Abstract
Trigeneric hybrids in Triticeae may help to establish evolutionary relationships among different genomes present in the same cellular genetic background and to transfer different alien characters into cultivated wheat. In the present study, a trigeneric hybrid involving species of Triticum, Secale, and Thinopyrum was synthesized by crossing hexaploid triticale with hexaploid trigopiro. The meiotic behaviour of chromosomes belonging to different genomes was analyzed, using routine and in situ hybridization techniques in F1, F2, and F3 generations of the trigeneric hybrid. The purpose of this study was to determine the chromosome number and genomic constitution and to discuss the mechanisms involved in the stabilization of the artificial tricepiro hybrids. The chromosome number of the trigeneric F1 hybrid was 2n = 42. Between 12 and 16 bivalents were observed in the central zone of the equatorial meiotic plate and between 9 and 18 univalents were found in the periphery of the MI equatorial plate. Seven of these univalents showed hybridization signals with rye DNA. Lagging rye and non-rye chromosomes and separation of sister chromatids were found in anaphase I. Tetrads with a maximum of six micronuclei, with and without hybridization signals of rye DNA, were observed. After three generations, meiotic cells revealed the presence of 42 chromosomes and 21 bivalents in diakinesis cells. The presence of 14 rye (Secale cereale) chromosomes and the complete pairing of chromosomes in F3 hybrids suggest that rye chromosomes would be preferentially transmitted to the progeny and that an elimination mechanism would act on chromosomes of Thinopyrum and wheat D genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum sp.) is one of the most important crops in the world and increasing its productivity and genetic variability became objective for the breeding plans. Artificial hybridization between wheat and wild-related species is a valuable method that allows improving agronomic characteristics (Wulff and Moscou 2014; Baker et al. 2020).
During 1972, an artificial hybrid of trigeneric origin (Triticum, Secale, and Thinopyrum) was obtained in Argentina for the first time, by crossing a hexaploid triticale (2n = 6x = 42, AABBRR; A, B from Triticum and R from Secale) with an octoploid trigopiro (2n = 8x = 56, AABBDDJJ; with A, B, D from Triticumand J fromThinopyrum). This hybrid was named “tricepiro”, was characterized by agronomic abilities superior to those of their parents, and its chromosome number was stabilized in 2n = 42 chromosomes (Covas 1976). After several generations of selection, in 1994, one of the most promising lines was registered as a cultivar called “Don René INTA”. In situ hybridization techniques GISH (genomic in situ hybridization) and FISH (fluorescence in situ hybridization) have shown that the genomic constitution of this allopolyploid (2n = 42) was AABBRR, with introgression of a Thinopyrum region into a pair of wheat chromosomes belonging to A genome and that the R genome was completely retained (Ferrari et al. 2005).
The productive potential, rusticity, and nutritional value of tricepiros have generated significant researches and developments (Covas 1976, 1985, 1989; Frecentese and Covas 1985; Ferreira and Szpiniak 1994; Brizuela et al. 1997; Tosso et al. 1997; Ruiz et al. 2000). In the 1990s, the Facultad de Agronomía y Veterinaria of Universidad Nacional de Río Cuarto (UNRC), Córdoba, Argentina, began to develop tricepiro germplasm using different hexaploid triticale lines (2n = 6x = 42) and trigopiro lines with different ploidy levels (2n = 6x = 42 and 2n = 8x = 56) (Ferreira et al. 1994, 1998; Galván et al. 2015). After seven generations of selection, tricepiros of different origin were all hexaploid (6x) and presented some meiotic irregularities, such as the presence of univalents and synapse failures, which would contribute to the formation of micronuclei in the microspores (Tosso et al. 2000; Ferreira et al. 2007).
Fradkin et al. (2009), using cytogenetic techniques, studied cells in meiotic metaphase I of some tricepiro F1 hybrids (triticale Don Santiago x trigopiro Don Noé). They found the presence of 49 chromosomes and the most frequent configuration was 14 bivalent and 21 univalent, suggesting the absence of homoeologous pairing.
In the F3 generation of tricepiros, some authors observed the presence of a high number of bivalents in the metaphase I cells (Covas 1976; Fradkin et al. 2009). Using molecular techniques, Fradkin et al. (2009) also determined the presence of the complete R genome (14 rye chromosomes) and almost all chromosomes formed bivalents (18 to 23).
The study of mitotic cells in advanced generations of tricepiros showed that they all have the same chromosome number (2n = 42 chromosomes) (Ferrari 2004; Ferrari et al. 2005; Ferreira et al. 2007; Fradkin et al. 2009). Molecular techniques showed retention of the complete R genome and loss of the D and J genomes. Tricepiros, like triticales, seem to tolerate the hexaploid level better (Ferreira et al. 2007; Li et al. 2015).
Different authors observed changes in chromosome behaviour and genomic composition during the first generations of some tricepiros (Ferrari et al. 2005; Ferreira et al. 2007; Fradkin et al. 2009).
In the present work, a tricepiro hybrid was synthesized by crossing triticale Kettu, as a maternal parent and trigopiro SH16, as a pollen donor. Previous studies indicated that both parents have 2n = 42 chromosomes (Fradkin et al. 2011; Estévez et al. 2016). In triticale Kettu, GISH using total genomic DNA of Secale indicated the presence of 14 rye chromosomes, and FISH, using the pSc119.2 probe, confirmed and identified the complete genome of rye. The use of specific microsatellites made it possible to recognize some introgressions of the D genome (Estévez et al. 2016). Fradkin et al. (2012) analyzed the artificial hybrid, trigopiro SH16, employing hybridization techniques and concluded that the chromosome number was 2n = 42 and the genome composition would be: 14 chromosomes of the B genome, 14 chromosomes of the J genome, 2 pairs of the D genome (the 2D and 4D), and the remaining 10 chromosomes probably belong to the A genome of wheat.
The aim of the present work was to analyze the meiotic behaviour of the first three generations of triticale Kettu x trigopiro SH16 INTA in order to discuss putative mechanisms involved in the stabilization of this artificial triticale hybrid.
Materials and methods
Plant materials
The F1 hybrids were obtained at the Universidad Nacional de Río Cuarto (UNRC), Argentina, by crossing triticale Kettu as female parent with trigopiro SH16 INTA as male parent. F1 and F2 plants were grown in the UNRC greenhouse controlling irrigation and fertilizers. F3 also grew in the UNRC but under field conditions.
Hybrid seeds were obtained by emasculating 32 flowers belonging to the same spike of a triticale Kettu and inseminating them with pollen of trigopiro SH16. In total, four small hybrid seeds were produced, sown in a germination chamber, and then transplanted into a phytotechnical cage. The four F1 hybrids obtained were grown under optimal irrigation and fertilization conditions and self-pollinated. It was possible to harvest 3 g of seeds from the F1 plants (approximately 90 seeds). Only 50 of these seeds were sown and 28 of them germinated.
The F2 hybrid plants were kept in adequate water and fertility conditions, and 15 g of seeds (about 450 seeds) was obtained. Approximately 200 seeds were sown, and the F3 plants were maintained under field conditions (without irrigation and fertilization). These F3 hybrids produced a total of 38 g of seeds (about 1200 seeds).
Chromosome preparations
Routine and molecular meiotic studies were done in a pool of immature flowers from each generation (F1, F2, and F3 hybrids). Flowers were fixed in absolute ethanol:acetic acid (3:1 vol/vol) and stored at − 20 °C until use.
For molecular cytogenetic analysis of mitosis, 1-cm-long roots of F3 hybrid seeds were pre-treated in ice-cold water for 48 h and fixed in absolute ethanol:acetic acid (3:1 vol/vol) for 24 h at room temperature and stored at − 20 °C until use.
Cytogenetic analysis of meiosis and mitosis
For routine meiotic studies, immature anthers were squashed in 2% propionic hematoxylin and 1% ferric citrate as mordant (Nuñez 1968).
Probes and labeling
GISH was carried out in F1, F2, and F3 hybrids using rye genomic DNA labeled as a probe and unlabeled DNA of hexaploid wheat as blocking agent. Rye genomic DNA was isolated from adult leaves of Secale cereale cv. Quehue, and wheat genomic DNA from adult leaves of Triticum. aestivum cv. Klein Estrella. Genomic DNAs were obtained using Wizard® Genomic DNA Purification Kit (Promega, USA. Cat#A1620).
According to Fradkin et al. (2013), rye genomic DNA probes were labeled with digoxigenin DIG High Prime (ROCHE, Germany. Cat #11585606910) or biotin nick-translation kit (Invitrogen, USA.Cat#18247–015). To detect digoxigenin-labeled probes, slides were treated with sheep antidigoxigenin FITC (fluorescein isothiocyanate) (Anti-digoxigenin AP-conjugate. ROCHE, Germany. Cat#11093274910) (green); for biotin-labeled probes, they were treated with conjugate streptavidine–Cy3 (GE Healthcare, UK. Cat#PA53022) (red).
FISH study was done in meiotic F2 cells, using pTa71 probe in order to be detected rDNA zone. This probe contains 9 kilobases (kb) EcoRI repeated unit of 18S-5.8S-25S rDNA genes and spacer and is isolated from wheat, Triticum aestivum (Gerlach and Bedbrook 1979). In the present work, pTa71 probe was labeled with biotin using the biotin nick-translation kit (Invitrogen, USA), and slides were treated with conjugate streptavidine-Cy3.
For molecular cytological analysis of mitotic cells, cells were obtained from roots of F3 hybrids seeds, and GISH and FISH techniques were used. Rye genomic DNA labeled with digoxigenin DIG High Prime (ROCHE, Germany) was used as a probe and unlabeled wheat genomic DNA as a block for GISH. The pSc119.2 probe from Secale cereale (McIntyre et al. 1990) was used for FISH, labeled with biotin using the biotin nick-translation kit (Invitrogen, USA).
Both probes pTa71 and pSc119.2 were kindly supplied by A. Cuadrado Department of Cell Biology and Genetics, University of Alcalá de Henares, Spain.
Fluorescence in situ hybridization
Fixed roots and anthers were washed in 0.01 mol/L citric acid sodium—citrate buffer (pH 4.6) to remove the fixative and transferred to an enzyme solution containing 2 mL of 2% (w/v) cellulase Onozuka RS (Yakult pharmaceutical, Japan) and 20% (v/v) liquid pectinase (Sigma, USA and Canada. Cat#P4716). The incubation was performed at 37 °C during 3 h, and then the sample was squashed in a drop of 45% acetic acid. Slides were selected by phase-contrast light microscopy. After removal of coverslips by freezing, the slides were air dried. Slide preparations were incubated in 100 mg/mL DNase-free RNase in 2 × SSC (100 g /mL) for 1 h at 37 °C in a humid chamber and then washed three times in 2 × SSC for 5 min at room temperature. The slides were post-fixed in freshly prepared 4% (w/v) paraformaldehyde in water for 10 min, washed three times in 2 × SSC for 5 min, dehydrated in a graded ethanol series, and air dried. The hybridization mixture consisted of 50%w/w deionized formamide, 10% w/v dextran sulphate, 0.1% w/v SDS, and salmon sperm DNA 0.3 ng /mL in 2 × SSC. A volume containing 50 ng of the labeled probe was then added to 30 µL of this hybridization mixture for each slide. The hybridization mixture was denatured for 15 min at 75 °C, loaded onto the slide preparation, and covered with a plastic coverslip. The slides were placed on a thermocycler for 7 min at 75 °C (denaturation), 10 min at 45 °C, and 10 min at 38 °C, and subsequently incubated overnight at 37 °C to allow hybridization. Following this hybridization step, coverslips were carefully floated off by placing the slides in 2 × SSC for 3 min at 42 °C. The slides were then given a stringent wash in 20% formamide (v/v) in 0.1 × SSC for 10 min at 42 °C. The slides were washed in 0.1 × SSC for 5 min at 42 °C, followed by 2 × SSC for 5 min at 42 °C, transferred to detection buffer (4 × SSC/0.2% v/v Tween 20) for 5 min at 42 °C, and finally treated in detection buffer for 1 h at room temperature. Slides were treated with 2.5% w/v bovine serum albumin (BSA) in detection buffer, incubated in a solution of 1/20 of the corresponding antibody in detection buffer containing 2.5% w/v BSA for 1 h at 37 °C, and washed three times in detection buffer for 10 min at room temperature. Slides were counterstained with 1 mg/mL 4′, 6-diamidino-2-phenylindole (DAPI) in distilled water for 15 min at room temperature and then mounted in antifade.
The images were captured with a Leica LMDB epifluorescence microscope equipped with a digital camera (Leica DFC 350 FX) using the Leica IM50 version 4.0 program (Leica Microsystem, Cambridge, UK). Images were analyzed using Adobe Photoshop CS6 software.
Analysis statistics
The Kruskal–Wallis test (p < 0.05) was used to evaluate the differences in the number of univalents present in meiotic metaphase I of the F1 hybrid and the F2 and F3 segregants. Statistical analyses were performed using the Infostat program, FCA, National University of Córdoba, Argentina (Di Rienzo et al. 2020).
Results
To obtain a trigeneric hybrid combining the Triticum-Secale-Thinopyrum genomes, tetraploid triticale (cv. Kettu), used as maternal material, was crossed with hexaploid trigopiro (cv. SH16 INTA), as male parental, as shown in Table 1 where the number of chromosomes and genomic composition in the parental and in the expected F1 is represented.
F1 routine cytogenetic studies
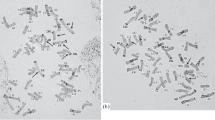
Meiotic metaphase I showed the presence of open and closed bivalents ranging from 10 to 16. These bivalents had a compact distribution, and their identification was difficult in some cases. Univalents were present in the 66 metaphases I studied and were found outside the equatorial plate (Fig. 1a and b). The number of univalents per cell ranged from 10 to 18 and 97% of the meiotic metaphases I found a number of univalents greater than or equal to 10 (Fig. 2, Table 2). The presence of a quadrivalent (IV) was observed at a low frequency of around 4% (Fig. 1b).
Meiotic and mitotic chromosome configurations. Multivalents, univalents, and abnormal configurations in three generations of trigeneric hybrid (triticale Kettu x trigopiro SH16-INTA). a–h Analysis of F1 trigeneric hybrid. a Meiotic metaphase I with 18 univalents. b Meiotic metaphase I. Arrowhead shows a quadrivalent. c Early meiotic anaphase I. d Mitotic metaphase of tapetum cell, observed 42 chromosomes. e, f Later meiotic anaphase I, observed lagging chromosomes. e Arrows show sister chromatid separation precociously. f Arrowhead shows the presence of a bridge not associated with fragment. g Meiotic telophase I with lagging chromosomes. h–k Meiotic cells of F2 trigeneric hybrid. h, i Diakinesis. h Arrowhead shows a quadrivalent. j Meiotic metaphase I. k Meiotic telophase I with lagging chromosomes. Arrow shows sister chromatid separation precociously. l–o Meiotic analysis of F3 trigeneric hybrid. l, m Diakinesis. n, o Metaphase I. Scale bar = 10 µm
At meiotic early anaphase I, the chromosome number was determined as 2n = 42 (Fig. 1c) and was corroborated in metaphase mitotic cells present in the anther, probably in the tapetum (Fig. 1d). In meiotic anaphase I, delayed univalents could be observed in almost all the cells analyzed, and some of them showed early disjunction of their two chromatids (Fig. 1e). The presence of a bridge, which was not associated with a fragment, was observed in some meiotic late anaphase I cells (Fig. 1f).
At the beginning of meiotic telophase I, the number of delayed chromosomes decreased (Fig. 1g).
F1 molecular cytogenetic studies
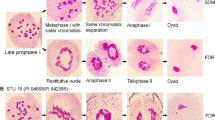
GISH detection was performed to know the genomic composition of the F1 hybrid Kettu x SH16, using total genomic DNA labeled of rye as a probe and unlabeled total genomic DNA of hexaploid wheat as blocking. Meiotic metaphase I showed that seven univalents corresponded to rye chromosomes, and the number of non-rye univalents was variable, reaching a maximum of eleven univalents (Fig. 3a and b). A polar view of metaphase I showed univalents in the periphery of the equatorial plate (Fig. 3b). As can be seen in early telophase I, some rye chromosomes began to migrate to the poles while the rest of the rye chromosomes remained behind (Fig. 3d). Many univalents, which showed no signs of rye hybridization, also remained close to the equatorial plate (Fig. 3c). Some univalents from different genomes were observed to separate their sister chromatids at the beginning of telophase I (Fig. 3c and d).
Meiotic and mitotic analysis in three generations of trigeneric hybrid (triticale Kettu x trigopiro SH16-INTA) by in situ hybridization (FISH – GISH). a–d F1 generation. a, b Metaphase I. Arrowheads show seven rye univalents with hybridization signal of labeled rye DNA probe. c Anaphase I. Seven rye univalents with hybridization signal of labeled rye DNA probe. Arrowheads indicate that two univalents of rye showed early separation sister chromatids. d Early telophase I. Arrowheads show chromosomes with hybridization signal of labeled rye DNA probe. e–m F2 generation. e, g Diakinesis. Bivalents of rye showed hybridization signal with rye DNA probe (yellow arrowheads) and pTa71 probe of wheat (red arrowheads). f Same cell as image (e) counterstained with DAPI. h, i Metaphase I. h Arrowheads show chromosomes hybridization signal with rye DNA labeled probe. i DAPI counterstaining. Arrowheads indicate the presence of prominent telomeric heterochromatic DAPI + bands corresponding to rye chromosomes. j–m Tetrads with micronuclei. j, l Micronuclei show hybridization signal with the labeled rye DNA probe. k, m Same cells of j and l counterstaining with DAPI. Asterisks and starts marks micronuclei. Yellow asterisks correspond to micronuclei with hybridization with labeled rye DNA probe and yellow star corresponds to micronuclei without hybridization signal. n–r F3 generation. n, o Meiotic metaphase I. Arrowheads show seven hybridized chromosomes with labeld rye DNA probe. p Early meiotic anaphase I. Arrowheads show chromosomes with hybridization signal with labeled rye DNA probe. q Mitotic metaphase of tapetum cell arrowheads shows 14 chromosomes with hybridization signals of labeled rye DNA probe. r Mitotic metaphase of root tip hybridized with pSc119.2 probe of rye. Scale bar = 10 µm
F2 routine cytogenetic analysis
F2 progeny exhibited that most of the 108 diakinesis studied showed 21 bivalents (Fig. 1h and i), and the presence of a quadrivalent was detected very infrequently (Fig. 1h). Based on these observations, we can infer that the F2 progeny has a 2n = 42 chromosomes. The 16 meiotic metaphase I analyzed showed between 0 and 4 univalents. The frequency of cells without univalents was 12.50%, and the frequency of univalents equal to or less than 3 was observed in 87.5% of metaphase I cells (Figs. 1j and 2, Table 2). Late telophase I showed a drastic reduction in lagging chromosomes (Fig. 1k).
F2 molecular cytogenetic analysis
GISH analysis using labeled rye genomic DNA as a probe and unlabeled genomic DNA of hexaploidy wheat as a blocking agent was performed on diakinesis and metaphase I cells of the F2 hybrids Kettu x SH16, and it showed 7 bivalents with hybridization signals (Fig. 3e and g). The use of the pTa71 probe in this generation allowed us to observe the presence of 3 bivalents with hybridization signals, indicating that there were three nucleolar organizer regions in the F2 hybrid. One of them was a rye bivalent recognized by GISH (Fig. 3e and g).
Micronuclei with and without rye material were observed with GISH using rye DNA as a probe (Fig. 3j and i).
F3 routine cytogenetic analysis
The analysis of 20 diplotene cells and 16 diakinesis cells of the F3 hybrid showed the presence of 21 bivalents and none univalent was presented, while in 45 metaphase I cells, there were between 0 and 4 univalents (Fig. 1 l–o). Besides, 22.22% of metaphase I did not show univalents while only 2.22% of the cells showed 4 univalents (Fig. 2, Table 2). Figure 2 compares the presence of univalents in metaphase I of three generations, F1, F2, and F3.
F3 molecular cytogenetic analysis
GISH analysis using rye genomic DNA as a probe and wheat genomic DNA as a blocking agent was performed, confirming the presence of 7 rye bivalents in metaphase I cells (Fig. 3n and o). With the same probe, in early anaphase I, 14 rye chromosomes showed hybridization signals, suggesting a correct distribution of chromosomes (Fig. 3p).
The statistical analysis showed a significant difference (p < 0.0001) between F1, F2, and F3 in the number of univalents present in metaphase I. F1 was significantly different from F2 and F3, whereas the last two generations did not differ from each other.
Mitotic analysis in seeds of F3 hybrid
We confirmed the chromosome number of 2n = 42 in mitotic metaphase cells, and the presence of 14 rye chromosomes (Fig. 3q). FISH study with pSc119.2 probe allowed us to identify the seven chromosome pairs of rye in F3 generation (Fig. 3r). Mitotic cells were observed separately in the images given by FISH and DAPI to confirm each signal and each chromosome. DAPI + bands also contributed to the recognition of rye chromosomes because they are characterized by showing a prominent intensity in the telomeric zones and, in addition, the size of rye chromosomes is usually a little larger than that of wheat chromosomes.
Discussion
Synthetic tricepiros (2n = 6x = 42) are forage crops with valuable characteristics of Secale, Triticum, and Thinopyrum; they have a marked resistance to disease and high tolerance to freezing and drought (Ferreira et al. 2007). By crossbreeding different accessions of hexaploid triticales (2n = 6x = 42, AABBRR) and octoploid trigopiros (2n = 8x = 56, AABBDDJJ), previous studies showed that the F1 hybrids had 2n = 49 chromosomes, and in advanced generations, the chromosome number was 2n = 42, retaining the R genome and completely losing all chromosomes of J and D genomes (Ferrari et al. 2005; Ferreira et al. 2007; Fradkin et al. 2009).
In this work, we analyzed for the first time, using routine and molecular cytogenetic techniques, a tricepiro whose parents, triticale Kettu and trigopiro SH16, both have the same ploidy level (2n = 6x = 42). Mitotic and meiotic observations in this work indicated that the chromosome number of this F1 hybrid was 2n = 42 as it was expected, according to the chromosome number of their parents. When crossing different genera and/or species, the F1 hybrids usually have a chromosome number that is the mean value of the chromosome number of their parents (Lima-Brito et al. 1996; Li et al. 2006; Kang et al. 2016; Dai et al. 2017).
Routine and molecular cytogenetic analysis carried out in the present work allowed us to recognize, in metaphase I cells of the F1 generation, the presence of univalent chromosomes in a range from 10 to 18 and bivalent chromosomes from 10 to 16. However, the number of univalents theoretically expected would be 18, and the number of bivalents would be 12 as was show in the Table 1. The presence of meiotic cells with a number of univalents lower than 18 and bivalents higher than 12 suggested the presence of homoeologous associations.
GISH results, using DNA from rye as a probe, provided a reliable approach to discriminate Secale chromosomes from Triticum and Thinopyrum chromosomes in the F1 hybrids Kettu x SH16. Seven univalents of rye were clearly recognized in all the cells, and they were distributed outside the equatorial plate in metaphase I cells. Therefore, chromosomes belonging to R genome would not be involved in the homoeologous pairing but, J, A, and D genomes could be associated.
Extensive studies made in polyploid wheat showed that mainly the Ph1 gene, located on chromosome 5B, determine the exclusive homologous pairing by suppressing the homoeologous pairing (Dvorak et al. 2006; Naranjo and Benavente 2015). However, homoeologous chromosome pairing can be achieved if the chromosome 5B is absent or if the mutant ph1 gene is present (Koebner and Shepherd 1985; Aghaee-Sarbarzeh and Dhaliwal 2000).
Numerous F1 hybrids of wheat and related species have been studied by several authors, and some of them showed the presence of homoeologous associations during the meiotic process (Rey et al. 2021; Aghaee-Sarbarzeh and Dhaliwal 2000). Epistatic genes with Ph1 were found in Ae. speltoides and homoeologous mating was observed in F1 plants of Ae. speltoides x wheat crosses (Aghaee-Sarbarzeh and Dhaliwal 2000). According to Jauhar (1995), the homoeologous pairing in F1 hybrids between Thinopyrum ponticum and Triticum aestivum showed associations that could involve wheat and wheatgrass chromosomes, and he proposed that a high homoeologous pairing would be obtained if Ph1 was somehow inactivated. We hypothesize that the presence of the J genome in the trigopiro SH16 parent (Fradkin et al. 2012) would be related to the homoeologous association phenomena observed in the F1 hybrid studied in the present work.
The F1 tricepiros obtained in previous works, crossing octoploid trigopiro with hexaploid triticale, did not show homoeologous pairing in the meiotic cells (Ferrari 2004; Fradkin et al. 2009). We suggest that the presence of different alleles of a Ph1 gene could generate tricepiro F1 plants with and without homoeologous pairing, as was suggested in studies carried out on hybrids between wheat and species of the genus Aegilops (Ozkan et al.2001; Dvorak et al.2006).
Distribution of univalents and separation of sister chromatids in meiotic anaphase I of the F1 hybrids were random phenomena, not limited to a single species, according to observations made in cells hybridized with a labeled rye genomic DNA.
Statistically significant differences between F1 and F2 and F3 hybrids in the number of univalents in meiotic metaphase I suggest rapid changes in genomic organization. Although the chromosome number was the same in all generations (2n = 42), the chromosome composition was very different. GISH technique indicated the presence of seven chromosomes of rye in F1 hybrids. F2 and F3 generations showed 14 rye chromosomes forming seven bivalents, while the remaining 28 chromosomes formed a maximum of 14 bivalents with no signs of hybridization. These results suggest the loss of seven chromosomes and the duplication of rye chromosomes.
The use of the pSc119.2 probe allowed the recognition of the entire rye genome in mitotic cells, confirming the retention of this genome in the F3 hybrids studied in the present work. The pattern of hybridization signals we found with this probe is specific to recognize the seven rye chromosomes pairs in hybrids (Ferrari et al. 2005; Fradkin et al. 2009, 2013) Likewise, the use of the pTa71 probe allowed the recognition of a rye bivalent, in the F2 generation, with two nucleolar organizer regions (NOR). This result confirmed the presence of chromosomes 1R, since NOR regions are characteristic of this chromosome (Sánchez-Morán et al. 1999; Fradkin et al. 2016).
The trend towards normalization of the meiotic process observed in the F3 generation in the present study has been suggested by previous work in which were observed high seed fertility (Covas 1976) and high number of bivalents in metaphase I (Fradkin et al. 2009). Also, in advanced generations, preferential retention of rye chromosomes and total elimination of the D and J genomes was observed (Ferrari et al. 2005; Fradkin et al. 2009).
The loss of entire genomes of interspecific hybrids was analyzed by different authors (Brasileiro-Vidal et al. 2005; Sanei et al. 2011; Evtushenko et al. 2019). Several mechanisms were suggested to explain chromosome elimination in interspecific and intergeneric hybrids (Hao et al. 2013; Moreno et al. 2014). Now, the incompatibility of centromeres from different species seems to be the main cause of chromosome elimination from one of the parental genomes in hybrids (Sanei et al. 2011; Xie et al. 2013; Evtushenko et al. 2019). The stability of the wheat genome could be altered by the presence of the R genome in hybrids of wheat and rye and, according to Evtushenko et al. (2017), the stability of the D genome is more strongly affected by the R genome than A and B genomes. The differences in the molecular structure of the centromeric histone CENH3 could account for differences in the stability of hybrid genomes in triticales (Evtushenko et al. 2017; 2019). We suggest that the loss of non-rye chromosomes observed in F2 and F3 generations could be related to centromere incompatibility as described for triticales (Evtushenko et al. 2017; 2019).
We do not have a clear understanding of the mechanism that could explain such an abrupt retention of the R genome in tricepiro. We have not found explanations in the literature. However, different authors observed a preferential transmission of rye chromosomes in hybrids involving different species of the tribe triticeae (Li et al. 2006; Kang et al. 2012, 2016). Bernardo et al. (1988) propose the behaviour of the rye chromosome in the progeny of AABBR hybrids and proposed that there was elimination of rye chromosomes when they were present individually, but the whole genome was retained.
Micronuclei can have different origins. They can be generated in acentric fragments resulting from double-strand breaks that are incorrectly repaired or they could also be produced from whole chromosomes that do not bind to the spindle in metaphase (Kang et al.; 2016). Fluorescence in situ hybridization is a useful technique to analyze the origin of micronuclei (Kwasniewska and Bara 2022). In the present work, GISH using rye genomic DNA allowed us to observe in F2 tetrads and micronuclei with and without rye genetic material. Therefore, the origin of these micronuclei was not restricted to a single genome.
The presence of quadrivalents in metaphase I and bridges not associated with fragments in late anaphase I were observed with a very low frequency in the hybrid studied in the present work. Quadrivalents are related to chromosomal rearrangement (Kang et al. 2012; He et al. 2017) and bridges not associated with fragments were described in many animal and plant species as “side-arm bridges” (Palermo et al. 2001). These bridges would be the product of spontaneous breaks during recombination and subsequent incorrect resolution (Stockert 1994), but we do not discard that the lack of fragments observed could be due to oversquashing. Both anomalies can be related to “genomic shock”, and according to many authors, following interspecific or intergenic hybridization, genomic changes may occur, including gain and loss of chromosomal segments, gene activation and repression, changes in the epigenome, and activation of transposons (Li et al. 2015; Wang et al. 2014).
In the present study, we conclude that the hybrid obtained crossing two tetraploid parents showed 2n = 42 chromosomes in all generations analyzed. In F3 generation, the complete conservation of R genome of rye was observed. These results are like those obtained in previous studies, crossing different tetraploid triticales and octoploid trigopiros.
Analysis of the meiotic behaviour of the F1 hybrid indicated the presence of pairing of homoeologous chromosomes, suggesting the presence of an inhibitor of Ph gene activity. Previous studies of tricepiros from different origins did not show homoeologous chromosome pairing. Allelic differences in a Ph inhibitor gene could be the origin of the variability in the homoeologous paring between tricepiros with different origins.
The genomic and chromosome composition of triticale Kettu x trigopiro SH16 hybrids has relevance in establishing evolutionary relationships among different genomes sharing the same genetic background. Furthermore, it turns out to be a potential material to enrich the genetic base in wheat breeding programs.
References
Aghaee-Sarbarzeh M, Dhaliwal HS (2000) Brief communication. Ph′ gene derived from Aegilops speltoides induces homoeologous chromosome pairing in wide crosses of Triticum aestivum. J Hered 91:417–421. https://doi.org/10.1093/jhered/91.5.417
Baker SL, Grewal S, Yang C, Hubbart-Edwards S, Scholefield D, Ashling S, Burridge AJ, Przewieslik-Allen AM, Wilkinson PA, King IP, King J (2020) Exploiting the genome of Thinopyrum elongatum to expand the gene pool of hexaploid wheat. Theor Appl Genet 133:2213–2226. https://doi.org/10.1007/s00122-020-03591-3
Bernardo A, Luengo P, Jouve N (1988) Chromosome constitution in G2 and G3 progenies of 6x-triticale × T. turgidum L. hybrids. Euphytica 37:157–166. https://doi.org/10.1007/BF00036853
Brasileiro-Vidal AC, Brammer S, Puertas MJ, Zanatta AC, Prestes A, Moraes-Fernandes MI, Guerra M (2005) Mitotic instability in wheat x Thinopyrum ponticum derivatives revealed by chromosome counting, nuclear DNA content and histone H3 phosphorylation pattern. Plant Cell Rep 24:172–178. https://doi.org/10.1007/s00299-005-0913-4
Brizuela M, Passarotti J, Cseh S (1997) Rendimiento del forraje y valor nutritive de tricepiro (Triticum x Secale x Thinopyrum) con diferente manejo de defoliación. Rev Arg Prod Animal 17:385–393
Covas G (1976) Tricepiro, un nuevo verdeo sintético que involucra al trigo, centeno y agropiro. Inf Tecnol Agrop para la Reg Semiár Pampeana 68:5
Covas G (1989) Pampa Semiárida: Nuevos Cultivos. Cienc Hoy 1:75–77
Covas G (1985) El género Elytrigia (=Agropyron s. lat La Pampa). Apuntes para la Flora de La Pampa. INTA Anguil:398–404
Dai Y, Duan Y, Chi D, Liu H, Huang S, Cao W, Gao Y, Fedak G, Chen J (2017) Chromosome identification by new molecular markers and genomic in situ hybridization in the Triticum–Secale–Thinopyrum trigeneric hybrids. Genome 60:687–694. https://doi.org/10.1139/gen-2017-0025
Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW. InfoStat version 2020. Argentina: Grupo InfoStat, FCA, Universidad Nacional de Córdoba. URL http://www.infostat.com.ar.
Dvorak J, Deal KR, Luo MC (2006) Discovery and mapping of wheat Ph1 suppressors. Genetics 174:17–27. https://doi.org/10.1534/genetics.106.058115
Estévez D, Fradkin M, Lopez C, Poggio L, Ferrari MR, Greizerstein E (2016) Genomic and chromosome characterization of Kettu triticale by Cytogenetic and Molecular Techniques. In Cytogenetic and Genome Research (Vol. 148, No. 2–3, pp 143–143). Allschwilerstrasse 10, Ch-4009 Basel, Switzerland: Karger. https://doi.org/10.1159/000446523
Evtushenko EV, Elisafenko EA, Gatzkaya SS, Lipikhina YA, Houben A, Vershinin AV (2017) Conserved molecular structure of the centromeric histone CENH3 in Secale and its phylogenetic relationships. Sci Rep 7:17628. https://doi.org/10.1038/s41598-017-17932-8
Evtushenko EV, Lipikhina YA, Stepochkin PI, Vershinin AV (2019) Cytogenetic and molecular characteristics of rye genome in octoploid triticale (× Triticosecale Wittmack). Comp Cytogenet 13:423–434. https://doi.org/10.3897/CompCytogen.v13i4.39576
Ferrari MR, Greizerstein EJ, Paccapelo H, Naranjo CA, Cuadrado A, Jouve N, Poggio L (2005) The genomic composition of Tricepiro a synthetic forage crop. Genome 48:154–159. https://doi.org/10.1139/g04-081
Ferrari MR. (2004). Estudio de la composición genómica de forrajeras mediante técnicas electroforéticas y de citogenética clásica y molecular. Doctoral Thesis, Fac. Ciencias Exactas y Naturales, Universidad de Buenos Aires. Buenos Aires. Argentina. https://bibliotecadigital.exactas.uba.ar/download/tesis/tesis_n3716_Ferrari.pdf
Ferreira V, Szpiniak B, Grassi E, Croatto D (1998) Tricepiros forrajeros (Triticale x Agrotriticum): obtención y mejora. Rev Arg Prod Animal 18:182
Ferreira V, Scaldaferro M, Grassi E, Szpiniak B (2007) Nivel de ploidía, estabilidad citológica y fertilidad en cruzas de triticale x trigopiro (tricepiros). J Basic Appl Genet 18:15–26
Ferreira V, Szpiniak B (1994) Mejoramiento de triticale y tricepiro para forraje en la U.N. de Río IV. En: Semillas Forrajeras: producción y mejoramiento. Orientación Gráfica. Editores, Buenos Aires, Argentina, pp 110–120
Fradkin M, Greizestein EJ, Paccapelo H, Ferreira V, Grassi E, Poggio L, Ferrari MR (2009) Cytological analysis of hybrids among triticale and trigopiro. Genet Mol Biol 32:797–801. https://doi.org/10.1590/S1415-47572009005000070
Fradkin M, Greizerstein EJ, Ferrari MR, Ferreira V, Grassi E, Poggio L (2011) Comportamiento meiótico y análisis genómico en generaciones tempranas del híbrido triticale Kettu x trigopiro SH16. En el XL Congreso Argentino de Genética y III Simposio Latinoamericano de Citogenética y Evolución, y I Jornadas Regionales SAG-NEA, Corrientes (Argentina). BAG 22(suppl.1):130–131
Fradkin M, Ferrari MR, Ferreira V, Grassi EM, Greizerstein EJ, Poggio L (2012) Chromosome and genome composition of a Triticum× Thinopyrum hybrid by classical and molecular cytogenetic techniques. Genet Resour 59:231–237. https://doi.org/10.1007/s10722-011-9679-4
Fradkin M, Ferrari MR, Espert S, Ferreira V, Grassi E, Greizerstein E, Poggio L (2013) Differentiation of triticale cultivars through FISH karyotyping of their rye chromosomes. Genome 56:267–272. https://doi.org/10.1139/gen-2012-0117
Fradkin M, Greizerstein EJ, Ferrari MR, Poggio L (2016) Nucleolar activity in Triticum x Thinopyrum hybrids with different ploidy level. Plant Biosyst 151:996–1001. https://doi.org/10.1080/11263504.2016.1218973
Frecentese M, Covas G (1985) Comportamiento de nuevos verdeos en la región pampeana semiárida. Inf Tecnol Agrop Para La Reg Semiár Pampeana 84:6–7
Galván B, Castillo E, di Santo H, Grassi E, Ferreira A, Ferreira V (2015) Estudio citogenético en la generación F6 de Tricepiros primarios. J Basic Appl Genet 26:89
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885. https://doi.org/10.1093/nar/7.7.1869
Hao M, Luo J, Zhang L, Yuan Z, Yang Y, Wu M, Chen W, Zheng Y, Zhang H, Liu D (2013) Production of hexaploid triticale by a synthetic hexaploid wheat-rye hybrid method. Euphytica 193:347–357. https://doi.org/10.1007/s10681-013-0930-2
He F, Xing P, Bao Y, Ren M, Liu S, Wang Y, Li X, Wang H (2017) Chromosome pairing in hybrid progeny between Triticum aestivum and Elytrigia elongata. Front Plant Sci 8:2161. https://doi.org/10.3389/fpls.2017.02161
Jauhar PP (1995) Meiosis and fertility of F1 hybrids between hexaploid bread wheat and decaploid tall wheatgrass (Thinopyrum ponticum). Theor Appl Genet 90:865–871. https://doi.org/10.1007/BF00222024
Kang H, Zhong M, Xie Q et al (2012) Production and cytogenetics of trigeneric hybrid involving Triticum, Psathyrostachys and Secale. Genet Resour Crop Evol 59:445–453. https://doi.org/10.1007/s10722-011-9694-5
Kang HY, Huang J, Zhu W, Li DY, Diao CD, Tang L, Wang Y, Xu LL, Zeng J, Fan X, Sha LN, Zhang HQ, Zheng YL, Zhou YH (2016) Cytogenetic behavior of trigeneric hybrid progeny involving wheat, rye and Psathyrostachys huashanica. Cytogenet Genome Res 148:74–82. https://doi.org/10.1159/000445793
Koebner RMD, Shepherd KW (1985) Induction of recombination between rye chromosome 1RL and wheat chromosomes. Theor Appl Genet 71:208–215
Kwasniewska J, Bara AW (2022) Plant cytogenetics in the micronuclei investigation—the past, current status, and perspectives. Int J Mol Sci 23:1306. https://doi.org/10.3390/ijms23031306
Li XF, Song ZQ, Liu SB, Gao JR, Wang HG (2006) Cytogenetic study of a trigeneric (triticale x tritileymus) hybrid. Euphytica 150:117–122. https://doi.org/10.1007/s10681-006-9099-2
Li H, Guo X, Wang C, Ji W (2015) Spontaneous and divergent hexaploid triticales derived from common wheat× rye by complete elimination of D-genome chromosomes. PLoS ONE 10(3):e0120421. https://doi.org/10.1371/journal.pone.0120421
Lima-Brito J, Guedes-Pinto H, Heslop-Harrison JS (1996) Molecular cytogenetics analysis of triticale X tritordeum F1 hybrids. In: Guedes-Pinto, H., Darvey, N., Carnide, V.P. (eds) Triticale: today and tomorrow. Developments in Plant Breeding, vol 5. Springer, Dordrecht, pp 183–188
McIntyre CL, Pereira S, Moran LB, Appels R (1990) New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 33:635–640. https://doi.org/10.1139/g90-094
Moreno P, Caetano C, Olaya C, Agrono T, Torres E (2014) Chromosome elimination in intergeneric hybrid of Oryza sativa×Luziola peruviana. Agric Sci 5:1344–1350. https://doi.org/10.4236/AS.2014.513144
Naranjo T, Benavente A (2015) The mode and regulation of chromosome pairing in wheat-alien hybrids (Ph genes, an updated view). In: Molnár-Láng M, Ceoloni C, Doležel J, (ed) Allien introgression in wheat. Cytogenetics, Molecular Biology, and Genomics. Springer, Berlin/Heidelberg, Germany, pp 133–162
Nuñez O (1968) An acetic-haematoxilin squash method for small chromosomes. Caryologia 21:115–119. https://doi.org/10.1080/00087114.1968.10796290
Ozkan H, Levy AA, Feldman M (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13:1735–1747. https://doi.org/10.1105/TPC.010082
Palermo AM, Ferrari MR, Naranjo CA (2001) The male meiotic system in three South American species of Vicia (Fabaceae). Cytologia 66:261–267. https://doi.org/10.1508/cytologia.66.261
Rey MD, Ramírez C, Martín AC (2021) Wheat, rye, and barley genomes can associate during meiosis in newly synthesized trigeneric hybrids. Plants 10:113. https://doi.org/10.3390/plants10010113
Ruiz M, Paccapelo H, Covas G (2000) Tricepiro. Cultivo y usos. Forrajes Granos Agribusiness J 57:50–52
Sánchez-Morán E, Benavente E, Orellana J (1999) Simultaneous identification of A, B, D and R genomes by genomic in situ hybridization in wheat-rye derivatives. Heredity (edinb) 83:249–252. https://doi.org/10.1038/sj.hdy.6885570
Sanei M, Pickering R, Kumke K, Nasuda S, Houben A (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci 108:E498–E505. https://doi.org/10.1073/pnas.1103190108
Stockert JC (1994) Mecanismos Moleculares de Recombinación Genética General. In: de Citogenética S (ed) Gosalvez and García de la Vega C. Universidad Autónoma de Madrid, Madrid, pp 59–78
Tosso H, Paccapelo HA, Covas GF (1997) Caracterización de líneas avanzadas de tricepiro. II. Producción deforraje, producción de grano y evaluación de componentes de rendimiento. Rev Invest Agrop 28:47–54
Tosso H, Paccapelo HA, Covas GF (2000) Caracterización de líneas avanzadas de Tricepiro. I Descripción morfológica y citológica. Rev Invest Agrop 29:39–51
Wang H, Jiang J, Chen S, Qi X, Fang W, Guan Z, Teng N, Liao Y, Chen F (2014) Rapid genetic and epigenetic alterations under intergeneric genomic shock in newly synthesized Chrysanthemum morifolium x Leucanthemum paludosum hybrids (Asteraceae). Genome Biol Evol 6:247–259. https://doi.org/10.1093/gbe/evu008
Wulff BB, Moscou MJ (2014) Strategies for transferring resistance into wheat: from wide crosses to GM cassettes. Front Plant Sci 5:692. https://doi.org/10.3389/fpls.2014.00692
Xie Q, Kang H, Sparkes DL, Tao S, Fan XM, Xu L, Fan X, Sha L, Zhang H, Wang Y, Zeng J, Zhou Y (2013) Mitotic and meiotic behavior of rye chromosomes in wheat - Psathyrostachys huashanica amphiploid x triticale progeny. Genet Mol Res 12:2537–2544. https://doi.org/10.4238/2013.January.4.16
Acknowledgements
This research was carried out in Argentina and supported by grants from Universidad Nacional de Lomas de Zamora (LOMASCYT V FCA31).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Handling Editor: Sergey Mursalimov
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dr. Greizerstein deceased in January 2021.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fradkin, M., Greizerstein, E.J., Grassi, E. et al. Cytogenetic analysis of meiotic behaviour and stability in a trigeneric hybrid (triticale x trigopiro). Protoplasma (2024). https://doi.org/10.1007/s00709-024-01964-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00709-024-01964-9