Abstract
A significant level of root elongation was induced in rice (Oryza sativa) grown under phosphorus-deficient conditions. The root elongation clearly varied among a total of 62 varieties screened under two different phosphorus levels. Two contrasting varieties, ‘Gimbozu’, with a low elongating response and ‘Kasalath’, with a high elongating response, were chosen and crossed to produce a hybrid population for QTL analyses. QTLs for the phosphorus deficiency-induced root elongation were detected by two linkage maps, i.e., one with 82 F3 families constructed by 97 simple sequence repeat (SSR) and sequence-tag site markers and another with 97 F8 lines by 790 amplified fragment length polymorphism and SSR markers. A single QTL for the elongation response was detected on chromosome 6, with a LOD score of 4.5 in both maps and explained about 20% of total phenotypic variance. In addition, this QTL itself, or a region tightly linked with it, partly explained an ability to reduce accumulation of excess iron in the shoots. The identified QTL will be useful to improve rice varieties against a complex nutritional disorder caused by phosphorus deficiency and iron toxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phosphorus is essential for normal cell growth and cell division in living organisms, being a component of energy pathways and nucleic acids. Consequently, plants have developed various adaptation mechanisms for phosphorus uptake and transport to ensure their growth under phosphorus-deficient conditions, as reviewed by Rausch and Bucher (2002).
Phosphorus deficiency is a characteristic feature of problem soils under which crop yield is limited. Even when phosphorus is present, its deficiency may also be caused by low availability of soluble phosphorus under strong acidity or high pH. On the other hand, soil phosphorus becomes available under flooded conditions. In this context, rice (Oryza sativa) is potentially adapted to withstand phosphorus deficiency, as it is inherently adapted to flooded conditions. At the same time, rice is a staple food throughout the world. Among the poorer populations of tropical and subtropical countries, it supplies more than two thirds of the total caloric intake (Maclean et al. 2002). Hence, rice will become a key crop to ensure sustainable agriculture in phosphorus-deficient soils.
Among responses by plants to adverse conditions, changes in root architecture are considered adaptations that enhance phosphorus uptake (Lópes-Bucio et al. 2003; Lynch 1995). Interestingly, root elongation is observed in phosphorus-deficient soils in various plant species, including rice (He et al. 2003; Kirk and Du 1997). The elongated rice root under such conditions has higher porosity and oxygen-release ability (Kirk and Du 1997). This trait will therefore be involved in tolerance to multiple nutritional disorders caused by both phosphorus deficiency and iron toxicity (Ottow et al. 1983). In previous studies on the tolerance of phosphorus deficiency in rice (Ni et al. 1998; Wissuwa et al. 1998), some QTLs were identified for phosphorus uptake and shoot biomass reduction. The objective of this study was to analyze varietal differences and QTLs for the root elongation under phosphorus deficiency.
Materials and methods
Screening of root elongation induced by phosphorus deficiency
Screening of rice seedlings was conducted using Yoshida’s nutritional solution (Yoshida et al. 1976), with minor modifications to the phosphorus concentration and the addition of 50 mg/l of silicate. Pre-germinated seeds were placed into a polystyrene floating plate on a plastic container containing 80 l nutritional solution. A strip-plot design (Gomez and Gomez 1984) was employed with 62 varieties (see Electronic Supplementary Material 1), since it was not realistic to assign different levels of nutritional solutions for several tens of varieties at random as required in a randomized complete block design. In a strip-plot design, which is also called a split-block design (see Electronic Supplementary Material 2), the vertical- and the horizontal-strip plots are perpendicular, and the precision to detect the interaction effect is improved at the sacrifice of the degrees of precision of main effects. Each variety was assigned as horizontal treatments to a one-row plot consisting of seven plants as a bulk, and two phosphorus levels (0.5 and 10 mg/l P) were assigned as vertical treatments, in duplicate. The nutritional solutions were changed weekly, and the pH of the solution was adjusted daily to 5.0 with 1 N NaOH/HCl. After a 4-week growth, the root length of each plant was measured.
Statistical procedure for assessing varietal differences
ANOVA for a strip-plot design was conducted according to Gomez and Gomez (1984). To examine differential responses of varieties to the phosphorus (P) levels, the sum of the squares (SS) of the interaction between variety (V) and phosphorus level (V×P SS) was partitioned into several orthogonal components, according to Gomez and Gomez (1984). Based on a ratio of the root length in the phosphorus-deficient conditions to that in the control (C) (P/C ratio), the varieties tested were classified into the following three groups: (1) >1.36 (elongating, G1); (2) 1.16–1.35 (moderately elongating, G2); and (3) less than 1.16 (non-elongating, G3) (Fig. 1; Electronic Supplementary Material 1). This classification showed the highest partitioning ratio of the interaction (V×P SS).
Varietal differences of root length under phosphorus deficiency (P deficiency) and control. Rice (Oryza sativa) varieties ‘Gimbozu’ and ‘Kasalath’ indicate their positions, respectively. Grouping of varieties were conducted as outlined in “Materials and methods”: G1 group of elongating ratio >1.36, G2 group of moderately elongating ratio 1.16–1.35, G3, group of non-elongating ratio <1.16. Variety names and values are listed in Electronic Supplementary Material 1
Dose–response curves of parental lines
From the varieties tested, a japonica, ‘Gimbozu’, and an indica, ‘Kasalath’, were chosen, respectively, from the non-elongating group G3 and from the elongating group G1. The two varieties, each with a plot containing 16 plants, were tested at four different phosphorus concentrations (0.5, 1.36, 3.68, and 10 mg/l) in Yoshida’s nutritional solution, with each phosphorus level replicated four times. The root length was measured after 4 weeks’ growth to construct dose–response curves for both varieties. These two varieties were crossed to raise a hybrid population for QTL analyses.
QTL analysis for root elongation ratio in phosphorus deficiency using F3 families
Eighty-two F3 families from the cross between ‘Gimbozu’ and ‘Kasalath’ were screened with Yoshida’s nutritional solution (Yoshida et al. 1976) and genotyped with 81 simple sequence repeat (SSR) markers (Chen et al. 1997) and 16 sequence-tag site (STS) markers (Robeniol et al. 1996). Four plants per family were assigned to a strip-plot design (Gomez and Gomez 1984), replicated twice with the F3 families as horizontal treatments and two phosphorus levels (0.5 mg/l and 10 mg/l) as vertical treatments. The root length was measured after 4 weeks’ growth, and the P/C ratio was used for QTL analysis. For the QTL analysis, a mean phenotypic value per family was used as “F2 intercross lines,” according to the method of Hayashi and Ukai (1999).
QTL analysis under multiple nutritional disorders, using F8 lines
To provide a model of multiple nutritional disorders, a population of 97 F8 lines, which were derived by single-seed descent from the cross between ‘Gimbozu’ and ‘Kasalath’, were subjected to the following nutrition treatments. Four plants per line were assigned to a strip-plot design (Gomez and Gomez 1984) in duplicate, with the F8 lines as horizontal treatments and three different nutritional conditions as vertical treatments, i.e., (1) phosphorus deficient, (2) potassium deficient and (3) normal. The normal condition was provided by Yoshida’s nutritional solution (Yoshida et al. 1976), which included 10 mg/l phosphorus and 40 mg/l potassium. To simulate phosphorus and potassium-deficient conditions, solutions containing 1/20 of these levels, namely, 0.5 mg/l phosphorus and 2 mg/l potassium, respectively, were used.
The root lengths of 2-week-old seedlings in the three treatments were measured and then exposed to 600 mg/l of ferrous iron as FeSO4 and 5 mg/l of Fe-EDTA for 48 h. After the exposure to excess iron, shoot samples were dried at 80°C for 48 h, and the dry weights of the shoots were measured. Shoot samples were digested with mixed acids and filtered with Whatman filter No. 42 (Whatman International, Maidstone, UK). The concentrations of phosphorus, potassium, magnesium, and iron in the shoot samples were measured according to Yoshida et al. (1976). The phosphorus content was determined colorimetrically by a molybdate–vanadate method, and the others were determined by atomic absorption spectroscopy.
The P/C ratio was calculated using the measurements described above. The ratio of root length under potassium (K) deficient to that under normal (K/C) was similarly calculated. The ratio of iron (Fe) content in the shoots under phosphorus deficiency to that under the control phosphorus level [Fe(P/C)] was calculated to estimate effects of the root system affected by the phosphorus levels upon absorption of excess iron.
The F8 population was genotyped with 727 amplified fragment length polymorphism [(AFLP) Vos et al. 1995] markers and 63 SSR markers, using a high efficiency genome scanning system (Kawasaki et al. 2003). The nomenclature of AFLP marker bands, e.g., ACGCCA690A, indicated triplet selective nucleotides of EcoRI+3 and MseI+3 primers, fragment size, and origin of the fragment, namely, “A” from ‘Gimbozu’ and “B” from ‘Kasalath’. The suffix “H” indicated a co-dominant marker.
Construction of linkage maps and QTL scanning
Linkage maps were constructed using the program MAPMAKER (Lander et al. 1987). For the F3 generation, the total length of linkage map was 1,400 cM, covering 80% of the total genome. For the F8 generation, a saturated linkage map of 1,700 cM was constructed, with a mean interval of 2.3±3.1 (SD) cM (see Electronic Supplementary Materials 2 for the map details). SSR markers with known locations were used as landmarks, according to the rice genome maps which are available via the Gramene Web site (http://www.gramene.org/).
To detect QTLs, composite interval mapping (Zeng 1994) was applied using QTL Cartographer, version 1.17 (Basten et al. 1994). One thousand repeats of a permutation test (Churchill and Doerge 1994) were performed to estimate a threshold LOD score for each trait at a significance level of α=0.05. Then, composite interval mapping for multiple traits (Jiang and Zeng 1995) was conducted to jointly analyze associated traits with the P/C ratio, using QTL Cartographer (Basten et al. 1994) when a trait had a QTL around the LOD peak of the P/C ratio. The normality of the measured traits was assessed using the D’Agostino–Pearson K2-test (Zar 1996).
All the nutritional screenings were conducted in a daylight greenhouse at the Experimental Farm of Kyoto University, Kyoto, Japan. Mean temperatures during the experiments for varieties, parents, F3, and F8 progenies were 27.4, 24.7, 28.0, and 29.5°C, respectively. Marginal effects in the culture box on root length were equally given to all the entries not to lead to any biases.
Results
Varietal difference in phosphorus deficiency-induced root elongation
The P/Cs obtained under normal and phosphorus-deficient conditions varied according to variety (Fig. 1). The results of ANOVA showed highly significant differences among varieties and a highly significant interaction between variety and phosphorus levels (V×P, Table 1). The sum of squares of the interaction between variety and phosphorus levels (V×P SS) was partitioned to five orthogonal components with a significant difference (Table 1). The screened varieties were grouped into G1, G2, and G3 based on varietal differences in the P/C ratio, and this grouping brought the highest partitioning ratio of the interaction (V×P SS, Fig. 1). The interaction between the G1 group and the phosphorus levels, namely (Pdef vs Con) × G1 in the Table 1, had the highest partitioning of the total interaction, explaining 52.4% of the total interaction (V×P SS), although only nine varieties (15%) were classified into the G1 group. Of all the varieties screened, which included 17 Philippine, 12 Japanese, 8 Indian, and 5 Bangladeshi varieties, all the Japanese varieties tested here were grouped into the non-elongating group (G3), while the elongating group (G1) consisted of Philippine, Indian and Bangladeshi varieties (see Electronic Supplementary Material 1).
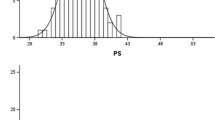
In particular, a contrasting response in the phosphorus level-dependent elongation was confirmed between japonica variety ‘Gimbozu’ from the non-elongating group (G3) and indica variety ‘Kasalath’ from the elongating group (G1) (Fig. 1), so that they were crossed to produce a hybrid population. The differential response of root length was dependent on phosphorus level. A threshold between 0.5 mg/l and 1.36 mg/l phosphorus was needed to stimulate the root elongation (Fig. 2).
Genetic analysis of the root elongation induced by phosphorus deficiency
In both F3 families and F8 lines, root lengths under the phosphorus-deficient and control conditions (Fig. 3a, c) and the P/C ratio distributed without significant deviation from normal (Fig. 3b, d). These traits were therefore analyzed in QTL without transformation.
Histograms, a for root length (cm) in F3 families under control (Con) and phosphorus deficiency (Pdef); b for relative root length [ratio of the root length in the phosphorus (P)-deficient conditions to that in the control (C) (P/C)] of F3 families; c for root length (cm) of F8 lines under control (Con), phosphorus deficiency (Pdef) and potassium deficiency (Kdef); d relative root length [P/C and the ratio of the root length in the potassium-deficient conditions to that in the control (C) (K/C) of F8 lines. Gim, Kas, and F 1 indicate the average values of ‘Gimbozu’, ‘Kasalath’, and F1 (‘Gimbozu’/‘Kasalath’), respectively. Data of F3 (a, b) were measured from 4-week-old seedlings and data of F8 (c, d) were measured from 2-week-old seedlings. By the D’Agostino-Pearson K2-test, the distributions of the eight traits were not deviated from normality with probabilities of larger than 0.11
Correlation coefficients (r) between the F3 families and F8 lines were calculated in terms of the root lengths under the control, the root length under phosphorus-deficient conditions, and the P/C ratio; the obtained coefficients (r) were 0.523, 0.362, and 0.271, respectively. They were confirmed to be significant at the 1% level. The P/C ratio in the F8 lines distributed with a mean of more than one (Fig. 3d), but the K/C ratio, which was calculated as a ratio of the root length under potassium-deficient conditions to that under control, distributed with a mean of less than 1 (Fig. 3d). A paired t-test between the root length under phosphorus deficiency and that under the control confirmed a significant increase in root length under the deficiency at a probability level of 7.3×10−13. Similarly, by a paired t-test between the root length under potassium deficiency and the control, a significant decrease in root length was confirmed under the deficiency at a probability level of 1.6×10−15.
Composite interval mapping of the P/C ratio showed a single LOD peak of 4.5 near an SSR marker, RM30, on chromosome 6 in the F3 map (Table 2). This explained 19.9% of the phenotypic variance, and there was no other LOD score higher than the threshold estimated by the permutation test. The results of QTL analysis in the F3 map were consistent with that in the F8 map, where a single QTL of the P/C ratio was also confirmed on the corresponding region of chromosome 6 (Table 2; Fig. 4). This explained 20.3% of the total phenotypic variance. The additive effects of those QTLs were contributed by ‘Kasalath’ (Table 2, the last column).
QTL scan on chromosome 6 for root elongation induced by phosphorus deficiency (P/C ratio) and associated traits such as dry weight of shoots under phosphorus deficiency [DWS(Pdef)], relative iron content in shoots [Fe(P/C)], and relative phosphorus content in shoots [P(P/C)]. Maximum threshold LOD score of 3.57 among the four traits (Table 2) is indicated as a dotted line
Genetic analysis of associated traits measured
The dry weights of the shoots (DWS) and their contents of phosphorus, potassium, magnesium, and iron after a 4-week growth under phosphorus-deficient conditions, were studied using F8 lines. All the traits showed a normal distribution by a normality test (data not shown) except for the iron content, for which the reciprocal transformation of 1/x was used according to Zar (1996). The numbers of QTLs for these associated traits varied from one to four, and their estimated positions varied throughout the linkage map (Table 2). Of these QTLs, those for DWS under phosphorus deficiency [DWS(Pdef)], the relative contents of phosphorus [P(P/C)], and the reciprocally transformed values of the relative contents of iron [Fe(P/C)] showed LOD peaks around that of the P/C ratio on chromosome 6 (Fig. 4).
These four associated traits and the P/C ratio were then re-examined by multiple-trait analysis, using QTL Cartographer. The joint LOD scores estimated at the LOD peak of the P/C ratio were shown (Fig. 5). Among the combined effects of two factors, the highest was jointly achieved with the P/C and Fe(P/C) ratios. The iron content of the shoots was highly dependent on the level of phosphorus and was significantly lower under the phosphorus deficiency than that under the control (Fig. 6).
Joint QTL on an LOD peak of root elongation induced by phosphorus deficiency (P/C ratio) with dry shoot weight under phosphorus deficiency [+DWS(Pdef)], relative iron content in shoots [+Fe(P/C)], and relative phosphorus content in shoots [+P(P/C)]. Joint LOD score of the four traits is indicated at the far right (+All)
Histograms of iron content (mg/g of shoot) for a ion contents under control, b ion contents under potassium deficiency, and c ion contents under phosphorus deficiency. The arrows indicate the average values of parents and F1. Paired t-tests between ion contents under the control and under potassium deficiency and between ion contents under the control and under phosphorus deficiency showed significant difference with a probability of 0.01 and 8.4×10−43, respectively
Discussion
Root elongation induced in native cultivars by phosphorus deficiency
There have been a number of reports on the root length under phosphorus-deficient conditions. Root elongation by phosphorus deficiency was observed in Arabidopsis (Ma et al. 2003), barley (Steingrobe et al. 2001), horsegram (Anuradha and Narayanan 1991), and rice (Kirk and Du 1997; He et al. 2003), whereas unchanged root length was observed in Phaseolus vulgaris (Borch et al. 1999). A level of varietal difference was observed in maize (Gaume et al. 2001). The screening of rice varieties in the present study revealed an apparent varietal difference in the root elongation, induced by phosphorus deficiency. Since the ANOVA demonstrated a highly significant interaction between variety and phosphorus levels (Fig. 1), partitioning of the interaction sum square (V×P SS) was useful in understanding the varietal response to phosphorus levels. Varieties in the group G1 were found to elongate their roots specifically under conditions of phosphorus deficiency.
There was a clear difference in this response, according to the origin of the variety. All the Japanese varieties tested here were classified into the non-elongating or insensitive group (G3). Japanese varieties may have lost their ability to respond to phosphorus deficiency, since modern varieties have been selected for cultivation in phosphorus-sufficient fields.
The differential response of root length between japonica variety ‘Gimbozu’ and indica variety ‘Kasalath’ was dependent on phosphorus level with a threshold between 0.5 mg/l and 1.36 mg/l phosphorus (Fig. 2). There is at present no information concerning a possible receptor protein that is responsive to low phosphorus levels. Cloning of related genes will therefore be important to analyze and utilize this adaptive mechanism, as any increase of root surface would enhance phosphorus uptake (for a review, see Raghothama 1999). Since the whole genomic sequence has been analyzed in rice, the newly identified variation for response to phosphorus deficiency may provide a basis for isolating related genes using genomic approaches (Feng et al. 2002; Sasaki et al. 2002).
QTL of root elongation induced by phosphorus deficiency
Generally, deficiency of an essential element causes a reduction of total plant biomass. The root length of rice was reduced under potassium deficiency, but increased under phosphorus deficiency. The P/C ratio showed a normal segregation in the progeny population resulting from a cross between two contrasting varieties. The correlation coefficient, r, significant at 1% for the P/C ratio between F3 and F8 lines, also confirmed that the P/C ratio is a heritable trait. The reason that r was not so high, with the r2 of less than 50%, was in part due to the difference in the duration of screening for F8 and F3. The duration was shorter by 2 weeks for F8 than for F3. It was also partly due to the genetic fixation advanced to the F8 generation. The heritable nature of the P/C ratio was also confirmed by the consistency in the number and position of detected QTLs in the F3 and F8 maps. The common QTL had an LOD peak of around 4.5 on long arm of chromosome 6 and explained about one fifth of the total phenotypic variance. ‘Kasalath’ alleles increased root length under phosphorus deficiency. There were previous reports of QTLs for tolerance for biomass reduction under phosphorus deficiency and QTLs for the efficiency of phosphorus uptake on chromosome 6 (Ni et al. 1998; Wissuwa et al. 1998). The latter was mapped on the short arm, i.e., on the opposite side of our QTL. The former for relative tillering ability, relative shoot dry weight, and relative root dry weight was mapped on the region approximately 120 cM and 134 cM far from landmarks on short arm, i.e., RG123 and RG64, respectively. So it was not possible to determine whether Ni’s QTLs are close to our QTL or not. However, both were on long arm of chromosome 6. We have tentatively designated this locus REP (root elongation under phosphorus deficiency).
Analysis of associated traits with the locus
There were other traits that showed LOD peaks around REP (126.4 cM on chromosome 6). These were DWS(Pdef), P(P/C), and Fe(P/C). Of these, LOD peak of Fe(P/C) was mapped on a closer position to REP than the others (Fig. 4), and these two showed the highest joint LOD score among the three (Fig. 5). Considering the reciprocal transformation (1/x) applied to the QTL analysis of Fe(P/C), the ‘Kasalath’ allele at the LOD peak on chromosome 6, where the REP was located, may have reduced the accumulation of excess iron in the shoots. The simultaneous positive effect may be explained by a tight linkage of these loci or by a pleiotropic effect of REP.
Lower accumulation of iron in shoots induced by phosphorus deficiency
The iron content of shoots is indicative of root metabolism for excluding excessively supplied iron; therefore, this was chosen for detailed examination out of the associated traits. For plants grown under phosphorus deficiency, the iron content of shoots was significantly lower than the control. The mean reduction rate was seven times greater than the rate in potassium deficiency and in normal condition (data not shown). Since the plant was exposed to excess iron (600 mg/l Fe) for only 48 h after the screening for phosphorus deficiency, the iron content of the shoots may have been affected by a metabolism changed in response to the preceding nutritional conditions. The higher iron-excluding power, which was indirectly observed as reductions of iron content in the phosphorus-deficient plants, was analyzed as a QTL of Fe(P/C). This result indicates that the iron-excluding power is partly contributed by REP itself or tightly linked with the REP locus. In addition, a large contribution was provided by the highest LOD peak on chromosome 8 (Table 2), which suggests another adaptive mechanism to phosphorus deficiency and iron toxicity. Since Kirk and Du (1997) reported a twofold increase in root-released oxygen under 0.5-mg/l phosphorus sand culture, the trait Fe(P/C) might reflect indirectly these metabolic changes induced by phosphorus deficiency. These results again indicate the importance of REP locus in the breeding rice for tolerance to phosphorus deficiency and iron toxicity, both of which are widespread in acid sulfate soils or in flooded Oxisols.
References
Anuradha M, Narayanan A (1991) Promotion of root elongation by phosphorus deficiency. Plant Soil 136:273–275
Basten CJ, Weir BS, Zeng ZB (1994) Zmap-a QTL cartographer. In: Smith C, Gavora JS, Benkel B, Chesnais J, Fairfull W, Gibson JP, Kennedy BW, Burnside EB (eds) Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: computing strategies and software, vol 22. The Organizing Committee, 5th World Congress on Genetics Applied to Livestock Production, Ontario, pp 65–66
Borch K, Bouma TJ, Lynch JP, Brown KM (1999) Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ 22:425–431
Chen X, Temnykh S, Xu Y, Cho YG, McCouch SR (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor Appl Genet 95:553–567
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Feng Q, et al (2002) Sequence and analysis of the rice chromosome 4. Nature 420:316–320
Gaume A, Mächler F, De León C, Narro L, Frossard E (2001) Low-P tolerance by maize (Zea mays L.) genotypes: significance of root growth, and organic acids and acid phosphatase root exudation. Plant Soil 228:253–264
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research, 2nd edn. Wiley, New York
Hayashi T, Ukai Y (1999) Method of QTL mapping in an F2 population using phenotypic means of F3 lines. Breed Sci 49:105–114
He Y, Lian H, Yan X (2003) Localized supply of phosphorus induces root morphological and architectural changes of rice in split and stratified soil cultures. Plant Soil 248:247–256
Jiang C, Zeng ZB (1995) Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140:1111–1127
Kawasaki S, Murakami Y, Imaizumi-Anraku H, Shimizu A, Mikami I (2003) Construction of high-density map, genome library, and saturation mapping of nodulation genes. In: Nagata T, Tabata S (eds) Biotechnology in agriculture and forestry, vol 52. Springer, Berlin Heidelberg New York, pp 183–202
Kirk GJD, Du LV (1997) Changes in rice root architecture, porosity, and oxygen and proton release under phosphorus deficiency. New Phytol 135:191–200
Lander ES, Green P, Abrahamson J, Barow A, Daly MJ, Lincoln SE, Newbug L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lópes-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6:280–287
Lynch J (1995) Root architecture and plant productivity. Plant Physiol 109:7–13
Ma Z, Baskin TI, Brown KM, Lynch JP (2003) Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol 131:1381–1390
Maclean JL, Dawe DC, Hardy B, Hettel GP (2002) Rice almanac, 3rd edn. IRRI, Manila
Ni JJ, Wu P, Senadhira D, Huang N (1998) Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.) Theor Appl Genet 97:1361–1369
Ottow JCG, Benckiser G, Watanabe I (1983) Is iron toxicity of rice the result of a multiple nutritional stress? Plant Res Dev 17:96–109
Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50:665–693
Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216:23–37
Robeniol JA, Constantino SV, Resurreccion AP, Villareal CP, Ghareyazie B, Lu BR, Katiyar SK, Menguito CA, Angeles ER, Fu HY, Reddy S, Park W, McCouch SR, Khush GS, Bennett J (1996) Sequence-tagged sites and low-cost DNA markers for rice. In: Rice genetics 3. IRRI, Manila, pp 293–306
Sasaki T et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420:312–316
Steingrobe B, Schmid H, Claassen N (2001) Root production and root mortality of winter barley and its implication with regard to phosphate acquisition. Plant Soil 237:239–248
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wissuwa M, Yano M, Ae N (1998) Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet 97:777–783
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice, 3rd edn. IRRI, Manila
Zar JH (1996) Biostatistical analysis, 4th edn. Prentice Hall, Upper Saddle River
Zeng Z (1994) Precision mapping of quantitative trait loci. Genetics 196:1457–1468
Acknowledgements
This research was funded by the Japan-IRRI Shuttle Research Project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Q. Zhang
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Shimizu, A., Yanagihara, S., Kawasaki, S. et al. Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Theor Appl Genet 109, 1361–1368 (2004). https://doi.org/10.1007/s00122-004-1751-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1751-4