic Resources Institute, Rome, Italy
Ozkan H, Brandolini A, Schafer-Pregl R, Salamini F (2002) AFLP analysis of a collection of tetraploid wheat indicated the origin of emmer and hard wheat domestication in southeastern Turkey. Mol Biol Evol 19:1797–1801
Ozkan H, Brandolini A, Pozzi C, Effgen S, Wunder J, Salamini F (2005) A reconsideration of the domestication geography of tetraploid wheat. Theor Appl Genet 110:1052–1060
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci 12:357–358
Pritchard JK, Wen W (2004) Documentation for structure software, http://www.pritch.bsd.uchicago.edu
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385
Ryan WBF, Pitman WC (2000) Noah’s flood. Simon and Schuster, New York
Sears ER (1966) Nullisomic-tetrasomic combinations in hexaploid wheat. In: Riley R, Lewis KR (eds) Chromosome manipulations and plant genetics. Oliver & Boyd, Edinburgh, pp 29–44
Thuillet AC, Bru D, David J, Roumet P, Santomi S, Sourdille P, Bataillon T (2002) Direct estimation of mutation rate for 10 microsatellite loci in durum wheat, Triticum turgidum (L.) Thell. ssp durum Desf. Mol Biol Evol 19:122–125
van Zeist W, Bakker-Heeres JAH (1975) Archaeobotanical studies in the Levant. 1. Neolithic sites in the Damascus basin: Aswad, Ghoraife, Ramad. Palaeohistoria 24:165–256
van Zeist W, de Roller GJ (1991/1992) The plant husbandry of aceramic Cayonu, SE Turkey. Palaeohistoria 33/34:65–96
Weir BB (1996) Genetic data analysis. Sinauer Assoc, Sunderland
Willcox G (1998) Archaeobotanical evidence for the beginnings of agriculture in Southwest Asia. In: Damania AB, Valkoun, J, Willcox G, Qualset CO (eds) The origins of agriculture and crop domestication, ICARDA, Aleppo, Syria, ICARDA, IPGRI, FAO and UC/GRCP
Willcox J (1991) Cafer Hoyuk (Turquie): Les Charbons de bois neolothiques. Cahiers de l’Euphrate 5–6:139–150
Acknowledgments
We thank O.D. Anderson, M.D. Gale, A. Graner, G.E. Hart, A. Kleinhofs, M.E. Sorrells, and M.K. Walker-Simmons for sharing clones with us, H.E. Bockelman, S. Jana, B.L. Johnson, C.O. Qualset, R. Papa, and J. Valkoun for supplying the seeds of plants used in this study. We also thank E.D. Akhunov for valuable assistance with statistical analyses and P. Morrell for valuable discussions. Financial support from USDA/ACSREES/NRICGP by grant 99-35301-7905 to J. Dvorak is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Lübberstedt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, MC., Yang, ZL., You, F.M. et al. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor Appl Genet 114, 947–959 (2007). https://doi.org/10.1007/s00122-006-0474-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0474-0