Abstract
Camelina sativa L. Crantz (Brassicaceae family), known as camelina, has gained new attention as a re-emerging oil seed crop. With a unique seed oil profile, with the majority of the fatty acids consisting of linolenic (C18:3), oleic (C18:1), linoleic (C18:2), and eicosenoic (C20:1), camelina oil is reported to be useful as a food oil and biofuel. However, there are still many unknown factors about the structure and genetic variability of this crop. Chromosomal localization of ribosomal DNA was performed using fluorescence in situ hybridization (FISH) with 5S rDNA and 25S rDNA sequences as molecular probes on mitotic chromosomes of enzymatically digested root-tip meristematic cells. Here, we present for the first time a comparative analysis of selected genotypes (cultivars, breeding lines and mutants) of C. sativa with the use of cytogenetic techniques. The main aim of the study was to determine the intraspecific and interspecific polymorphisms in the structure of chromosomes of selected accessions using conserved 5S and 25S rDNA repetitive sequences as molecular probes. The results were compared with C. microcarpa (closely related to C. sativa) rDNA gene loci distribution. The presence of minor rDNA sites was discussed and compared with other Brassicaceae species. In addition, demonstration karyograms of C. sativa and C. microcarpa mapped with rDNA probes were prepared based on the cv. “Przybrodzka” and GE2011-02 genotype, respectively. The use of 5S and 25S rDNA probes provided an insight on the genome structure of C. sativa at the cytogenetic level and can help to understand the genome organization of this crop. The putative role of cytogenetic markers in phylogenetic analyses of camelina was discussed, as well.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Camelina (Camelina sativa L. Crantz), known as a ‘false flax’ or ‘gold of pleasure,’ is one of the oldest cultivated plants in Europe belonging to the Brassicaceae family. Camelina comes from central and southeastern Europe where it has been cultivated since 4000 years BC (Zohary et al. 2012). This species shows the ability to adapt to adverse environmental conditions, is vigorous, and has a high content of polyunsaturated fatty acids (30–40%) in seeds (Kurasiak-Popowska and Stuper-Szablewska 2020). An interest in this crop grew up rapidly, when it was recognized, that camelina seeds and oil are rich in ω-3 acids (α-linolenic acid (C18:3 ω-3), ω-6 acids (linoleic acid (C18:2 ω-6), phytosterols, and phenolic compounds (Berti et al. 2016). This is what makes it an attractive raw material for the production of food and biofuels (Belayneh et al. 2015; Walia et al. 2018; Yang et al. 2016). What is more, high levels of long-chain hydrocarbons in C. sativa oil are commonly considered as an aviation biofuel and have been reported to reduce CO2 emissions compared with traditional petroleum jet fuels (Shonnard et al. 2010).
Although camelina has high potential as an energy and food raw material, there is a growing interest in the structure and evolution of the C. sativa genome, as well as intraspecific variability of this plant. Kagale et al. (2014) published the first chromosome-scale high-quality reference genome sequence of homozygous, doubled-haploid line DH55, derived from C. sativa genotype SRS 933. Research conducted so far has shown the high complexity of the camelina genome, indicating that camelina is an allohexaploid organism (Mandáková et al. 2019). Considering narrow genetic variability, which was reported mainly in spring camelina cultivars (Luo et al. 2019; Vollmann et al. 2005), there is a huge need to explore all available genepools, including winter cultivars, regional ecotypes, breeding lines, mutants, and related species among Camelina species. For example, winter camelina genotypes show significantly lower average percentage content of erucic acid, which is crucial, considering the utilization of the camelina’s oil for food purposes (Kurasiak-Popowska and Stuper-Szablewska 2020). Moreover, valuable gene configurations can be found in mutant lines. Łuczkiewicz and Błaszczyk (1998) developed a dwarf mutant by gamma radiation of winter cultivar Przybrodzka II. Mutant plants showed lower height and more branches than control plants, but the number of silicles, number of seeds, and weight of seeds per plant were higher. Gamma ray irradiation of seeds of cv. “Przybrodzka” revealed several mutants, including a “clavate” mutant which had compact inflorescences (Kurasiak-Popowska et al. 2018). This mutant was a starting point for development of new breeding lines followed by cultivars, “Maczuga” and “Luna,” with the potential to yield reaching 3.0 t/ha and yielding 25–30% higher in comparison with cv. Przybrodzka (Kurasiak-Popowska et al. 2018). Recently, CRISPR-Cas9 system was harnessed to produce camelina breeding lines with increased oleic acid (Jiang et al. 2017; Morineau et al. 2017), decreased long-chain fatty acids (Ozseyhan et al. 2018), and increased oil content (Waltz 2018). Furthermore, C. sativa is closely related to Arabidopsis (both Arabidopsis and Camelina are classified within Camelineae tribe) (Nikolov et al. 2019) which gives the opportunity to explore the genome by comparative studies using C. sativa and A. thaliana genome sequences (Lysák et al. 2016).
Taking into consideration the growing importance of C. sativa, surprisingly, little is known about the chromosome constitution and karyotype structure of this crop. Tepfer et al. (2020) used cytology to study chromosome behavior of progeny produced by interspecific hybridization between Camelina sativa and C. microcarpa during meiosis. Karyotype analysis is crucial for comparative studies considering the exploration of camelina’s related species. Karyotyping is widely used to reveal the number and characteristics of chromosomes and can be used to elucidate the origin, ploidy, and phylogenetic relationships among crop plants, including, among others: Triticeae tribe (Kwiatek et al. 2013; Lysák et al. 1999; Ruban and Badaeva 2018); Brassicaceae (Hasterok et al. 2006; Xiong and Pires 2011) Zea asp. (Albert et al. 2010; Kato et al. 2004), and Glycine sp. (Findley et al. 2010). Routinely, karyotyping is performed on mitotic or meiotic chromosomes and uses several staining methods including Giemsa staining, C-banding, CMA3/DAPIor/and fluorescence in situ hybridization (FISH) with repetitive DNA sequences or BAC clones and flow karyotyping (Badaeva et al. 2017; Doležel et al. 2004). 5S and 18S-5.8S-26S (45S) rDNA clusters are considered as the most common chromosome landmarks. Their number, position, and structure in chromosomes can be characteristic of a given species or genus and used for comparative purposes, including studies in evolutionary biology and systematics, as well as in crop science and plant breeding.
In the Camelina genus, only C. microcarpa chromosomes have been karyotyped using rDNA markers (Ali et al. 2005). To fill this gap, this work for the first time focuses on the use of cytogenetic techniques in the comparative analysis of selected varieties and genotypes of C. sativa. The main aim of the study was to map the chromosomes bearing rDNA gene loci and to determine intraspecific polymorphisms in the structure of chromosomes of selected varieties and breeding lines of camelina with the use of conserved 5S and 25S rDNA repetitive sequences as molecular probes.
Material and methods
Plant material
The research material included ten genotypes (both spring and winter forms; Table 1), originating from Poland, Georgia, Austria, Ukraine, and the area of the former Soviet Union. Three genotypes were obtained from the US Department of Agriculture (USDA), Research, Education, and Economics Agricultural Research Service, Midwest Area, Plant Introduction Station, Iowa State University, Ames (IA, USA). Seven genotypes were obtained from the collection of the Department of Genetics and Plant Breeding at the Poznan University of Life Sciences (PULS). Three of them: cv. Maczuga and 14/3 and K9 breeding lines were developed through mutation breeding (initiated by gamma irradiation) from cv. Przybrodzka (Kurasiak-Popowska et al. 2018) at the Department of Genetics and Plant Breeding at the PULS.
Chromosome preparation
Seeds of C. sativa were germinated on moist filter paper in Petri dishes at 20–22 °C. Further treatment is according to Hasterok et al. (2001). Briefly, seedlings with roots, 1–2 cm long, were treated with 2 mM 8-hydroxyquinoline for 1.5–2 h at room temperature (RT), fixed in ethanol-acetic acid (3:1) fixative, and stored at − 20 °C until use. Fixed roots were washed in citrate buffer (0.01 M citric acid with 0.01 M sodium citrate, pH 4.8) and digested enzymatically in a mixture of 1% (w/v) cellulase from Aspergillus niger (Calbiochem), 1% (w/v) cellulase “Onozuka R-10” (Serva), and 20% (v/v) pectinase (Sigma) for 1.5 h at 37 °C. The root tips were squashed in a drop of 60% acetic acid. After freezing using liquid nitrogen, the coverslips were removed, and the slides were air-dried.
DNA probes and fluorescence in situ hybridization
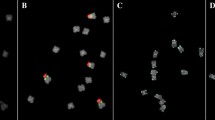
A coding region of 25S rDNA of Arabidopsis thaliana (Unfried and Gruendler 1990), used for detection of 45S rDNA loci, was labeled with digoxigenin-11-dUTP (Roche) by nick translation following the protocol provided by the kit’s manufacturer (Roche). The 5S rDNA from the clone pTa-794 (Gerlach and Dyer 1980) was amplified and labeled with tetramethyl-rhodamine-5-dUTP (Roche) using PCR with universal M13 “forward” (5′-CAG GGT TTT CCC AGT CAC GA-3′) and “reverse” (5′-CGG ATA ACA ATT TCA CAC AGG A-3′) sequencing primers. Conditions for PCR labeling were as follows: 94 °C × 1 min, 35 cycles of 94 °C × 40 s, 55 °C × 40 s, 72 °C × 1 min, 1 cycle of 72 °C × 5 min. Chromosome preparations were pretreated with RNase in 2 × SSC (DNase-free, 100 μg/ml, 1 h at 37 °C) followed by 2 washing runs in 2 × SSC. The hybridization mixture consisted of 50% deionized formamide, 10% dextran sulfate, 2 × SSC, and 0.5% SDS, and probe DNA was mixed to a concentration of 100–130 ng per slide. The hybridization mixture was denatured at 75 °C for 10 min. Next,10 μl of hybridization mixture was applied to the slides and denatured at 70 °C for 5 min using heating plate (Slide Warmer XH2002, C & A Scientific—Premiere). The slides were incubated overnight in a humid chamber at 37 °C. Next day, four runs of washing with 2 × SSC and 0.1 × SSC were performed at 42 °C. The immunodetection of digoxigenated DNA probe was carried out by FITC-conjugated anti-digoxigenin antibodies (Roche). Finally, preparations were mounted in 2 μg/ml DAPI in Vectashield (Vector Laboratories). Images were acquired with a DLT-CAM Pro 12MP CCD (Delta Optical) camera attached to Delta Optical L-1000 epifluorescence microscope then processed using Delta Optical DLT-Cam Viewer software. Karyograms of C. microcarpa (GE2011-02: Fig. 1) and C. sativa (cv. Przybrodzka: Fig. 2) were prepared using IdeoKar software (ver. 1.2) according to Mirzaghaderi and Marzangi (2015).
Results
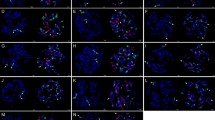
The mitotic chromosome sets of ten genotypes of camelina were evaluated using 3 slides per genotype and based on 20 digital images per slide. In total, 600 images were screened. The chromosomes with 5S, 45S, and 5S + 45S gene loci were counted (Table 1). The rDNA landmarks were divided into major and minor signals according to Ali et al. (2005). All genotypes possessed 40 chromosomes. The morphology of the mitotic chromosomes during metaphase was mostly identical. The only chromosomes of the complement that can be easily recognized, but not confirmed as strictly homologous, were the NOR-bearing chromosomes with usually noticeable secondary constrictions. C. microcarpa (GE2011-02) was used as a control genotype, which is closely related to C. sativa. Cytogenetic analysis revealed that six chromosome pairs of GE2011-02 carried 45S rDNA sites (6 major and 6 minor signals), and five chromosome pairs showed 5S rDNA landmarks (6 major and 4 minor signals) (Fig. 1). Moreover, one pair (no. 10: Fig. 1) revealed a co-localization of 5S and 45S rDNA loci. Chromosomes of cv. Przybrodzka, used as a representative genotype for rDNA mapping on chromosomes of C. sativa, were arranged into a karyogram, as well (Fig. 2). The cytomolecular analysis showed that chromosomes of this cultivar carried one more chromosome pair with 5S rDNA signals. The number of 45S rDNA signals was comparable with C. microcarpa (GE2011-02) line, but only two chromosome pairs showed strong signals. What is more, Przybrodzka revealed two extra chromosome pairs with co-localization of 5S and 45S rRNA loci. The numbers of 5S and 45S rRNA loci vary among the genotypes (Table 1, Supplementary material 1-10). The number of 5S rDNA bearing chromosomes ranged between 8 and 12, with an exception of genotype “Ukrainskij,” which showed eighteen 5S rDNA signals (Table 1). The detailed analysis of distribution of these loci revealed that the number of major 5S rDNA signals ranged between 6 and 10. The number of 45S rDNA loci was diversified. What is interesting, cv. Przybrodzka and Ukrainskij revealed only four chromosomes with strong signals. The highest number (20) of chromosomes with 45S rDNA signals was observed in K9 and 14/9 genotypes. Moreover, four genotypes (GE2011-02, VNIIMK17, 11025, and cv. Przybrodzka) had identical number (12) of chromosomes bearing 45S rDNA loci (Table 1). Co-localization of 5S and 45S rRNA gene loci was observed, as well. The number of chromosomes carrying both types of signals ranged from 2 to 6 (Table 1). What is interesting is that the number of this type of chromosomes was species and habit specific, more precisely: C. microcarpa (GE2011-02) carried two chromosomes carrying both signals, spring cultivars of C. sativa showed 4 chromosomes with co-localization of rDNA probes, and winter cultivars of C. sativa possessed six chromosomes with both sites (Table 1).
Discussion
Comparative analysis of 5S and 45S rDNA gene loci distribution performed on camelina chromosomes indicated the occurrence of intraspecific and interspecific polymorphisms in the number, size (major and minor sites), and localization of rDNA loci. This may indicate chromosomal rearrangements that could occur during species evolution or the breeding process.
A similar analysis was made for the mustard family (Ali et al. 2005), including C. microcarpa Andrz. Ex DC. (2n = 40), which is closely related to C. sativa (Mandáková et al. 2019). Using multicolor fluorescence in situ hybridization with 5S and 25S rDNA probes, Ali et al. (2005) identified 14 chromosomes carrying 45S rDNA signals, six chromosomes with 5S rDNA sites, and four chromosomes bearing both landmarks. The number of C. microcarpa rDNA gene loci is not equal to any observation made in this study considering both C. microcarpa and C. sativa. The number of chromosomes (four) carrying both 45S and 5S rDNA sites reported by Ali et al. (2005) in C. microcarpa is also not the same as the present study (two chromosomes). Moreover, Ali et al. (2005) did not report any minor sites of rDNA loci on chromosomes of C. microcarpa. The differences in the number of minor rDNA sites may reflect their origin. Here, the probe signals were classified into two groups, including minor (weak) and major (strong) signals. This kind of signal categorization is quite common in cytogenetic studies—and can be associated with copy number of particular rDNA loci. For example, Taketa et al. (1999) observed the differences in distribution of weak and strong 5S signals on chromosomes of H. spontaneum which gave an evidence for variation in its copy number among different accessions of this species. It was also hypothesized that the minor rDNA sites are NOR residue remains of ancestral forms. What is more, it was reported that the activity of rRNA genes is correlated with the length of the intergenic spacer and to the level of the methylation of cytosine residues in regulatory sequences (Abou-Ellail et al. 2011). Moore et al. (1993) showed that 45S rDNA loci, mostly at minor loci and in chromosomal positions more proximal than those of the normally active NORs, are more methylated and become deleted during evolution. The presence of minor rDNA sites has also been described in other Brassicaceae species, such as Arabidopsis suecica, Brassica oleracea, Olimarabidopsis pumila, O. cabulica, Rorippa palustris, Neslia paniculata (Ali et al. 2005), and B. rapa (Maluszynska and Heslop-Harrison 1993).
Another issue concerns the number chromosomes carrying both signals (5S and 45S rDNA) of cv. Przybrodzka, which was similar to 14/9, K9, and cv. Maczuga (developed from cv. Przybrodzka through mutation breeding; Kurasiak-Popowska et al. 2018). Noticeably, cv. Przybrodzka and other winter accessions showed higher number of chromosomes with co-localization of 5S and 45S rDNA than spring accessions (four chromosomes) and C. microcarpa (two chromosomes). Generally, it is said that nucleolar architecture and rDNA transcription respond to cellular stresses, including irradiation or chemical mutation (Kus et al. 2018; Stimpson et al. 2014). The changes in rDNA gene loci distribution induced by irradiation were observed in other species, such as barley (Juchimiuk-Kwasniewska et al. 2011), Brachypodium (Kus et al. 2019), or wheat (Mukai et al. 1993). Moreover, Amosova et al. (2019) reported that rapeseed breeding by chemical mutagenesis can result in cytogenetic instability in the mutant progeny. Significant differences in karyotypes were observed in Brassica napus mutant lines using 5S and 45S rDNA gene loci visualization (Amosova et al. 2019).
An increased number of chromosomes carrying rDNA gene loci are characteristic for polyploids (Kovarik et al. 2008). However, there are many examples of biased elimination of those loci related to asymmetric transcription and epigenetic modifications caused by the polyploid formation (McStay 2006). Schranz et al. (2006) proposed a speciation theory of the ancestral genome of Camelina, called CAM (2n = 14), which has evolved from an ancestral crucifer karyotype (ACK). Mandáková et al. (2019) elucidated the following evolution pathway of Camelina species. Bacterial artificial chromosome–based chromosome painting together with genomic in situ hybridization and multi-gene phylogenetics revealed that CAM (2n = 14) was an ancestor of diploid camelinas (Camelina hispida, 2n = 14; Camelina laxa, 2n = 12; and Camelina neglecta, 2n = 12). It was also reported that diploid species C. neglecta (2n = 12, N6N6) and C. hispida (2n = 14, H7H7) participated in the origin of allotetraploid Camelina rumelica (2n = 26, N6N6H7H7). Recently, Mandáková et al. (2019) reported that allohexaploid C. sativa (2n = 40) originated through hybridization between an auto-allotetraploid C. neglecta–like genome (2n = 13, N6N6N7N7) and C. hispida (2n = 14, H7). What is interesting, the same model of origin and chromosome number (2n = 40) was assigned to C. microcarpa; however, this species appears to have different cytotypes (2n = 12; 26 or 40) (Martin et al. 2017). Moreover, considering differences in 45S rDNA loci distribution between chromosome complements of C. sativa and C. microcarpa, it could be hypothesized that it is possible that both species have evolved from a common allohexapliod ancestor and followed separate speciation pathway. Considering similar ploidy level and origin pathways, C. microcarpa can be used as a primary genepool for improvement of C. sativa genetic diversity.
In conclusion, depending on genotype, rDNA loci landmarks allowed us to discriminate 11 to 16 pairs of camelina chromosomes, which indicate significant intraspecific polymorphisms and varied chromosome composition and structure within this species. Moreover, similar but not identical numbers of rDNA gene loci in C. microcarpa can be one of selective markers for breeding purposes, such as cytogenetic evaluation of C. sativa × C. microcarpa hybrids. Minor rDNA sites can be also used as a cytogenetic marker to map chromosomes of the diploid ancestors in order to confirm the putative parental species as well as the origin of camelina.
Data availability
Data and material are available upon reasonable request.
References
Abou-Ellail M, Cooke R, Sáez-Vásquez J (2011) Variations in a team: major and minor variants of Arabidopsis thaliana rDNA genes. Nucleus 2:294–299
Albert PS, Gao Z, Danilova TV, Birchler JA (2010) Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet Genome Res 129:6–16
Ali HBM, Lysak MA, Schubert I (2005) Chromosomal localization of rDNA in the Brassicaceae. Genome 48:341–346
Amosova AV, Zoshchuk SA, Volovik VT, Shirokova AV, Horuzhiy NE, Mozgova GV, Yurkevich OY, Artyukhova MA, Lemesh VA, Samatadze TE, Muravenko OV (2019) Phenotypic, biochemical and genomic variability in generations of the rapeseed (Brassica napus L.) mutant lines obtained via chemical mutagenesis. PLoS One 14:e0221699
Badaeva ED, Ruban AS, Aliyeva-Schnorr L, Municio C, Hesse S, Houben A (2017) In situ hybridization to plant chromosomes. In: Liehr T (ed) Fluorescence In Situ Hybridization (FISH): Application Guide. Springer, Berlin Heidelberg, pp 477–494
Belayneh HD, Wehling RL, Cahoon E, Ciftci ON (2015) Extraction of omega-3-rich oil from Camelina sativa seed using supercritical carbon dioxide. J Supercrit Fluids 104:153–159
Berti M, Gesch R, Eynck C, Anderson J, Cermak S (2016) Camelina uses, genetics, genomics, production, and management. Ind Crop Prod 94:690–710
Doležel J, Kubaláková M, Bartoš J, Macas J (2004) Flow cytogenetics and plant genome mapping. Chromosom Res 12:77–91
Findley SD, Cannon S, Varala K, Du J, Ma J, Hudson ME, Birchler JA, Stacey G (2010) A fluorescence in situ hybridization system for karyotyping soybean. Genetics 185:727–744
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res 8:4851–4865
Hasterok R, Jenkins G, Langdon T, Jones RN, Maluszynska J (2001) Ribosomal DNA is an effective marker of Brassica chromosomes. Theor Appl Genet 103:486–490
Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, Heneen WK, Maluszynska J (2006) Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann Bot 97:205–216
Jiang WZ, Henry IM, Lynagh PG, Comai L, Cahoon EB, Weeks DP (2017) Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol J 15:648–657
Juchimiuk-Kwasniewska J, Brodziak L, Maluszynska J (2011) FISH in analysis of gamma ray-induced micronuclei formation in barley. J Appl Genet 52:23–29
Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C, Higgins EE, Huebert T, Sharpe AG, Parkin IAP (2014) The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun 5:3706
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci U S A 101:13554–13559
Kovarik A, Dadejova M, Lim YK, Chase MW, Clarkson JJ, Knapp S, Leitch AR (2008) Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Ann Bot 101:815–823
Kurasiak-Popowska D, Stuper-Szablewska K (2020) The phytochemical quality of Camelina sativa seed and oil. Acta Agric Scand Sect B Soil Plant Sci 70:39–47
Kurasiak-Popowska D, Tomkowiak A, Człopińska, Bocianowski J, Weigt D, Nawracała J (2018) Analysis of yield and genetic similarity of Polish and Ukrainian Camelina sativa genotypes. Ind Crop Prod 123:667–675
Kus A, Kwasniewska J, Szymanowska-Pułka J, Hasterok R (2018) Dissecting the chromosomal composition of mutagen-induced micronuclei in Brachypodium distachyon using multicolour FISH. Ann Bot 122:1161–1171
Kus A, Szymanowska-Pułka J, Kwasniewska J, Hasterok R (2019) Detecting Brachypodium distachyon chromosomes Bd4 and Bd5 in MH- and X-ray-induced micronuclei using mcFISH. Int J Mol Sci 20(11):2848
Kwiatek M, Wiśniewska H, Apolinarska B (2013) Cytogenetic analysis of Aegilops chromosomes, potentially usable in triticale (X Triticosecale Witt.) breeding. J Appl Genet 54:147–155
Łuczkiewicz T, Błaszczyk L (1998) Dwarf mutant of Camelina sativa L. Oilseed Crops XIX:615–620
Luo Z, Brock J, Dyer JM, Kutchan T, Schachtman D, Augustin M, Ge Y, Fahlgren N, Abdel-Haleem H (2019) Genetic diversity and population structure of a Camelina sativa spring panel. Front Plant Sci. https://doi.org/10.3389/fpls.2019.00184
Lysák MA, Číuhalíková J, Kubaláková M, Šimková H, Künzel G, Doležel J (1999) Flow karyotyping and sorting of mitotic chromosomes of barley (Hordeum vulgare L.). Chromosom Res 7:431–444
Lysák MA, Mandáková T, Schranz ME (2016) Comparative paleogenomics of crucifers: ancestral genomic blocks revisited. Curr Opin Plant Biol 30:108–115
Maluszynska J, Heslop-Harrison JS (1993) Physical mapping of rDNA loci in Brassica species. Genome 36:774–781
Mandáková T, Pouch M, Brock JR, Al-Shehbaz IA, Lysák MA (2019) Origin and evolution of diploid and allopolyploid Camelina genomes were accompanied by chromosome shattering. Plant Cell 31:2596–2612
Martin SL, Smith TW, James T, Shalabi F, Kron P, Sauder CA (2017) An update to the Canadian range, abundance, and ploidy of Camelina spp. (Brassicaceae) east of the Rocky Mountains. Botany 95:405–417
McStay B (2006) Nucleolar dominance: a model for rRNA gene silencing. Genes Dev 20:1207–1214
Mirzaghaderi G, Marzangi K (2015) IdeoKar: an ideogram constructing and karyotype analyzing software. Caryologia 68:31–35
Moore G, Gale MD, Kurata N, Flavell RB (1993) Molecular analysis of small grain cereal genomes: current status and prospects. Bio/Technology 11:584–589
Morineau C, Bellec Y, Tellier F, Gissot L, Kelemen Z, Nogué F, Faure J-D (2017) Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol J 15:729–739
Mukai Y, Friebe B, Hatchett JH, Yamamoto M, Gill BS (1993) Molecular cytogenetic analysis of radiation-induced wheat-rye terminal and intercalary chromosomal translocations and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma 102:88–95
Nikolov LA, Shushkov P, Nevado B, Gan X, Al-Shehbaz IA, Filatov D, Bailey CD, Tsiantis M (2019) Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol 222:1638–1651
Ozseyhan ME, Kang J, Mu X, Lu C (2018) Mutagenesis of the FAE1 genes significantly changes fatty acid composition in seeds of Camelina sativa. Plant Physiol Biochem 123:1–7
Ruban AS, Badaeva ED (2018) Evolution of the S-genomes in Triticum-Aegilops Alliance: evidences from chromosome analysis. Front Plant Sci 9:1756
Schranz ME, Lysák MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:535–542
Shonnard DR, Williams L, Kalnes TN (2010) Camelina-derived jet fuel and diesel: sustainable advanced biofuels. Environ Prog Sustain Energy 29:382–392
Stimpson KM, Sullivan LL, Kuo ME, Sullivan BA (2014) Nucleolar organization, ribosomal DNA Array stability, and acrocentric chromosome integrity are linked to telomere function. PLoS One 9:e92432
Taketa S, Harrison G, Heslop-Harrison J (1999) Comparative physical mapping of the 5S and 18S-25S rDNA in nine wild Hordeum species and cytotypes. Theor Appl Genet 98:1–9
Tepfer M, Hurel A, Tellier F, Jenczewski E (2020) Evaluation of the progeny produced by interspecific hybridization between Camelina sativa and C. microcarpa. Ann Bot 125(6):993–1002
Unfried I, Gruendler P (1990) Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res 18:4011
Vollmann J, Grausgruber H, Stift G, Dryzhyruk V, Lelley T (2005) Genetic diversity in camelina germplasm as revealed by seed quality characteristics and RAPD polymorphism. Plant Breed 124:446–453
Walia MK, Wells MS, Cubins J, Wyse D, Gardner RD, Forcella F, Gesch R (2018) Winter camelina seed yield and quality responses to harvest time. Ind Crop Prod 124:765–775
Waltz E (2018) With a free pass, CRISPR-edited plants reach market in record time. Nat Biotechnol 36:6–7
Xiong Z, Pires JC (2011) Karyotype and identification of all Homoeologous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics 187:37–49
Yang J, Caldwell C, Corscadden K, He QS, Li J (2016) An evaluation of biodiesel production from Camelina sativa grown in Nova Scotia. Ind Crops Prod 81:162–168
Zohary D, Hopf M, Weiss E (2012) Domestication of Plants in the Old World: The origin and spread of domesticated plants in Southwest Asia, Europe, and the Mediterranean Basin, 4th edn. Oxford University Press, Oxford
Acknowledgments
We thank Candice Gardner and Stacey Estrada at the USDA/ARS Midwest Area—Plant Introduction Research Unit, North Central Regional Plant Introduction Station, Iowa State University, Ames (IA, USA), for providing the seeds of camelina accessions. In addition, we would like to thank all of the reviewers and manuscript editor for their careful review of the manuscript and for their excellent suggestions for improving our initial work.
Funding
This publication is being co-financed by the framework of Ministry of Science and Higher Education program as “Regional Initiative Excellence” in years 2019–2022, project no. 005/RID/2018/19.
Author information
Authors and Affiliations
Contributions
MK initiated the project. ZD, MK, and AN made the experiments and analyses, wrote the first draft, and incorporated all inputs from co-authors. MK, DKP, and JN revised the draft. MK wrote the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Code availability
Not applicable.
Additional information
Communicated by: Izabela Pawłowicz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary material 1-10 Variability of 5S rDNA (red) and 45S rDNA (green) loci in Camelina genotypes demonstrated by the FISH results for selected ten genotypes: (1) GE2011–02 (C. microcarpa); (2) VNIIMK17; (3) CSS-CAM38; (4) 11025; 5) 7; (6) Ukrainskij; (7) 14/3; (8) K9; (9) ‘Maczuga’ and (10) Przybrodzka. Scale bar: 5 μm.
ESM 1
(PNG 3234 kb)
ESM 2
(PNG 3424 kb)
ESM 3
(PNG 3069 kb)
ESM 4
(PNG 3227 kb)
ESM 5
(PNG 4593 kb)
ESM 6
(PNG 3274 kb)
ESM 7
(PNG 3264 kb)
ESM 8
(PNG 3022 kb)
ESM 9
(PNG 2538 kb)
ESM 10
(PNG 2613 kb)
Rights and permissions
About this article
Cite this article
Kwiatek, M.T., Drozdowska, Z., Kurasiak-Popowska, D. et al. Cytomolecular analysis of mutants, breeding lines, and varieties of camelina (Camelina sativa L. Crantz). J Appl Genetics 62, 199–205 (2021). https://doi.org/10.1007/s13353-020-00600-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-020-00600-5