Abstract
Rice (Oryza sativa L.) plants develop vertically with shoot elongation and horizontally with tillering. The purpose of this study was to identify and characterize genomic regions influencing the rice plant architecture by quantitative trait locus (QTL) analysis for the component traits: culm length (CL), panicle length (PnL), panicle number (PnN) and tiller number (TN). For this QTL analysis, 191 recombinant inbred lines (F7) derived from a cross of Milyang 23 (M23) and Akihikari (AK) were grown in 1995, 1996 and 1997 (May–Oct) in Joetsu, Japan (temperate climate), and in the 2000 dry season (Jan–Apr), the 2000 wet season (Jun–Oct) and the 2001 dry season in Los Baños, The Philippines (tropical climate). Results showed that rice plant architecture was influenced by 19 genomic regions categorized into five groups. In Group I, two regions (on chrs. 6 and 11) affected shoot elongation (CL and PnL) and tillering (PnN and TN) in opposite directions more significantly in Los Baños than in Joetsu. In Group II, two regions (chrs. 3 and 12) affected shoot elongation, whereas in Group III, five regions [chrs. 1 (two), 2, 3 and 9] affected only culm length (CL). Expressions of four regions of Group III were influenced by either tropical or temperate environments. In Group IV, seven regions (chrs. 1, 2, 4, 5, 6, 8 and 9) controlled panicle development (PnN or PnL), and in Group V, three regions (chrs. 1, 2 and 3) regulated tillering (PnN or TN). Characterizing these 19 genomic regions provided a detailed analysis of rice plant architecture with emphasis on the multiple effect and environmental responsive regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) plants develop vertically with shoot elongation and horizontally with tillering. Shoot elongation and tillering are usually represented by plant height (PH) and panicle number (PnN), respectively. PH consists of the length of the culm and panicle. Hence, culm length (CL), panicle length (PnL) and PnN are major traits composing rice plant architecture, and ultimately the grain yield (Sasahara 1997; Tanisaka 1997). Past studies have reported that these component traits are inherited quantitatively, related genetically to one another, and influenced by growing environments (Gravois and McNew 1993; Sasahara 1997; Tanisaka 1997). Because of these trait characteristics, it has been difficult to identify the genetic factors of each trait and to comprehend the genetic mechanisms of plant architecture in rice.
The recent development of molecular markers enabled researchers to identify gene loci affecting quantitative traits, such as CL, PnL and PnN, on a linkage map in rice (Yano and Sasaki 1997). Using the IR64/Azucena double-haploid population, Yan et al. (1999) identified nine and six quantitative trait loci (QTLs) for PH and PnN, respectively, and found that QTLs for both traits were detected in the same region of chr. 1. The QTLs of this region increased PH and decreased PnN with the Azucena allele. Yamamoto et al. (2001) likewise found two regions (chrs. 3 and 6) that increased CL and PnL through QTL analysis using the Koshihikari/Kasalath BC1F3 population. These genomic regions with multiple effects possibly show how the component traits are related to one another (Li et al. 1995; Lin et al. 1996; Yan et al. 1999). Their localization and characterization would help to understand the genetic mechanisms controlling rice plant architecture. However, QTL analyses that examined the three component traits simultaneously and focused on the multiple effect regions are not abundant.
The component traits change dramatically with growing environments; for example rapid culm elongation in floating varieties (Hamamura 1997). The changes are partly attributable to the genes (QTLs) expressed in a particular environment, in other words the interaction between QTLs and the environment (Lu et al. 1996; Yan et al. 1999; Yamamoto et al. 2000). Past QTL analyses measured CL, PnL and PnN at a fixed location (environment), thereby providing little acknowledgment of the QTL × environment interaction. Exceptionally, Lu et al. (1996), Zhuang et al. (1997) and Yan et al. (1999) examined small mapping populations at a few locations in China (temperate and subtropical climates) and statistically quantified the effect of the QTL × environment interaction on the component traits. Further analyses must be necessary to fully locate the QTLs interacting with the environments, especially the tropical dry and wet seasons, and to qualify the interactions.
The objective of this study was to clarify genomic regions affecting the component traits of plant architecture, and their interactions with environments. For this objective, 191 recombinant inbred lines (RILs) derived from a cross of Milyang 23 (M23) and Akihikari (AK) (Fukuta et al. 1999) were repeatedly grown under two major rice-growing climates, temperate (Japan) and tropical (Philippines), to locate QTLs for CL, PnL, PnN and tiller number (TN). M23 is an indica variety with heavy panicles, whereas AK is a japonica variety with a large number of panicles. Both varieties are high yielding and have been used in actual breeding programs (Yokoo et al. 1994). QTL analyses using the M23/AK cross are possible to disclose a typical genetic nature (background) underlying the different types of high yielding varieties (Yano and Sasaki 1997).
Materials and methods
Plant materials and phenotypic data collection
One hundred and ninety one RILs (F7 generation) derived from a cross between M23 and AK (Fukuta et al. 1999) were grown in irrigated field conditions during May–Oct in 1995 (95 J), 1996 (96 J) and 1997 (97 J) at the former Hokuriku National Agricultural Experiment Station (HNAES, currently the Hokuriku Research Center, 138° E; 38° N), Joetsu, Japan, and during the 2000 dry season (Jan–Apr, 00DS), the 2000 wet season (Jun–Oct, 00WS), and the 2001 dry season (01DS) at the International Rice Research Institute (IRRI, 121°E; 14°N), Los Baños, The Philippines. Joetsu and Los Baños belong to the temperate and tropical zones, respectively, both having annual rainfall of >2,000 mm per year (see Table 1 for detailed climatic data). Thirty day-old seedlings were transplanted with a single plant per hill at locally accepted densities: 15.4 plants m−2 in Joetsu (22.2 plants m−2 for 1996) and 16.7 plants m−2 in Los Baños. Nitrogen, phosphorus and potassium fertilizers were applied 70, 70 and 70 kg ha−1, respectively, at HNAES, and at 134, 66 and 66 kg ha−1 at IRRI, following the experimental-farm recommendations based on soil fertility.
More than five plants were harvested 35 days after heading (when the plants were mature) to measure CL, PnL and PnN. TN was also examined in Los Baños since tillering continued in many RILs after heading under high air temperature and abundant rainfall. Based on planting density, PnN and TN were converted to a per-square-meter basis for data comparison between the two locations.
QTL analysis
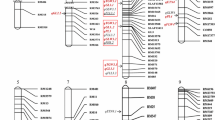
Using 182 restriction fragment length polymorphism (RFLP) markers, Fukuta et al. (1999) determined the genotype of marker loci and constructed a complete linkage map (see Fig. 1). With the genotypic data, interval mapping was performed using the computer software QGENE ver. 3.05 (Nelson 1997) and QTLs were identified by the following two steps. For the first step, all visible LOD peaks >2.0 were located, including the one near the highest peak. This threshold is suitable for detecting relatively minor-effect genes, according to Yano and Sasaki (1997). For the second step, the LOD peaks (>2.0), of which the nearest RFLP marker showed P<0.01, were accepted as QTLs. This threshold has also been used commonly in recent single-point QTL analyses (Yano and Sasaki 1997). To further confirm our analysis, we focused on only the genomic regions that repeatedly showed QTLs for a trait across crop seasons. These regions were designated tentatively with the letter R based on each chromosome and region number.
Linkage map sowing QTLs for culm length (CL), panicle length (PnL), panicle number (PnN) and tiller number (TN) in the Milyang 23/Akihikari RILs grown during 1995 (95 J), 1996 (96 J) and 1997 (97 J) in Joetsu, Japan, and during the 2000 dry season (00DS), the 2000 wet season (00WS) and the 2001 dry season (01DS) in Los Baños, The Philippines. Vertical bars show the genomic region recording LOD >2.0. Open and filled arrows present the highest LOD peak and the neighboring one, respectively. Upward and downward arrows indicate that the Milyang 23 and Akihikari alleles increased the value, respectively. The genomic regions that repeatedly detected QTLs for a trait across seasons were shown by bold vertical bars and designated tentatively with the letter R, and the chromosome and region number
Results and discussion
Trait segregation and correlation
The four traits examined showed a continuous frequency distribution with transgressive segregants for all seasons (Table 2). For CL, the mean value of the RILs was higher in 95 J, 96 J, 97 J and 00WS than in 00DS and 01DS. Parental differences were only observed in the latter two seasons where the mean value of M23 was larger than that of AK. The former four seasons were from May to Oct, whereas the latter two were the dry seasons from Jan to Apr. For PnL, the mean value of the RILs was larger in Joetsu (95 J, 96 J and 97 J) than in Los Baños (00DS, 00WS and 01DS), whereas for PnN, the mean value was larger in Los Baños than in Joetsu. These data showed that CL differed with cropping time, whereas PnL and PnN differed with cropping location.
CL was positively correlated between seasons (r=0.49–0.79). The correlation coefficients were especially high between different years in Joetsu (0.76–0.79) and between different seasons in Los Baños (0.68–0.72), and between 00WS and 95 J, 96 J and 97 J (0.67–0.75). Such a trend was also observed for the three other traits. PnL had higher coefficients (r=0.78–0.61) than CL, whereas PnN and TN had similar or lower coefficients (r=0.78–0.49 and r=0.70–0.48).
CL was positively correlated with PnL (r=0.20–0.61). The correlation coefficients were higher between 00DS and 01DS (0.43–0.61). Positive correlations were also observed between PnN and TN (r=0.36–0.93), whereas negative ones were observed between CL and PnN (r=−0.38 to −0.08) and between PnL and PnN (r=−0.54 to −0.19). These results showed that the two vertical growth components, CL and PnL, were positively correlated especially in the tropical dry season, and that these two were negatively correlated with the two horizontal growth components, PnN and TN.
QTL mapping
Throughout six crop seasons (three for TN), a total of 31, 59, 38 and 14 QTLs were detected for CL, PnL, PnN and TN, respectively. The total phenotypic variance explained by these QTLs ranged from 17.0–44.4%, 37.4–52.2%, 24.1–47.2% and 19.7–34.0% in CL, PnL, PnN and TN, respectively. Most QTLs were mapped onto 19 genomic regions (Fig. 1, Table 3). Based on the effects of these QTLs, the 19 regions were characterized and categorized into five groups.
Group I: QTLs affecting both shoot elongation (CL and PnL) and tillering (PnN and TN)
The M23 allele for R6-2 (G2140C-XNpb12) resulted in increased CL and PnL (shoot length) and decreased PnN and TN (tillering) (Fig. 1). The LOD score peak indicated that the QTL for these four traits was present near XNpb12. This QTL showed high r 2, suggesting a large genetic effect (Table 3). Comparison of the r 2 between Joetsu and Los Baños showed that the effects of R6-2 on CL and PnN were elevated by the tropical climate. A similar region was also found near the centromere on chromosome 11 (R11, C477-XNpb257). QTLs for CL and PnL were identified near N099K in the Los Baños dry seasons (00DS and 01DS), whereas two different QTLs for PnN were located near C477 and XNpb257 in all crop seasons. These results showed that R11 possibly consists of three loci: one functions for shoot elongation in the tropical dry season (Jan–Apr) and two for tillering regardless of cropping season. The above results demonstrated that R6-2 and R11 affected the balance between shoot elongation and tillering, especially in the tropics. These two regions well explained the observed positive genetic relations between CL and PnL, or between PnN and TN, and the negative genetic relations of PnN with CL and PnL.
Past QTL analyses examined CL, PnL and PnN individually in different crop seasons or mapping populations, thereby providing limited information on genomic regions affecting both shoot elongation and tillering. Exceptionally, Yan et al. (1999) reported a region that increased PH and decreased PnN near RZ801 on the long arm of chr. 1. However, the effects of R6-2 and R11 on these traits were identified only in this study.
Group II: QTLs affecting shoot elongation (CL and PnL)
The M23 allele for R3-2 (C198-R250) resulted in increased CL and PnL. In R3-2, QTLs appeared at two different positions, C198 and R250. A similar type of region was found in R12 (XNpb304–C1069). Two different LOD score peaks were observed near XNpb304 and between XNpb307–C1016, although the peaks of PnL were <2.0 in LOD (ranging from 1.5 to 2.0 for most seasons). These results indicate that R3-2 and R12 could each have two loci affecting shoot elongation (CL and PnL). These two regions explained the observed positive genetic relations between CL and PnL.
Group III: QTLs affecting culm elongation (CL) only
A powerful QTL for CL was identified near C86 (R1-4). Using near-isogenic lines, Cho et al. (1994) located the semidwarf gene sd1, which reduces PH markedly, near the RFLP marker RG220 on chromosome 1. Since RG220 is close to C86, the powerful CL QTL near C86 could be identical to the sd1 locus (Fukuta 1995; Tabuchi et al. 2000). Because of the large sd1 effect, other genes with a minor effect on CL have not been investigated in detail by past genetic analyses, although they play an important role in fine-tuning CL toward the desired PH (Liang et al. 1994; Li et al. 1995). Our 6-season investigation found QTLs with a minor or intermediate effect on CL in four regions, R1-1 (C970), R2-1 (XNpb227), R3-3 (XNpb48) and R9-2 (G1445). QTLs were detected in R1-1 for the Los Baños crop seasons, in R2-1 and R9-2 for the Joetsu crop seasons, and in R3-3 for 95 J, 96 J, 97 J and 00WS. These detection patterns indicated that the QTLs of these four regions interacted with the seasonal environments. Since R6-2 and R11 affected CL more significantly in Los Baños, a total of six CL-affecting regions (QTLs) were expressed differently in temperate and tropical environments.
Group IV: QTLs affecting panicle development (PnL and PnN)
QTLs for PnL were detected in five regions, R1-3 (R210A), R5 (C734B), R6-1 (C235), R8 (XNpb56) and R9-1 (XNpb13). No seasonal detection pattern was observed for these five regions, suggesting relatively stable expression of PnL QTLs. Generally, the heritability of PnL is higher than that of CL (Sasahara 1997; Tanisaka 1997). The observed stable QTL expression may explain the high PnL heritability. QTLs for PnL and PnN were also detected in R2-3 (XNpb317B) and R4 (XNpb331). The M23 allele of R2-3 resulted in increased PnL and decreased PnN, whereas that of R4 resulted in the opposite effect. In R4, QTLs for PnN were detected only in the tropical location of Los Baños. Previously published literature reported QTLs for panicle traits, such as spikelet fertility and spikelet number per panicle, in the vicinities of R5, R6-1 and R8 (Lin et al. 1996; Lu et al. 1996; Xiao et al. 1996; Ishimaru et al. 2001). However, QTLs for PnL or PnN were not reported previously in R1-3, R9-1, R2-3 and R4, and the effect of temperate climate on R4 was not shown.
Group V: QTLs affecting tillering (PnN and TN)
It has been difficult to locate QTLs controlling PnN or TN because of the large environmental error (Sasahara 1997). The 6-season investigation repeatedly detected QTLs for PnN or TN in three regions, R1-2 (R210B-XNpb90), R2-2 (XNpb353-XNpb132) and R3-1 (C74-XNpb129). In R1-2, two high LOD peaks were observed near R210B and XNpb90, suggesting two loci of large effect were involved. The AK allele of R1-2 resulted in increased PnN and TN. Yagi et al. (2001) reported that the M23 allele of this region resulted in an increased number of spikelets per panicle. R1-2 corresponded to the QTL of Yagi et al. (2001). These allelic effects agree well with the varietal descriptions of AK as having a large number of panicles, and M23 as having a large number of spikelets per panicle (heavy panicle). These past and current results from our research group, for the first time, shed light on R1-2 as a genetic factor that differentiates rice varieties, such as AK and M23, into panicle-number and heavy panicle types. It was also noteworthy that R3-1 showed only the TN QTLs in Los Baños. In other words, R3-1 has a locus (loci) that stimulates tillering only in the tropics.
Conclusions
The plant architectures of M23 and AK were differentiated by at least 19 genomic regions. The 19 regions were characterized and categorized into five groups: Group I (two regions) promoted shoot elongation and suppressed tillering, Group II (two regions) affected shoot elongation, Group III (five regions) culm elongation, Group IV (seven regions) panicle development and Group V (three regions) tillering. The multiple effects of two regions in Group I were enhanced in a tropical climate. Such a genetic response to the tropical climate also occurred with two regions in Groups IV and V. Moreover, the effects of four regions in Group III were different in a tropical climate, a temperate climate, or the Joetsu and Los Baños wet seasons. Therefore, a total of eight regions interacted with the environments. These findings demonstrate that: (1) more than a dozen genes are involved in the complex genetic control of plant architecture, (2) a few genes control the balance between shoot elongation and tillering, and (3) several genes interact with environments and result in differentiated genetic mechanisms among locations. The actual number of genes in each region, and their accurate locations and effects, are being investigated by our subsequent experiments using the substitution lines in the M23 or AK background.
References
Cho YG, Eun MY, McCouch SR, Chae YA (1994) The semidwarf gene, sd-1, of rice (Oryza sativa L.). II. Molecular mapping and marker-assisted selection. Theor Appl Genet 89:54–59
Fukuta Y (1995) RFLP mapping of a shattering-resistance gene in the mutant line, SR-1, induced from an indica rice variety, Nan-jing 11. Breeding Sci 45:15–19
Fukuta Y, Tamura K, Sasahara H, Fukuyama T (1999) Genetic and breeding analysis using molecular marker 18: variations of gene frequency and the RFLP map of the hybrid population derived from the cross between the rice variety Milyang23 and Akihikari. Breeding Res (Suppl 2):176
Gravois KA, McNew RW (1993) Genetic relationships among and selection for rice yield and yield components. Crop Sci 33:249–252
Hamamura K (1997) Floating rice as a specific ecotype. In: Matsuo T, Futsuhara Y, Kikuchi F, Yamaguchi H (eds) Science of the rice plant. Vol III genetics. Food and Agriculture Policy Research Center, Tokyo, Japan, pp 131–135
Ishimaru K, Yano M, Aoki N, Ono K, Hirose T, Lin SY, Monna L, Sasaki T, Ohsugi R (2001) Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor Appl Genet 102:793–800
Li Z, Pinson SRM, Stansel JW, Park WD (1995) Identification of quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor Appl Genet 91:374–381
Liang CZ, Gu MH, Pan XB, Liang GH, Zhu LH (1994) RFLP tagging of a new semidwarfing gene in rice. Theor Appl Genet 88:898–900
Lin H-X, Qian H-R, Zhuang J-Y, Lu J, Min S-K, Xiong Z-M, Huang N, Zheng K-L (1996) RFLP mapping of QTLs for yield and related characters in rice (Oryza sativa L.). Theor Appl Genet 92:920–927
Lu C, Shen L, Tan Z, Xu Y, He P, Chen Y, Zhu L (1996) Comparative mapping of QTLs for agronomic traits of rice across environments using a double-haploid population. Theor Appl Genet 93:1211–1217
Nelson JC (1997) QGENE: software for marker-based genomic analysis and breeding. Mol Breed 3:239–245
Sasahara T (1997) Panicles. In: Matsuo T, Futsuhara Y, Kikuchi F, Yamaguchi H (eds) Science of the rice plant. Vol III genetics. Food and Agriculture Policy Research Center, Tokyo, Japan, pp 285–294
Tabuchi H, Hashimoto N, Takeuchi A, Terao T, Fukuta Y (2000) Genetic analysis of semidwarfism of the japonica rice cultivar Kinuhikari. Breeding Sci 50:1–7
Tanisaka T (1997) Culm. In: Matsuo T, Futsuhara Y, Kikuchi F, Yamaguchi H (eds) Science of the Rice Plant. Vol. III Genetics. Food and Agriculture Policy Research Center, Tokyo, Japan, pp 270–277
Xiao J, Li J, Yan L, Tanksley SD (1996) Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet 92:230–244
Yagi T, Nagata K, Fukuta Y, Tamura K, Ashikawa I, Terao T (2001) QTL mapping of spikelet number in rice (Oryza sativa L.). Breeding Sci 51:53–56
Yamamoto T, Lin H, Sasaki T, Yano M (2000) Identification of heading-date quantitative trait locus Hd6 and characterization of its epistatic interaction with Hd2 in rice using advanced backcross progeny. Genetics 154:885–891
Yamamoto T, Shiobara FT, Ukai Y, Sasaki T, Yano M (2001) Mapping quantitative trait loci for days-to-heading, and culm, panicle and internode lengths in a BC1F3 population using an elite rice variety, Koshihikari, as the recurrent parent. Breeding Sci 51:63–71
Yan J, Zhu J, He C, Benmoussa M, Wu P (1999) Molecular marker-assisted dissection of genotype x environment interaction for plant-type traits in rice (Oryza sativa L.). Crop Sci 39:538–544
Yano M, Sasaki T (1997) Genetic and molecular dissection of quantitative traits in rice. Plant Mol Biol 35:145–153
Yokoo M, Saito S, Higashi T, Matsumoto S (1994) Use of a Korean rice cultivar Milyang23 for improving Japanese rice. Breeding Sci 44:219–222
Zhuang J-Y, Lin H-X, Lu J, Qian H-R, Hittalmani S, Huang N, Zheng K-L (1997) Analysis of QTL × environment interaction for yield components and plant height in rice. Theor Appl Genet 95:799–808
Acknowledgments
This study was partly conducted under the IRRI-Japan Collaborative Research Project. The climatic data of Joetsu and Los Baños were kindly provided by Dr. K. Yokoyama, National Agricultural Research Center, and Mr. Nestor Fabellar, IRRI, respectively. We wish to thank Messrs. T. Aota, H. Shimizu and I. Nozaki, HNAES, and Mr. L.A. Ebron, IRRI, for their skilled management of the paddy fields, and Dr. H. Yamagishi, Hokkaido University, for his valuable suggestions on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Mackill
Rights and permissions
About this article
Cite this article
Kobayashi, S., Fukuta, Y., Sato, T. et al. Molecular marker dissection of rice (Oryza sativa L.) plant architecture under temperate and tropical climates. Theor Appl Genet 107, 1350–1356 (2003). https://doi.org/10.1007/s00122-003-1388-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1388-8