Abstract
Low temperatures in summer bring about drastic reduction in seed yield of soybean [Glycine max (L.) Merr.]. To identify quantitative trait loci (QTL) associated with chilling tolerance during the reproductive growth in soybean, a recombinant inbred line (RIL) population consisting of 104 F6-derived lines was created from a cross between two cultivars, chilling-tolerant Hayahikari and chilling-sensitive Toyomusume. The RIL were genotyped with 181 molecular and phenotypic markers and were scored with regard to chilling tolerance, which was evaluated by comparison of seed-yielding abilities in two artificial climatic environments at chilling and usual temperatures. Three QTL were detected for chilling tolerance in seed-yielding ability. Two of them, qCTTSW1 and qCTTSW2, were mapped near QTL for flowering time, and the latter had an epistatic interaction with a marker locus located near another QTL for flowering time, where no significant QTL for chilling tolerance was detected. The analysis of an F2 population derived from the cross between Hayahikari and an RIL of the Hayahikari genotype at all QTL for flowering time confirmed the effect of the third QTL, qCTTSW3, on chilling tolerance and suggested that qCTTSW1 was basically independent of the QTL for flowering time. The findings and QTL found in this study may provide useful information for marker-assisted selection (MAS) and further genetic studies on soybean chilling tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cultivation of soybean has continued its expansion into cool areas including the eastern and northern Europe, Canada, Siberia, and Hokkaido, the northernmost part of Japan, where soybean plants have frequently suffered from low temperatures, especially in summer. Despite the general tendency of the global warming, we have recently experienced abnormal local drops in temperature. The most economically important legume crop is now at a higher risk of chilling injury, which increases the importance of breeding cultivars tolerant to low temperatures as well as heat and drought.
It is known that considerable genetic variation for chilling tolerance exists in soybean (Raper and Kramer 1987). Some cultivars from Hokkaido and the islands of the Okhotsk Sea, for instance, have a somewhat lower minimum temperature for good flowering and pod set than 17°C, which is required for standard cultivars (Raper and Kramer 1987). The identification of such genes and/or loci associated with chilling tolerance is of major interest for soybean breeders in cool regions.
Several genes associated with chilling tolerance have been identified by comparing cultivars and near-isogenic lines (NIL) (Sanbuichi 1979; Yumoto and Tsuchiya 1991; Morrison et al. 1994; Takahashi and Asanuma 1996; Morrison et al. 1997). The lines with the genes, p1 (pubescent surface) and T (tawny pubescence), were more tolerant to low temperatures for seed-yielding ability when compared to those with other alleles at those loci. However, the possibility of finding novel genes conferring chilling tolerance through this approach is limited because the candidate genes need to influence visible traits.
Very recently, the possible involvement of maturity genes, which affect both flowering and maturity times, in chilling tolerance for seed-yielding ability has been pointed out (Kurosaki et al. 2004; Takahashi et al. 2005). In the former study, the effect of the maturity locus E1 was examined using isolines with a Japanese genetic background, and the latter study compared two genotypes of Harosoy isolines with different alleles at three maturity loci. Through these approaches, comprehensive genetic dissection including the epistatic interaction analysis could not be conducted although epistatic interactions among the maturity loci have been reported for flowering time (Saindon et al. 1989a, b; Saindon et al. 1990; Cober et al. 1996; Watanabe et al. 2004). In addition, fluctuating, natural temperatures were used in the control environments and in the growth periods of chilling plants before and/or after chilling treatment, which confounded the effects. The effects of maturity loci on chilling tolerance need to be verified using a population segregating at multiple maturity loci and under completely controlled environment.
The quantitative trait locus (QTL) analysis is a powerful tool in genomic study. In rice, QTL for chilling tolerance with regard to seed reproduction have recently been mapped (Saito et al. 2001; Takeuchi et al. 2001; Andaya and Mackill 2003; Saito et al. 2004). They utilized numerous molecular markers and evaluation methods for chilling tolerance under controlled environments. However, there is little information on QTL analysis in soybeans, even though numerous molecular markers at high density are available for such a study (Cregan et al. 1999; Yamanaka et al. 2001; Song et al. 2004). The lack of reliable and convenient evaluation methods is one of the reasons why QTL studies have not been reported in the literature (Harada and Xia 2004).
In the present study, we conducted QTL analysis of soybean chilling tolerance using a newly developed evaluation method (Funatsuki et al. 2004) and recombinant inbred lines (RIL) derived from a cross between cultivars differing in chilling tolerance. The objectives of this study were (1) to identify QTL for chilling tolerance and (2) to clarify the association between these QTL and maturity loci. We report the genomic location of QTL for chilling tolerance and their association with QTL for flowering time. Epistatic interaction between these loci are also presented.
Materials and methods
Plant materials
A total of 104 F6 RIL were created from the cross between the soybean cultivars Toyomusume and Hayahikari. Hayahikari is among the most chilling-tolerant cultivars, while Toyomusume is relatively chilling-sensitive (Kurosaki and Yumoto 2003; Funatsuki et al. 2003). An F2 population derived from a cross between an RIL HC1-F7-50 and Hayahikari was also used. This RIL flowered 0.75 day later than Hayahikari on average and had an averaged LCTI of 2.75. The RIL was presumed to be of the Hayahikari genotype at all significant QTL for flowering time and have the alleles from Toyomusume at the T locus and the third QTL for chilling tolerance, qCTTSW3, as described below.
Plant growth conditions
The F6:7 RIL were grown in growth chambers to estimate chilling tolerance for seed-yield. Since the limited space in growth chambers precluded the possibility of growing all the RIL at the same time, the RIL were randomly divided in two groups I and II. Group I consisted of 76 RIL and the set was grown twice in 2003, while Group II was composed of the other 28 RIL and was grown twice in 2004. In the first experiment season of each year, seeds were sown in March, while the second seasons started in July. In each season, a pair of pots for each RIL was used for chilling evaluation, with each pot containing two plants. One pot of the pair was subjected to chilling treatment, while the other was used as a control. Parental cultivars were planted in two pots in each temperature regime in every experiment season. A set of 80 pots were placed in each environment in each season; 76 RIL plus four parental pots in 2003, 28 RIL plus four parental pots and 48 pots for another study in the first experiment of 2004, and 28 RIL plus four parental pots and 48 pots of the F2 population in the second experiment of 2004. Comparison of trait values of parental cultivars and those averaged for RIL between the years for each experimental season revealed no significant difference, while there could be a difference between experimental seasons. Consequently, the data obtained in the first experiments in both years were combined and dealt with as a block, while the second experiments were regarded as the other. Thus, randomized block design was employed.
The temperature regimes were set according to method III described by Funatsuki et al. (2004). Plants were grown until first flowering at 22°C/17°C with 15 h light supplied from metal-halide lamps (ca. 300 μmol m−2 s−1 at canopy height) in walk-in growth chambers A and B. In these growth chambers, the pairs of pots from RIL were placed randomly, but the paired pots were kept side by side. To minimize the effect of possible heterogeneous climate within these growth chambers, the positions of the paired pots were changed within the chambers every week, as recommended by Satake et al. (1969). Upon flowering of each RIL, one pot of the pair was moved to the chilling environment (15°C/15°C) and the other was transferred to the control environment (24°C/17°C). For these different environments, another type of two walk-in growth chambers C and D, which used the natural light source and supplementary lamps that kept 15 h light, were used. Growth chamber C and D were used for chilling and control environments, respectively, throughout all experiments. Since the variation in plant height among the RIL increased with time, pots were not placed randomly to avoid severe shading by adjacent plants which could occur to a small plant surrounded by tall plants. RIL were grouped in four sub-sets for a growth chamber according to their plant heights, and the sub-sets of pots were randomly placed and rotated every week. Within sub-sets, the positions of pots were also changed. The placement of pots and the rotation were carried out in the same way in growth chambers C and D so that the relative spatial positions of RIL will be preserved in the different environments.
Flowering dates were scored at first flowering corresponding to the developmental stage of R1.
In the F2 population, 192 plants were used. A half of them were exposed to chilling treatment, while the remainders were grown under the control environment. These plants were grown as described for RIL except that all pots were moved from growth chambers A and B to C and D when 50% plants of the population reached flowering.
Genotyping of RIL
DNA was isolated by taking a single leaf from the plants. The FastDNA Kit (Qbiogene, Irvine, CA, USA) was used to obtain purified DNA from individual F6 plants. For F2 plants, a simpler DNA isolation procedure was employed (Edwards et al. 1991).
Polymerase chain reactions (PCR) for single sequence repeat (SSR) markers were performed in a 96-well thermal cycler (GeneAmp PCR System 9600, Applied Biosystems, Foster City, CA, USA). The 10 μl PCR mix consisted of 40 ng template DNA, 1×PCR Buffer, 0.2 mM dNTP mixture (Pharmacia, Piscataway, NJ, USA), 0.2 μM forward and reverse primer, and 0.25 units of HotStarTaq (Qiagen, Hilden, Germany). The amplification conditions consisted of 95°C for 15 min followed by 40 cycles of 92°C denaturation for 30 s, 47°C annealing for 30 s, and 68°C extension for 30 s, ending with one cycle of 72°C for 5 min. PCR products were separated using 3.5% (w/v) agarose, 6% (w/v) non-denaturing polyacrylamide (Wang et al. 2003), and 6% (w/v) denaturing polyacrylamide gel electrophoreses (Echt et al. 1996) according to the size difference between amplicons for the parental cultivars. Ethidium bromide or Sybr Gold (Molecular Probes, Eugene, OR, USA) was used for staining. The RIL were scored based on visualization of the DNA bands.
The F2 plants derived from the cross between the RIL HC1-F7-50 and Hayahikari grown in the chilling environment were genotyped at the T locus by PCR-RFLP analysis as described by Toda et al. (2002). Briefly, amplified fragments of genomic DNA with primers specific to the candidate sequence for the T gene were incubated with EcoNI, which digested only the amplicon from the allele from Hayahikari due to a nucleotide deletion in Toyomusume.
Map construction
Toyomusume and Hayahikari were tested with 968 SSR markers previously placed on the integrated soybean genetic linkage map (Song et al. 2004). One hundred and seventy-eight polymorphic markers were chosen to achieve a reasonable coverage of all 20 molecular linkage groups. The 104 RIL were genotyped with these 178 SSR markers, two phenotypic markers (W1, T) and one In/Del marker (Funatsuki et al. 2003). Map Manager QTX (Manly et al. 2001) was used to estimate genetic distances between markers. The analysis employed the default values including linkage criterion at P=0.05, and Haldane map function. The nomenclature of linkage groups was according to Cregan et al. (1999), based on the common markers mapped.
Data analysis
Traits associated with the seed-yielding ability were analyzed. Total seed weight per plant was resolved into its components, seed number per plant, and single seed weight as follows:
Chilling tolerance index (CTI) was calculated according to Sanbuichi (1979) as follows:
Chilling tolerance index was calculated for each pair of pots grown at the same time, with the averaged values for the two plants of each pot being used. Furthermore, CTI were logarithmically transformed (LCTI) prior to statistical and QTL analyses. When LCTI were less than 0, corresponding to CTI<1, these values were substituted for a 0 for further analyses to avoid the overestimation of variance derived from these sample data. One LCTI value for seed number per plant and six values for total seed weight per plant were substituted. Trait values for flowering time (day to flowering) were used in further analyses without transformation.
Two-way analysis of variance (ANOVA) was carried out using the PROC GLM procedure (SAS Institute 1996) to evaluate the differences between blocks and between RIL with regard to LCTI.
Heritability (h2) of LCTI for each trait was defined as:
where r is the number of replications (Falconer 1976). Genotype and error variance components (σ2 g and σ 2 e) were computed from ANOVA using the PROC GLM procedure. Dominance variance was assumed to be negligible in the calculation of h2 because of the inbred nature of the population (F6:7). The PROC GLM procedure with the Tukey-Kramer method (option/tukey) was also used to estimate the difference in phenotypic values among marker classes. One-way ANOVA for the analysis of the F2 population was also carried out using PROC GLM.
Three-way ANOVA by PROC GLM was performed to reveal epistatic interactions among putative QTL, as represented by the nearest marker loci. The statistical model used was:
where X ijk = mean phenotypic value for the genotype in the ith, jth, and kth marker classes at the M1, M2 and M3 loci, μ = population mean, M1 i = the effect of the ith marker class at the M1 locus, M2 j = the effect of the jth marker class at the M2 locus, M3 k = the effect of the kth marker class at the M3 locus, (M1 i *M2 j ) = the effect of the interaction between the ith and jth marker classes at the M1 and M2 loci, (M1 i *M3 k ) = the effect of the interaction between the ith and kth marker classes at the M1 and M3 loci, (M2 j *M3 k ) = the effect of the interaction between the jth and kth marker classes at the M2 and M3 loci, (M1 i *M2 j *M3 k ) = the effect of the interaction among the ith, jth and kth marker classes at the M1, M2 and M3 loci, and e ijk = the residual error term. The P value to declare a significant epistatic interaction was defined to be 0.05, since epistatic interaction was examined only among the markers close to putative QTL for flowering time. Because we utilized an F6-derived RIL population, heterozygotes were dropped from the data set in three-way ANOVA.
QTL analysis
Quantitative trait loci analysis was performed using the mean phenotypic trait values and genotypic scores on the 104 RIL planted in the growth chamber environments. Composite interval mapping (CIM) was performed using QTL Cartographer (Basten et al. 2001). For CIM, model 6 in the Zmapqtl program module was used for detecting QTL and their effects. The genome was scanned at 2-cM intervals, with a window size of 10 cM. One thousand permutation tests were performed on each trait to establish empirical LOD thresholds at the 5% α level for experimentwise type I error (Churchill and Doerge 1994).
Results
Phenotypic variation
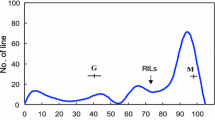
The mean LCTI of total seed weight of the parental cultivars, and the RIL population in the first and second experiment seasons each year are shown in Table 1. Hayahikari exhibited consistently and significantly superior chilling tolerance for seed-yielding ability. Since no significant differences were found between years in the mean values for parental cultivars or for the RIL, the data of 2003 and 2004 were combined for each experiment season and dealt with as blocks. Therefore data analyses were performed without adjustment for any experimental season. As shown in Fig. 1a, in the F6:7 population, LCTI for seed-yielding ability showed a large variation. ANOVA confirmed the genotypic difference in LCTI among RIL (df=103, F-value=7.19, P<0.0001). Entry-mean heritability was computed to be 0.83.
Frequency distributions of 104 recombinant inbred lines and heritabilities for logarithmically transformed chilling tolerance indices (LCTI) of total seed weight per plant (a) and for flowering time (b). Parental mean values are indicated with arrows (T: Toyomusume, A: Hayahikari). The vertical axes represent number of RIL, while the horizontal ones indicate LCTI in a and days to first flowering in b, respectively. Open, solid, and shaded boxes in b represent the frequencies of RIL of the Toyomusume, the Hayahikari and the heterozygous genotypes at the Satt100, respectively
Linkage map construction
The resulting genetic linkage map covered 1,794 cM. Linkage groups (LG) E, K, and N were divided into two groups for each, since no polymorphic markers were found in some regions. Consequently, the markers constituted 23 linkage groups.
QTL analysis for chilling tolerance
Composite interval mapping at the empirical threshold value of LOD score 3.8 identified three QTL, qCTTSW1, qCTTSW2, and qCTTSW3, for LCTI of total seed weight per plant (Fig. 2, Table 2). The QTL with the highest LOD score, qCTTSW1, was located in the vicinity of the phenotypic marker T on LG C2. The second QTL, qCTTSW2, was discovered on LG L. The remaining minor QTL, qCTTSW3, was detected on LG H. At all the QTL the Hayahikari allele had beneficial effects. To examine associations between these QTL and chilling tolerance in the components of total seed weight per plant, i.e., seed number per plant and single seed weight, QTL analysis was performed also for these two traits. In the region on LG C2 including qCTTSW1, a major QTL for LCTI of single seed weight and a minor QTL for LCTI of seed number were detected (Fig. 2, Table 2), suggesting that the Hayahikari allele at this QTL has a large effect to alleviate the drop in single seed weight along with some beneficial effect to maintain seed number at low temperatures. As for qCTTSW2, only QTL for LCTI of seed number was detected in the region adjacent to this QTL. In the region near qCTTSW3, a LOD peak with the score of 3.1 for LCTI of seed number was observed although it did not exceed the empirical threshold value of 4.0.
Linkage map and locations of QTL for chilling tolerance in traits associated with seed-yielding ability and for flowering time. Horizontally striped, vertically striped, black and white bars represent putative regions of QTL for chilling tolerance in total seed number per plant, single seed weight, total seed weight per plant and flowering time, respectively. A reduction of one LOD value from the LOD peaks was used to define borders of the confidence intervals. Linkage groups (LG) were designated according to Cregan et al. (1999). Linkage groups with no QTL are omitted from this figure
QTL analysis for flowering time
We examined whether Hayahikari and Toyomusume had different maturity genes by identifying QTL for flowering time in the RIL population. The flowering times of the RIL showed a large variation, and the frequency distribution of the RIL was discontinuous (Fig. 1b), which divided the population in two groups, Group E with days to flowering of 29–49 days and Group L with days to flowering of 53–76 days. QTL analysis resolved three significant loci for flowering time (Fig. 2, Table 2). The QTL with the largest effect, qFLT1, was found in the vicinity of the locus T and qCTTSW1 on LG C2. The second one, qFLT2, was located between SSR markers Satt229 and Satt373 on LG L, where qCTTSW2 was identified. The third one, qFLT3, was found on LG I.
Epistatic interaction among QTL for flowering time and chilling tolerance
To investigate the epistatic interactions among the QTL for flowering time, we conducted three-way ANOVA based on the genotypic classification at the nearest marker loci (Table 3). All combinations of the marker loci exhibited significant interaction effects. In the RIL carrying the Hayahikari allele at Satt100, the loci Satt229 and Satt496 had larger effects, but their effects were not additive but synonymous (Table 4).
The re-examination of the frequency distribution based on the genotyping at the marker loci closest to the QTL confirmed the epistatic interaction (Fig. 1b). All the lines homozygous for the allele from Toyomusume (RIL groups V–VIII in Table 4) were assigned to group E and they displayed a continuous distribution. On the other hand, the RIL with the allele from Hayahikari including the heterozygous genotype were divided into group E and L. All the lines of the Hayahikari genotype at all the markers (RIL group I in Table 4) belonged to group E, while the others (RIL group II–IV) were assigned to group L with one exception of an RIL of group III, which was assigned to group E. This exceptional RIL, however, had the Hayahikari allele at Satt373, the other marker flanking qFLT2, suggesting that a recombination occurred between qFLT2 and Satt229.
Subsequently, we examined epistatic interactions among these markers on LCTI for the traits associated with seed-yielding ability (Table 3). While no interaction effect between Satt100 and Satt229 or between Satt100 and Satt496 was evident, Satt229 and Satt496 displayed a significant interaction effect for LCTI in total seed weight per plant as well as for that in seed number per plant. The beneficial effect of the Hayahikari allele at Satt229 was pronounced when combined with the Hayahikari allele at Satt496 (Table 5).
Analysis of QTL for chilling tolerance independent of maturity loci
To verify whether QTL for chilling tolerance on total seed weight per plant independent of maturity loci are present, we conducted further examination on qCTTSW3 on LG H and the region around qCTTSW1 that did not include qFLT1, using the F2 population derived from a cross between an RIL HC1-F7-50 and Hayahikari. The marker genotype and its flowering time suggested that HC1-F7-50 had the alleles from Hayahikari at all QTL for flowering time (Table 6). The standard deviation (SD) of days to flowering for the F2 population was 1.2 days, which was slightly larger than those for the parental cultivars (0.6 day for Hayahikari and 0.9 day for Toyomusume) but was much smaller than 13.0 days for the RIL. Indeed, no significant difference in flowering time was detected in the F2 population among marker classes adjacent to qFLT1and qFLT2 (Table 7). The RIL was determined to be of the Toyomusume genotype at the marker loci of T and Satt277 (Table 6). In addition, HC1-F7-50 had the Toyomusume alleles at the marker loci around qCTTSW3 (Table 6).
As CTI cannot be calculated for F2 population, the populations grown in the chilling and the control environments were analyzed separately using T and Satt635, which were the markers closest to qCTTSW1 and qCTTSW3. One-way ANOVA on the F2 populations revealed the highly significant difference between marker classes at the T locus in seed number, single seed weight, and total seed weight per plant in the F2 population grown in the chilling environment, while no difference between the genotypes was seen in any trait in the control environment (Table 7). As for Satt635, there was a relatively little but significant difference between marker classes for seed number and total seed weight only in the population grown in the chilling environment. Furthermore, to explore the dominance effect of the T locus, the F2 population was genotyped using a co-dominance marker for this locus. The heterozygous genotypes exhibited almost the same seed-yielding ability in the chilling environment as did the homozygotes (Table 7), suggesting complete dominance effect at the T locus.
Discussion
Based on the previously mapped positions (Cregan et al. 1999; Yamanaka et al. 2001, 2005; Abe et al. 2003; Molnar et al. 2003) and the magnitudes of effect, qFLT1, qFLT2 and, qFLT3 are likely to correspond to maturity loci E1(Bernard 1971), E3 (Buzzell 1971), and E4 (Buzzell and Voldeng 1980), respectively. We observed significant epistatic interactions among qFLT1, qFLT2, and qFLT3. The results suggest that qFLT2 and qFLT3 have small effects in the RIL carrying the Toyomusume allele at qFLT1, but have large effects in the RIL with the Hayahikari allele at qFLT1, which resulted in the discontinuous frequency distribution of the latter RIL for flowering time. In addition, the effects of qFLT2 and qFLT3 are likely to be non-additive and synonymous. The epistatic interactions among qFLT1 3 are in agreement with previous studies that documented those among the E1, E3, and E4 loci (Saindon et al. 1989a, b, 1990; Cober et al. 1996; Watanabe et al. 2004) and those among the three loci have been first clearly demonstrated in the present study. Since the alleles with the effect of late flowering/maturity are dominant at these loci, the genotype of Hayahikari seems to be E1e3e4, while that of Toyomusume should be e1E3E4. This is consistent with the fact that Kitamishiro, a progenitor of Hayahikari, is of the genotype of E1e3e4 (Saindon et al. 1990).
The association of the T locus with chilling tolerance has been established (Yumoto and Tsuchiya 1991; Morrison et al. 1994; Takahashi and Asanuma 1996; Morrison et al. 1997; Kurosaki et al. 2004; Takahashi et al. 2005). However, it is not clear in the previous studies with NIL how large the introgressed regions were (Takahashi and Asanuma 1996; Kurosaki et al. 2004; Takahashi et al. 2005). In the present study, a QTL for chilling tolerance on total seed weight with the largest effect was detected at a location very close to the T locus. The E1 locus is located in the vicinity of the T locus (Cregan et al. 1999; Yamanaka et al. 2001). Since this maturity locus could also be associated with chilling tolerance as discussed below, we analyzed a population that segregated for the T locus, but not for qFLT1, the putative E1 locus. In the F2 population, a great difference in seed-yielding ability in the chilling environment between the genotypes at the T locus was observed, suggesting that a QTL linked with the T locus alone had a large effect on chilling tolerance at the seed-yielding level. In addition, the QTL seemed to have nearly complete dominance effect like that of the T locus on the pubescence color. These results may support the possibility of direct involvement of the T locus in chilling tolerance as proposed by Toda et al. (2002), who determined the putative nucleotide sequence and biochemical function of the T gene.
The advantage of E1 over e1 with regard to chilling tolerance was reported using NIL with a Japanese soybean background (Kurosaki et al. 2004), which were, more exactly, residual heterozygous lines, designated by Yamanaka et al. (2005). They applied the same period of four weeks of chilling treatment to the NIL with different growth periods from the beginning of flowering, which means that the NIL with E1 was subjected to chilling treatment for a shorter period on the biological scale. Their evaluation method may be useful for practical selection of breeder’s lines, but not for genomic study which needs the estimation of potential ability for chilling tolerance. Takahashi et al. (2005) reported that during the comparison between the Harosoy isolines with E1e3e4 and e1E3E4, the former genotype was more tolerant to 4-week chilling treatment at the flowering time, although the effect of each locus was unclear. This study employed the natural, fluctuating temperatures in the control environments. The chilled plants were also grown outside except for the period of chilling treatment. Since these NIL had relatively similar but significantly different flowering and maturing times, the NIL encountered different temperatures before and after chilling treatment, which might differentially affect the growth of the chilled NIL. Thus, the fixed period of chilling treatment and the employment of the natural environment may confound the effects of maturity loci on chilling tolerance. In the present study, temperatures were completely controlled and chilling treatment was applied during the same developmental stages, allowing us to reduce such secondary effects of different times of flowering/maturity. While the results in our study did not strongly support the effect of the E1 locus, our observations are in consistent with the study by Takahashi et al. (2005). A significant QTL for chilling tolerance was detected at nearly the same location as that for qFLT2, where the allele from Hayahikari, putative e3, had the beneficial effect on chilling tolerance, especially in combination with the allele from Hayahikari at qFLT3, putative e4. The present study provided more reliable evidence for the involvement of the genomic regions near the maturity loci E3 and E4 in chilling tolerance as well as their epistatic interaction. However, it remains to be clarified whether the effects are due to pleiotropy of the maturity loci or to the linkage with other loci, since our approach does not allow us to determine it and the NIL used in the previous study (Takahashi et al. 2005) were not originated from mutants and could contain more than 10 cM of introgressed regions around each loci (Molnar et al. 2003).
A minor but significant QTL associated with chilling tolerance, qCTTSW3, was found on LG H. This effect was confirmed in an independent F2 population. In this region, no QTL associated with flowering time/maturity was detected in our study or in literature. These facts indicate that this QTL was recognized for the first time. Although the effect was not large, it may be necessary to pyramid QTL with small effects since there seem not to be many major QTL associated with chilling tolerance. In addition, it is possible that the QTL plays an important role in other low-temperature conditions. It may be important to evaluate this locus in long-time field test.
We performed QTL analysis not only for total seed weight per plant but also its components, seed number per plant and single seed weight. The major QTL for chilling tolerance in total seed weight, qCTTSW1, was associated with QTL for chilling tolerance in seed number and single seed weight, suggesting the effect of this QTL on the growth of whole reproductive organs or plant at low-temperatures. In contrast, the other two QTL for chilling tolerance in total seed weight were only associated with seed number. They may affect the process of fertilization at low temperatures, which is a critical factor for the reduction in seed yield in chilling environments (Saito et al. 1970).
An isozyme polymorphism was found among soybean cultivars and lines developed in Hokkaido, and many chilling tolerant genotypes lacked an isozyme APX1 (Funatsuki et al. 2003). Therefore we focused our attention on this polymorphism and mapped the locus for APX1 on LG B1 (data not shown). However, no significant QTL was mapped on this linkage group in the present study.
Chilling tolerance index is based on the ratio of the seed-yielding ability in a chilling environment to that in a control environment and represents the degree how stable the seed-yielding ability is at low temperatures, which has been successfully used in soybean breeding for chilling tolerance (Sanbuichi 1979). This index is especially useful for comparison of various genetic resources that have different seed-yielding levels even at control temperatures. In the RIL population used in the present study, there was a significant variation in seed-yielding ability even in the control environments. Therefore we monitored CTI throughout the experiments, and we paid our best attention to provide an identical environment apart from temperature for each of the paired pots from a RIL as far as possible. Furthermore we logarithmically transformed CTI for data analysis for normalization since the use of the raw CTI data resulted in a severely skewed frequency distribution. We consider that this transformation is rather appropriate since the effects of QTL for chilling tolerance may be non-additive but multiplicative. Similar indices and transformation have been used in other genetic studies on tolerance to abiotic stresses (e.g., Ronnberg-Wastljung et al. 2005). However, there is a possibility that the experimental errors of sample data for RIL with very weak chilling tolerance such as the ones yielding few seeds in chilling environments are biased upwards. In addition, CTI should comprise a relatively large experimental error since this index is calculated from two variables. For identification of QTL with higher sensitivity and precision, it requires to create populations that do not largely segregate in seed-yielding ability in the control environment and to do analysis only with the trait values in chilling environment.
We used a relatively small population size of 104 RIL, which may lead to an upwards bias in the estimates of QTL effects (Beavis et al. 1994). Therefore it will be desired to confirm the effects of the qCTTSW2 in independent experiments as demonstrated for qCTTSW1 and qCTTSW3. More preferably, the employment of a larger population size would lead to the accurate estimation of the QTL effects and to the detection of more QTL.
To the best of our knowledge, this is the first report on QTL analysis of soybean chilling tolerance. We identified several QTL with genome-wide significance, suggesting that our approach is useful to elucidate the genetic mechanism underlying soybean chilling tolerance. The use of different populations and/or evaluation methods would allow us to detect novel QTL in further studies. Moreover, the SSR markers flanking QTL associated with chilling tolerance may be used in marker-assisted selection in breeding programs.
References
Abe J, Xu DH, Miyano A, Komatsu K, Kanazawa A, Shimamoto Y (2003) Photoperiod-insensitive Japanese soybean landraces differ at two maturity loci. Crop Sci 43:1300–1304
Andaya VC, Mackill DJ (2003) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica × indica cross. Theor Appl Genet 106:1084–1090
Basten CJ, Weir BS, Zeng Z-B (2001) QTL Cartographer version 1.15. Department of Statistics, North Carolina State University, Raleigh
Beavis WD, Smith OS, Grant D, Fincher R (1994) Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny from maize. Crop Sci 34:882–896
Bernard RL (1971) Two major genes for time of flowering in soybeans. Crop Sci 11:242–244
Buzzell RI (1971) Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can J Cytol 13:703–707
Buzzell RI, Voldeng HD (1980) Inheritance of insensitivity to long daylength. Soybean Genet Newsl 7:26–29
Churchill G, Doerge R (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cober ER, Tanner JW, Voldeng HD (1996) Genetic control of photoperiod response in early-maturing, near-isogenic soybean lines. Crop Sci 36:601–605
Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, Van Toai TT, Lohnaes DG, Chung T, Specht JE (1999) An integrated genetic linkage map of the soybean genome. Crop Sci 39:1464–1490
Echt CS, May-Marquardt P, Hseih M, Zahorchak R (1996) Characterization of microsatellite markers in eastern white pine. Genome 39:1102–1108
Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19:1349
Falconer DS (1976) Introduction to quantitative genetics. Longman Group, London, pp 365
Funatsuki H, Kurosaki H, Murakami T, Matsuba S, Kawaguchi K, Yumoto S, Sato Y (2003) Deficiency of a cytosolic ascorbate peroxidase associated with chilling tolerance in soybean. Theor Appl Genet 106:494–502
Funatsuki H, Matsuba S, Kawaguchi K, Murakami T, Sato Y (2004) Methods for evaluation of soybean chilling tolerance at the reproductive stage under artificial climatic conditions. Plant Breed 123:558–563
Harada K, Xia ZJ (2004) Soybean genomics: efforts to reveal the complex genome. Breed Sci 54:215–224
Kurosaki H, Yumoto S (2003) Effects of low temperature and shading during flowering on the yield components in soybeans. Plant Prod Sci 6:17–23
Kurosaki H, Yumoto S, Matsukawa I (2004) Correlation of cold-weather tolerance with pubescence color and flowering time in yellow hilum soybeans in Hokkaido. Breed Sci 54:303–312
Manly KF, Cudmore Jr RH, Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
Molnar SJ, Rai S, Charette M, Cober ER (2003) Simple sequence repeat (SSR) markers linked to E1, E3, E4, and E7 maturity genes in soybean. Genome 46:1024–1036
Morrison MJ, Voldeng HD, Guillemette RJD (1994) Soybean pubescence color influences seed yield in cool-season climates. Agronomy J 86:796–799
Morrison MJ, Voldeng HD, Guillemette RJD, Cober ER (1997) Yield of cool-season soybean lines differing in pubescence color and density. Agronomy J 89:218–22
Raper CD Jr, Kramer PJ (1987) Stress physiology. In: Wilcox JR (ed) Soybeans: improvement, production, and uses, 2nd edn. American Soc Agronomy, Inc. Crop Sci Soc America, Inc. Soil Sci Soc America Inc. Publishers, Madison, pp 589–642
Ronnberg-Wastljung AC, Glynn C, Weih M (2005) QTL analyses of drought tolerance and growth for a Salix dasyclados × Salix viminalis hybrid in contrasting water regimes. Theor Appl Genet 110:537–549
Saindon G, Beversdorf WD, Voldeng HD (1989a) Adjustment of the soybean phenology using the E4 locus. Crop Sci 29:1361–1365
Saindon G, Voldeng HD, Beversdorf WD, Buzzel RI (1989b) Genetic control of long day length response in soybean. Crop Sci 29:1436–1439
Saindon G, Voldeng HD, Beversdorf WD (1990) Adjusting the phenology of determinate segregants grown at high-latitude. Crop Sci 30:516–521
Saito M, Yamamoto T, Goto K, Hashimoto K (1970) The influence of cool temperature before and after anthesis, on pod-setting and nutrients in soybean plants. Proc Crop Sci Soc Jpn 39:511–519
Saito K, Miura K, Nagano K, Hayano-Saito Y, Araki H, Kato A (2001) Identification of two closely linked quantitative trait loci for cold tolerance on chromosome 4 of rice and their association with anther length. Theor Appl Genet 103:862–868
Saito K, Hayano-Saito Y, Maruyama-Funatsuki W, Sato Y, Kato A (2004) A Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor Appl Genet 109:515–522
Sanbuichi T (1979) Studies on cool weather tolerance in soybean breeding (in Japanese with English summary). Rep Hokkaido Prefect Agric Exp Stn 28:1–57
SAS Institute (1996) SAS/STAT user’s guide, vols 1 and 2, version 6, 4th edn. Cary
Satake T, Nishiyama I, Ito N, Hayase H (1969) Male sterility caused by cooling treatment at the meiotic stage in rice plants. I. Methods of growing rice plants and inducing sterility in the phytotron. Proc Crop Sci Soc Jpn 38:603–609
Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Takahashi R, Asanuma S (1996) Association of T gene with chilling tolerance in soybean. Crop Sci 36:559–562
Takahashi R, Benitez ER, Funatsuki H, Ohnishi S (2005) Soybean maturity and pubescence color genes improve chilling tolerance at high latitude regions. Crop Sci 45:1387–1393
Takeuchi Y, Hayasaka H, Chiba B, Tanaka I, Shimano T, Yamagishi M, Nagano K, Sasaki T, Yano M (2001) Mapping quantitative trait loci controlling cool-temperature tolerance at booting stage in temperate japonica rice. Breed Sci 51:191–197
Toda K, Yang D, Yamanaka N, Watanabe S, Harada K, Takahashi R (2002) A single-base deletion in soybean flavonoid 3′-hydroxylase gene is associated with gray pubescence color. Plant Mol Biol 50:187–196
Wang D, Shi J, Carlson SR, Cregan PB, Ward RW, Diers BW (2003) A low-cost, high-throughput polyacrylamide gel electrophoresis system for genotyping with microsatellite DNA markers. Crop Sci 43:1828–1832
Watanabe S, Tajuddin T, Yamanaka N, Hayashi M, Harada K (2004) Analysis of QTLs for reproductive development and seed quality traits in soybean using recombinant inbred lines. Breed Sci 54:399–408
Yamanaka N, Ninomiya S, Hoshi M, Tsubokura Y, Yano M, Nagamura Y, Sasaki T, Harada K (2001) An informative linkage map of soybean reveals QTLs for flowering time, leaflet morphology and regions of segregation distortion. DNA Res 8:61–72
Yamanaka N, Watanabe S, Toda K, Hayashi M, Fuchigami H, Takahashi R, Harada K (2005) Fine mapping of the FT1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor Appl Genet 110:634–639
Yumoto S, Tsuchiya T (1991) Response to low temperature and its genotypic variations in soybean cultivars (in Japanese). Rep Hokkaido Branch Jpn Soc Breed Crop Sci Soc Jpn 31:60
Acknowledgements
The authors wish to thank Drs. S. Shirai, S. Ohnishi and the staff of the Soybean Breeding Laboratory, Tokachi Agric. Exp. Stn. and Dr. H. Kurosaki, Kitami Agric. Exp. Stn. for their valuable suggestions and helpful discussion. The technical assistance of R. Narita, S. Sudo, K. Wada and R. Sugisawa is gratefully acknowledged. We are also grateful to Dr. H. Yamauchi (NARCH) for his encouragement. This work was supported by the Rice Genome Research Program (Section: Development of DNA Marker-aided Selection Technology for Plants and Animals) of the Ministry of Agriculture, Forestry and Fishery of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Bernardo
Rights and permissions
About this article
Cite this article
Funatsuki, H., Kawaguchi, K., Matsuba, S. et al. Mapping of QTL associated with chilling tolerance during reproductive growth in soybean. Theor Appl Genet 111, 851–861 (2005). https://doi.org/10.1007/s00122-005-0007-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0007-2