Abstract
Two virus resistance loci on linkage groups II and VI have provided the only sources of natural resistance against Pea seed-borne mosaic virus (PSbMV, Potyviridae) in the important crop plant Pisum sativum L. A combination of parallel approaches was used to collate linked markers, particularly for sbm-1 resistance on linkage group VI. We have identified sequences derived from the genes for the eukaryotic translation initiation factors eIF4E and eIF(iso)4E as being very tightly linked to the resistance gene clusters on linkage groups VI and II, respectively. In particular, no recombinants between sbm-1 and eIF4E were found amongst 500 individuals of an F2 cross between the BC4 resistant line (JI1405) and its recurrent susceptible parent ‘Scout’. In a different mapping population, the gene eIF(iso)4E was also shown to be linked to sbm-2 on linkage group II. A parallel cDNA-AFLP comparison of pairs of resistant and susceptible lines also identified an expressed tag marker just 0.7 cM from sbm-1. eIF4E and eIF(iso)4E have been associated with resistance to related viruses in other hosts. This correlation strengthens the use of our markers as valuable tools to assist in breeding multiple virus resistances into peas, and identifies potential targets for resistance gene identification in pea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pea seed borne mosaic virus (PSbMV), in the genus Potyvirus in the family Potyviridae, is a pathogen of peas, broad beans, lentils and chickpeas and causes major crop losses. The disease is currently managed largely through the elimination of transmitting insects from the crop and by identifying and discarding batches of infected seed (Khetarpal and Maury 1987). PSbMV is represented by a large number of isolates and strains. With respect to those that infect peas and lentils, these have been grouped on the basis of their reaction to pea indicator lines of varying susceptibility into pathotypes P1, P4 and L1 (sometimes referred to as P2).
In 1971, Hagedorn and Gritton identified two Ethiopian lines of pea (PI193586 and PI193835) that exhibited resistance to the common strains of PSbMV (pathotype P1). They also showed that this trait was conferred by a single recessive resistance gene (sbm) (Stevenson and Hagedorn 1971; Hagedorn and Gritton 1973). This locus was found to be linked to wlo and p on linkage group (LG) VI (Gritton and Hagedorn 1975; Hampton and Marx 1981). Later, Provvidenti and Alconero (1988) found that cultivar ‘Bonnevillle’ showed resistance to the lentil (L1) strain of PSbMV and that this was closely linked to mo, a gene conferring resistance to two other potyviruses, Bean yellow mosaic virus (BYMV) and Watermelon mosaic virus (WMV). They also showed that a second gene, linked to sbm on LG VI conferred resistance to PSbMV L1. Gene mo is also linked with k (winged keel). These and later studies (Provvidenti and Alconero 1988; Provvidenti and Muehlbauer 1990; Providenti et al. 1991) have provided a picture of potyvirus resistance in pea based upon two clusters of recessive resistance genes specifying resistance to a diverse range of potyviruses (reviewed in Provvidenti and Hampton 1991). The locus on LG VI gives resistance to PSbMV P1 (sbm-1), PSbMV L1 (sbm-3), PSbMV P4 (sbm-4), Clover yellow vein virus (ClYVV; cyv-2) and White lupin mosaic virus (wlv). The locus on LG II gives independent resistance to PSbMV L1 (sbm-2) and resistance to BYMV and WMV (mo), Bean common mosaic virus (bcm), ClYVV (cyv-1), and Pea mosaic virus (pmv).
Through linkage of these loci with morphological markers, the introgression of the resistances into commercial lines without virus testing was possible (Provvidenti et al. 1991). However, because of the genetic distances involved and the potential existence of a modifier-gene system, caution was expressed as to the reliability of this approach (Hampton 1980). Hence, many commercial varieties of pea remain susceptible to the more common strains (P1 and P4) of PSbMV, presumably reflecting problems in manipulating the LG VI locus in commercial breeding strategies.
In addition to the recessive resistance loci in pea, other plant species exhibit recessive resistance to potyvirus infection. In lentil, the sbv gene confers resistance to PSbMV (Haddad et al 1978). In Capsicum spp., a series of recessive genes (pvr1, pvr2 1, pvr2 2 and pvr3; reviewed in Kyle and Palloix 1997) provide resistance against Potato virus Y (PVY), Tobacco etch virus (TEV) and Pepper mottle virus (PepMoV). Also in the Solanaceae, the va gene in Nicotiana tabacum cv. TN 86 confers resistance to Tobacco vein mottling virus (TVMV) (Deom et al. 1997). In lettuce, the mo1 gene in lettuce confers resistance to Lettuce mosaic virus (LMV) (references in Nicaise et al. 2003). In the case of pvr and mo1, these resistances have been identified as separate allelic series for the eukaryotic translation initiation factor eIF4E (Ruffel et al. 2002; Nicaise et al. 2003).
As part of the larger goal of sbm-1 characterisation from pea, we aimed to identify new linked markers that would assist in the breeding of virus resistance and in gene identification. By taking a candidate gene approach, we showed that eIF4E and eIF(iso)4E were tightly linked to sbm-1 and mo, respectively. Our markers for sbm-1 have been converted into PCR primers that can be conveniently used to assist in the breeding of virus resistance in pea.
Materials and methods
Plant and virus materials
Genetic mapping of Pisum sativum L. (pea) was carried out using two pairs of PSbMV P1-resistant or -susceptible lines. Resistant (sbm-1) JI1405 is a BC4 line with the recurrent susceptible (Sbm-1) parent JI2009. These lines were originally received from Dr. F. Muehlbauer as X78122 (registered as PI595954) and ‘Scout’ (registered as W6 17534), respectively (Muehlbauer. 1983). Resistant (sbm-1) line 835 and its recurrent susceptible (Sbm-1) backcross parent, line 744 (also referred to as RCG1402), were obtained from Dr. R. Cousin, INRA, Versailles. The infection phenotype of these lines was confirmed. Segregation of markers with respect to Sbm-1 and other known markers was assessed using the progeny of an F2 cross between JI1405 and JI2009 and a series of recombinant in-bred lines (RILs) generated from cross between pea lines JI15 and JI399. Markers mapping to LG II were analysed using an RI family (16 lines) generated from a cross between JI281 and JI399. All of the RILs were susceptible to PSbMV P1. Plants were grown in a glasshouse with a 14-h photoperiod and a temperature of 18–25°C.
PSbMV infection of susceptible peas leads to a systemic chlorosis and reduced growth and seed set. This is sometimes associated with virus seed transmission so contaminating subsequent generations (Maule and Wang 1996). To avoid this problem, an alternative approach was developed based upon the visualisation of a colorimetric reporter of infection in detached leaves. Virus infections were carried out using a recombinant PSbMV isolate DPD1 (pathotype P1) expressing β-glucuronidase from within the viral genome (PSbMV-GUS, Borgstrom and Johansen 2001). The inoculum was in the form of a DNA vector containing the PSbMV-GUS cDNA cloned between the CaMV 35S promoter and the nopaline synthase termination signal. The GUS gene is positioned between the P1 and helper-component-protease (HC-Pro) genes of the PSbMV polycistronic genome. The GUS protein is released from the viral polyprotein during the early stages of virus replication by excision by viral encoded protease activities. This recombinant virus is unchanged from the wild type isolate in its response to sbm-1. The plasmid DNA was coated onto gold particles (0.97-μm diameter, Christou et al. 1991) and used to inoculate the second and third leaves of pea seedlings using a hand-held microprojectile bombardment gun, essentially as described by Gal-On et al. (1997). After inoculation, leaves were removed and maintained in moistened petri dishes. Infection was visualised at 3 days post-inoculation (dpi) by incubating detached leaves in the β-glucuronidase substrate, X-Gluc (0.25 mg/ml in assay solution at 37°C overnight) [assay solution: 100 mM NaPO4buffer, pH 7.0, 0.1% Triton X-100, 10 mM EDTA, 1% H2O2(30% solution)].
cDNA-AFLP
cDNA-AFLP analysis was carried out essentially as described in Bachem et al. (1996). Total RNA was extracted from young pea leaves using TRI reagent (Sigma), and mRNA was purified using the Dynabeads mRNA purification kit (Dynal AS, Oslo, Norway). First- and second-strand cDNA synthesis was carried out according to standard protocols (Sambrook and Russell 2001) using Superscript II reverse transcriptase (Invitrogen), RNase H and DNA polymerase I. First-strand cDNA synthesis was primed with a degenerate poly-T primer (GTCGACCTGCAGGCGT19V; V=A, G, C), carrying a PstI site. This provided a PstI site 3′to the poly A of synthesised cDNAs. The template for cDNA-AFLP was prepared using PstI and MseI. The sequences of primers and adapters used for AFLP reactions are as follows (N can be any nucleotide): PstI adapter: top strand 5′ CTCGTAGACTGCGTACATGCA3′, bottom strand 5′TGTACGCAGTCTAC3′; MseI adapter: top strand 5′GACGATGAGTCCTGAG3′, bottom strand 5′TACTCAGGACTCAT3′; PstI pre-amplification primer: 5′GACTGCGTACATGCAG3′; MseI pre-amplification primer: 5′GATGAGTCCTGAGTAA3′; PstI selective amplification primer (PstA-PstQ): 5′GACTGCGTACATGCAGNN3′; MseI selective amplification primer (MseI-MseXIV): 5′GATGAGTCCTGAGTAANN3′.
Amplification products were separated on a 5% polyacrylamide gel run at 100 W until the bromophenol blue reached the bottom of the gel. Gels were dried and exposed to Kodak Biomax film (Sigma). Polymorphic bands were cut out from the dried gel after alignment with the autoradiograph. The DNA fragments were eluted from the excised gel pieces and re-amplified following the same PCR conditions and primer combinations.
One polymorphic cDNA-AFLP band (ZG10) was used as a marker for mapping. The sequence of this cDNA has been submitted to GenBank as accession AY423376.
Isolation of eIF4E and eIF(iso)4E cDNA fragments
To PCR-amplify a DNA product for eIF4E, two pairs of primers were designed from the mRNA sequence for eIF4E from Medicago truncatula (TC71850, http://www.tigr.org). These and other primers used for marker identification are listed in Table 1. Primers eIF4E Med5′and eIF4E Med3′were used to amplify a partial cDNA from Medicago. Primers eIFexpress5′and eIFexpress3′were used to amplify the homologous pea cDNA. Pea-specific primers peaeIF4E5′and eIF4E3pea3′, corresponding to nucleotides (nt) 308–337 and nt 725–747 on the Medicago coding sequence, were used to amplify pea genomic DNA at the eIF4E locus.
To amplify eIF(iso)4E cDNA, two primers (eIF-1 and iso4E23′) were designed from an alignment of several mRNA sequences from Arabidopsis thaliana (accession AF538308, U62044, AY086315, NM_122953, AY054630, Y10547). The amplified cDNA fragments were cloned into pGEM-T Easy vector (Promega, Madison, Wis.) and sequenced in both orientations. The sequence of the pea cDNA has been submitted to GenBank as accession AY423377.
Molecular markers and mapping
An F2 population from a cross between JI1405 and JI2009 was generated. Individual F2 plants were scored for their infection phenotype on excised leaves, and the plants were left to grow to maturity for the collection of F3 seeds. Sixteen RILs each from crosses between JI281 × JI399 and JI 15 × JI399 were used for mapping new markers. Pea genomic DNA was prepared (Ellis 1994) and used either in southern analyses or as templates for genomic PCR assays.
Markers for sbm-1
For the JI15 × JI399 RILs, the ZG10 cDNA was used in an RFLP analysis of segregation. For the analysis of the JI1405 × JI2009 F2 population and to develop a practical marker for marker-assisted selection of resistant lines, PCR primers were designed from the sequence of the cDNA-AFLP fragment, ZG10, and for eIF4E. For ZG10, the design was based on a single nucleotide polymorphism (SNP) found between homologous cDNAs from JI1405 and JI2009. Primers ZG1035′and ZG10SNP identified the dominant allele, whereas ZG1035′and ZG10SNP2 identified the recessive allele. Segregation for the JI1405 × JI2009 F2 population was scored after visualising the PCR products on stained agarose gels. eIF4E cDNA provided a RFLP marker. This was also converted into a co-dominant PCR-based marker using primers 4Egenomic5′and 4Egenomic3′and used to score segregation in the same F2 population.
Bin mapping eIF(iso)4E
The cloned eIF(iso)4E cDNA fragment was used as an RFLP marker following HindIII digestion of pea genomic DNA. The marker was mapped onto 16 RILs from JI281 × JI399. When a genetic map has been created for a mapping population (in this case an RI population), only a limited number of segregants is required to correlate a new marker with a known one showing identical segregation. The number of segregants can be determined empirically from the marker data by determining for a given set of x RILs how many unlinked markers have the same scores.
In general, where m is the number of lines with identical scores, and n is the number of lines with differing scores, the likelihood of the data under linkage (L l) given a linkage value of r is given by:
From Haldane and Waddington (1931), we have r=R/2(1−R), where R=n/(m+n). The likelihood of the data under no linkage (L u) is the value of L l where r=0.5. We can calculate the LOD score as Log10 (L l/L u), so the maximum possible LOD score is where n=0 and r=0.
Where (m+n)=10, the maximum LOD score is just over 3. So, from series of known markers scored for these lines, linkage and location of a new marker can be obtained with as few as ten RILs. If the data have a single mismatch, then for 16 RILs the LOD score is 3.12. Thus, for 16 RILs it is possible to position markers where the map is not recombination limited (i.e. a new marker may lie within an interval between two crossover events in the subpopulation in which there is no mapped marker). This procedure places markers with respect to an existing genetic map, but does not improve the genetic map. In essence, the procedure assigns markers to ‘recombination bins’. These are sets of markers of identical scores and where the bin size is determined by the number and location of the crossover events in the selected lines.
For sbm-1, ZG10, eIF4E and eIF(iso)4E, segregation data were converted to map positions using the programme JoinMap (Stam 1993; Stam and van Ooijen 1995).
Results
Phenotypic analysis using a colourimetric reporter
Using PSbMV-GUS to assess the virus infection phenotype, the amount and distribution of GUS activity after 3 dpi clearly differentiated between susceptible and resistant leaves. The former showed large spreading lesions, whereas on resistant leaves no blue staining was visible (Fig. 1). All of the parental lines for the mapping populations were confirmed for their phenotype following infection with PSbMV P1. Lines JI2009, JI281, JI15, JI399 and 744 were susceptible; lines JI1405 and 835 were resistant.
cDNA-AFLP analysis of expressed genes in resistant and susceptible peas
Because of the large size of the pea genome and the abundance of repetitive DNA (Murray et al. 1978), we decided to concentrate upon expressed genes in the search for polymorphic markers. The strategy utilised PstI and MseI digestions and adapted primers for cDNA amplification. PstI was selected since it is relatively abundant in the small set of known pea gene sequences in the EMBL database. A PstI site was also included in the preamplification cDNA primer. Hence, all cDNAs had the potential to be recognised by the Pst series amplification primers and should have been identified as a discrete band if there was an internal MseI site. A total of 256 amplifications had the potential to give complete genome coverage.
To assist in the identification of resistance gene linked cDNAs, we compared two resistant lines (JI1405 and 835) with two susceptible lines (JI2009 and 744) with each differential pair coming from different breeding lineages. A ‘complete’ genome survey identified only two polymorphic bands (Fig. 2a), detected with primer combinations PstO/MseII and PstO/MseV, which showed consistent segregation with resistance or susceptibility. The cDNAs were excised, reamplified and sequenced. The sequences showed that the two bands were alleles of the same gene. The nucleotide and amino acid sequences showed 88% and 78% identities with a galactosyltransferase gene from chromosome 4 of A. thaliana (accession NP_193838). The cloned fragments were used in a hybridisation analysis of pea genomic DNA. Only a single genomic band was detected after separate digestion with EcoRI, EcoRV and HindIII, indicating that the cDNA called ZG10 corresponded to a single gene in pea (data not shown). Based upon the nucleotide sequences of ZG10 from the two alleles, SNPs were identified and used to design allele-specific PCR primers (Fig. 2b). Segregation of ZG10 in the F2 population identified a map position at 0.7 cM away from sbm-1.
Polymorphic marker ZG10 segregates in resistant and susceptible pea lines. a cDNA AFLP comparison between resistant (JI1405 and 835) and susceptible (JI2009 and 744) lines identified single polymorphic bands with each of two primer combinations, Pst0/MseV and Pst0/MseII. b Conversion of the ZG10 cDNA into a PCR-based co-dominant polymorphic marker. Segregation within the segregating F2 population identified resistant (A), susceptible (B) and heterozygous lines (H)
eIF4E co-localises with sbm-1 on LG VI
Prompted by the genetic and functional association between eIF4E and resistance to potyvirus infection (Wittmann et al. 1997; Duprat et al. 2002; Lellis et al. 2002; Ruffel et al. 2002; Nicaise et al. 2003), we also followed a candidate gene approach to the identification of markers linked to sbm-1. Because Medicago is a close relative of pea, two primers, eIF4E Med5′and eIF4E Med3′, were designed from the Medicago eIF4E mRNA sequence. A 745-bp cDNA fragment was amplified from Medicago; these primers failed to amplify pea cDNA. However, using the fragment as a probe for Southern hybridisation of restriction-digested pea DNA, a hybridising region of pea DNA was identified. Restriction analysis indicated that this was a single-copy gene (Fig. 3a). HindIII digestion also identified a polymorphism between resistant and susceptible lines (Fig. 3a). The Medicago cDNA sequence contains two HindIII sites. To identify the origin of the polymorphic bands in pea, the three fragments from the HindIII digestion of the Medicago cDNA PCR fragment were used separately to probe replicate blots. Only the central HindIII fragment identified the smaller polymorphic bands (data not shown). Another pair of primers, eIFexpress5′and eIFexpress3′, was successful in amplifying a 658 bp eIF4E cDNA from pea line JI2009. The pea cDNA sequence, which showed 84% identity with the Medicago sequence, failed to reveal any HindIII sites. Nevertheless, alignment of the pea and Medicago sequences permitted the identification of pea-specific primers (peaeIF4E5′and eIF4E3pea3′) approximately flanking the central HindIII fragment in Medicago cDNA. When used in the amplification of pea genomic DNA, these primers identified a band approximately 1.8 kbp in size from lines JI2009 and JI1405. The sequence of these fragments identified 5′and 3′co-ordinates, corresponding to nt 308 and nt 747 on the Medicago cDNA sequence. This exon was split by three introns inserted at co-ordinates nt 492, 621 and 688. In JI2009, these introns were 89, 1,257 and 85 bp, whereas in JI1405 the introns were 89, 1,151 and 85 bp, i.e. 106 bp shorter in intron 2. This difference in size of 106 bp, and the presence of HindIII sites in intron 2 accounted for the polymorphic banding pattern in HindIII-digested genomic pea DNA. New primers (4Egenomic5′and 4Egenomic3′) were designed to convert the difference in size between the genomic DNA for JI1405 and JI2009 to a PCR-based polymorphic marker. This PCR marker produced PCR fragments of 425 bp and 535 bp from JI1405 and JI2009, respectively, and provided a co-dominant assay for the screen of the F2 segregating progeny from the JI2009 × JI1405 cross (Fig. 3b).
Marker eIF4E shows tight linkage with sbm-1. a Southern hybridisation of pea genomic DNA from resistant line JI1405 (1) and susceptible line JI2009 (2) after digestion with EcoRI, EcoRV, HindIII or BamHI, using the Medicago eIF4E fragment cDNA fragment as a probe. b Conversion of eIF4E genomic DNA into a PCR-based co-dominant polymorphic marker. Segregation within the segregating F2 population identified resistant (A), susceptible (B) and heterozygous lines (H)
When eIF4E was mapped relative to PSbMV resistance in a segregating F2 population of 500 individuals, no recombinants were identified. This was confirmed in the F3 generations. Hence, eIF4E appears to co-segregate with sbm-1.
eIF(iso)4E maps to a recessive resistance gene cluster on pea LG II
A multiple alignment of the nucleotide sequences of the eIF(iso)4E mRNA from Arabidopsis revealed several conserved regions in these proteins. Primers eIF-1 and iso4E23′were based upon these conserved sequences. These primers amplified a 306-bp fragment from pea cDNA. No difference was found between resistant and susceptible pea lines. However, when the amplified fragment was used as a hybridisation probe for restriction-digested pea genomic DNA, eIF(iso)4E was found to be a single-copy gene (Fig. 4a), which showed polymorphic behaviour between the parental lines JI281 and JI399. These lines have been used to generate an RI family (Ellis et al.1992; Lacou et al. 1998) from which a subset of 16 RI lines were used for bin mapping, providing the opportunity to map eIF(iso)4E (Fig. 4b). The gene mapped close to several AFLP markers located in a region of LG II known to contain the mo locus and linked in turn to sbm-2 (Ellis and Poyser 2002).
Mapping of eIF(iso)4E. a Southern hybridisation of pea genomic DNA from pea lines JI281 and JI399 shows size polymorphism after digestion with EcoRV, HindIII or BamHI. b RFLP analysis of 16 recombinant inbred (RI) lines from a cross between JI281 and JI399 identified a chromosomal location on linkage group (LG) II
Discussion
Peas constitute an economically important crop in many temperate parts of the world. Many commercial pea lines (and lentil lines in the sub-tropical zones) are susceptible to PSbMV, and there is an urgent need to identify tightly linked markers that could facilitate the transfer of the recessive resistances within breeding programmes. The use of markers for recessive genes is particularly powerful as it removes the delays in breeding programmes associated with the restriction of the phenotypic analysis to homozygous progeny. This, and inconsistencies in scoring infection phenotypes, has significantly delayed the introgression of PSbMV into elite pea lines. In peas, at least, the use of markers has the added attraction of assisting the introgression of further diverse potyvirus resistances genetically linked to sbm. Hence, sbm-1and sbm-2 are associated with resistances to at least two and five other potyviruses, respectively.
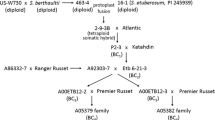
Progress towards identifying markers very tightly linked to sbm-1/-4 has been slow and may indicate a discrepancy between the physical and recombinational distances in this area. The previous closest markers to sbm-1 were GS185 (Timmerman et al. 1993) and sG05_2537 (Frew et al. 2002) at 8 cM and ~4 cM, respectively, and our cDNA-AFLP search yielded only one closer marker, ZG10, at 0.7 cM. Alignment of our data and the published (Frew et al. 2002) maps shows the relative positions of ZG10, eIF4E/sbm-1 and G05_2537 (Fig. 5). The determination of the orientation of eIF4E/sbm-1 and ZG10 with respect to Gsp was assisted by the location of a microsatellite marker (‘PSAC76a’, proprietary information from the ‘Agrogene Microsatellite Consortium’) on the opposite side of sbm-1 from ZG10 (data not shown).
Alignment of eIF4E and ZG10 relative to sbm-1 and selected linked reference markers on LG VI. The relationship between ZG10, sbm-1/eIF4E and G05_2537 was determined by mapping ZG10 onto the JI15 × JI399 RI family for which the marker Gsp provided an anchor for comparisons with the crosses used by Frew et al. (2002). Because of the limited size of the RI family, the approximate region to which ZG10 was mapped is indicated by the filled box. The scale indicates distance in centiMorgans
Recessive resistance to potyviruses in lettuce and pepper and the identification of susceptibility factors in Arabidopsis have implicated the viral genome-linked protein (VPg) as a virulence factor interacting with eIF4E or eIF(iso)4E. Further, a direct physical interaction between Turnip mosaic virus (TuMV) VPg and eIF(iso)4E has been demonstrated (Wittmann et al. 1997). Similarly, the VPg from PSbMV has been identified as the viral resistance determinant for sbm-1and sbm-4 on LG VI (Borgstrom and Johansen 2001). Interestingly, the avirulence determinant corresponding to sbm-2 on LG II corresponds to the PSbMV P3 protein (Johansen et al. 2001), although mo-mediated resistance to BYMV, linked to sbm-2 on LG II, does appear to involve VPg (Bruun-Rasmussen et al. 2003). It seems likely that resistances on LG II depend upon at least two functionally independent genes.
Using a segregating F2 population for sbm-1 resistance and a smaller RI family for mapping on LG II, we identified eIF4E and eIF(iso)4E as markers tightly linked to sbm-1 and mo, respectively. In the case of sbm-1, the polymorphism between resistant and susceptible lines was based upon the presence of a small intron with a fortuitous HindIII site within the single eIF4E gene. From the analysis of the F2, it appears that eIF4E was cosegregating with resistance. Our data does not show that eIF4E and eIF(iso)4E are functionally linked with these two resistance gene clusters, but they provide two useful tools for following the VPg-mediated resistances in breeding programmes and two strong potential candidates for the resistance gene products themselves. All of the sbm-linked sequences identified in this work have been converted to primers for PCR assays and are generally available for use.
References
Bachem CW, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RG (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J 9:745–753
Borgstrom B, Johansen IE (2001) Mutations in Pea seed-borne mosaic potyvirus genome-linked protein VPg alter pathotype specific virulence in Pisum sativum. Mol Plant Microbe Interact 14:707–714
Bruun-Rasmussen M, Moller IS, Cordea M, Sundelin T, Johansen IE (2003) Molecular resistance determinants of the Bean yellow mosaic virus in Pisum sativum. EMBO Workshop – Genomic approaches in plant virology. Abstract, p 6
Christou P, Ford TL, Kofron M (1991) Production of transgenic rice (Oryza sativa L.) plants from agronomically important indica and japonica varieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos. Bio technology 9:957–962
Deom CM, Murphy JF, Paguio OR (1997) Resistance to Tobacco etch virus in Capsicum annum: inhibition of virus RNA accumulation. Mol Plant Microbe Interact 10:917-921
Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32:927–934
Ellis THN (1994) Approaches to the genetic mapping of pea. In: Modern methods of plant analysis, vol 16. Springer, Berlin Heidelberg New York, pp 117–160
Ellis THN, Poyser SJ (2002) An integrated and comparative view of pea genetic and cytogenetic maps. New Phytol 153:17–25
Ellis THN, Turner L, Hellens RP, Lee D, Harker CL, Enard C, Domoney C, Davies DR (1992) Linkage maps in pea. Genetics 130:649–663
Frew TJ, Russell AC, Timmerman-Vaughan GM (2002) Sequence-tagged site markers linked to the sbm1 gene for resistance to Pea seed-borne mosaic virus. Plant Breed 121:512–516
Gal-On A, Meiri E, Elman C, Gray DJ, Gaba V (1997) Simple hand-held devices for the efficient infection of plants with viral-encoding constructs by particle bombardment. J Virol Methods 64:103–110
Gritton ET, Hagedorn DJ (1975) Linkage of the genes sbm and wlo in peas. Crop Sci 15:447–448
Haddad NI, Muehlbauer FJ, Hampton RO (1978) Inheritance of resistance to Pea seed-borne mosaic virus in lentils. Crop Sci 18:613–615
Hagedorn DJ, Gritton ET (1971) Registration of Wisconsin 7105 and 7106 pea germplasm. Crop Sci 11:945–946
Hagedorn DJ, Gritton ET (1973) Inheritance of resistance to the Pea seed-borne mosaic virus. Phytopathology 63:1130–1133
Haldane JBS, Waddington CH (1931) Inbreeding and linkage. Genetics 16:357–374
Hampton RO (1980) Pea seed-borne mosaic symptoms variation among Pisum plant introduction accessions: expression & variation in symptom expression among different genotypes of Pisum and the pathological implications. Pisum Newsl 12:29–30
Hampton RO, Marx GA (1981) Immunity to Pea seed-borne mosaic virus: a re-assessment. Pisum Newsl 13:16–17
Johansen IE, Lund OS, Hjulsager CK, Laursen J (2001) Recessive resistance in Pisum sativum and potyvirus pathotype resolved in a gene-for-cistron correspondence between host and virus. J Virol 75:6609–6614
Khetarpal RK, Maury Y (1987) Pea seed-borne mosaic virus: a review. Agronomie 7:215–224
Kyle MM, Palloix A (1997) Proposed revision of nomenclature for potyvirus resistance genes in Capsicum. Euphytica 97:183–188
Lacou V, Haurogné K, Ellis N, Rameau C (1998) Genetic mapping in pea. 1- RAPD-based genetic linkage map of Pisum sativum. Theor Appl Genet 97:905–915
Lellis AD, Kasschau KD, Whitham SA, Carrington JC (2002) Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus Infection. Curr Biol 12:1046–1051
Maule AJ, Wang D (1996) Seed transmission of plant viruses: a lesson in biological complexity. Trends Microbiol 4:153–158
Muehlbauer FJ (1983) Eight germplasm lines of pea resistant to Pea seed-borne mosaic virus. Crop Sci 23:1019
Murray MG, Cuellar RE, Thompson WF (1978) DNA sequence organization in the pea genome. Biochemistry 17:5781-5790
Nicaise V, German-Retana S, Sanjuán R, Dubrana M-P, Mazier M, Maisonneuve B, Candresse T, Caranta C, LeGall O (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol 132:1272–1282
Provvidenti R, Alconero R (1988) Inheritance of resistance to a lentil strain of Pea seed-borne mosaic virus in Pisum sativum. J Hered 79:45–47
Provvidenti R, Hampton RO (1991) Chromosomal distribution of gene for resistance to seven potyviruses in Pisum sativum. Pisum Genet 23:26–28
Provvidenti R, Muehlbauer FJ (1990) Evidence of a cluster of linked genes for resistance to Pea seed-borne mosaic virus and Clover yellow vein virus on chromosome 6. Pisum Newsl 22:43–45
Provvidenti R, Hampton RO, Muehlbauer FJ (1991) G-1000, Marx’s pea breeding line for multiviral resistance. Pisum Genet 23:50–52
Ruffel S, Dussault MH, Palloix A, Moury B, Bendahmane A, Robaglia C, Caranta C (2002) A natural recessive resistance gene against Potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J 32:1067–1075
Sambrook J, Russell DW (2001) Molecular cloning—a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Springer Harbor, N.Y.
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J 3:739–744
Stam P, van Ooijen JW (1995) JoinMap, ver 2.0: software for the calculation of genetic linkage maps. CPRO-DLO, Wageningen, The Netherlands
Stevenson WR, Hagedorn DJ (1971) Reaction of Pisum sativum to the Pea seed-borne mosaic virus. Plant Dis Rep 55:408–409
Timmerman GM, Frew TJ, Miller AL, Weeden NF, Jermyn WA (1993) Linkage mapping of sbm-1, a gene conferring resistance to Pea seed-borne mosaic virus, using molecular markers in Pisum sativum. Theor Appl Genet 85:609–615
Wittmann S, Chatel H, Fortin MG, Laliberte J-F (1997) Interaction of the viral protein genome linked of Turnip mosaic potyvirus with the translational eukaryotic initiation factor(iso)4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 234:84–92
Acknowledgements
We thank Elisabeth Johansen, Roger Cousin, Boulos Chaloub, Delphine Malayah, Maggie Knox and Linda Turner for constructive discussions, and particularly Elisabeth Johansen and Roger Cousin for making available to us PSbMV cDNA clones and pea lines 735 and 844, respectively. Z.G. and S.E. were supported directly from a research grant (208/P13344) provided by the UK Biotechnology and Biological Sciences Research Council (BBSRC). The John Innes Centre is also grant-aided by the BBSRC. The work was carried out under the UK Department of Environment, Food and Rural Affairs licence numbers PHL 185/4192 and 185A/4512.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Möllers
Rights and permissions
About this article
Cite this article
Gao, Z., Eyers, S., Thomas, C. et al. Identification of markers tightly linked to sbm recessive genes for resistance to Pea seed-borne mosaic virus . Theor Appl Genet 109, 488–494 (2004). https://doi.org/10.1007/s00122-004-1652-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1652-6