Abstract
Thioredoxins fulfill a number of different important cellular functions in all living organisms. In bacteria, thioredoxin genes are often regulated by external factors. In turn, thioredoxins influence the expression of many other genes. The multiple and important functions of thioredoxins in cells necessitate to appropriately adjust their level. This review outlines different strategies that have evolved for the regulation of bacterial thioredoxin genes. It also summarizes effects of thioredoxins on gene regulation and presents a recent model for a redox-dependent gene regulation that is mediated by thioredoxins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thioredoxins are small ubiquitous proteins with a highly conserved active site sequence [(Cys-Gly-Pro-Cys) (Holmgren 1985, 1995a; Martin 1995)]. These proteins share a common 3-D architecture known as the thioredoxin motif, consisting of four α-helices and five β-sheets (Eklund et al. 1991; Holmgren 1995b; Martin 1995; Capitani et al. 2000). Thioredoxins are part of the thioredoxin system, in which electrons are transferred from NADPH to thioredoxin reductase and finally to the thioredoxin (Trx). Because of their low redox potential [−270 to −330 mV in Escherichia coli (Krause et al. 1991; Aslund et al. 1997)], thioredoxins are efficient thiol-disulfide reductants. Thus, thioredoxins, together with the glutaredoxins, are responsible for maintaining a cellular reducing environment and, thereby, can regulate the activity of enzymes. Over the last years, thiol switches have emerged as a major regulatory mechanism in redox-dependent signal transduction (reviewed in, e.g., Paget and Buttner 2003). Apart from the function as thiol-disulfide reductases, thioredoxins also interact with other proteins to form functional protein complexes (reviewed in, e.g., Holmgren 1989).
The thiol-reducing activities of thioredoxins have been best characterized in E. coli, which comprises two thioredoxins, Trx 1 and Trx 2, encoded by the trxA and trxC genes, respectively (Laurent et al. 1964; Miranda-Vizuete et al. 1997; reviewed in Carmel-Harel and Storz 2000). In E. coli, a number of unique structural and regulatory features distinguish the thioredoxin 2 subfamily from the much larger thioredoxin 1 family. Trx 2 contains an additional N-terminal domain of 32 amino acids including two additional Cys-X1-X2-Cys motives compared to Trx 1 (Miranda-Vizuete et al. 1997). The four cysteines of these two Cys-X1-X2-Cys motives function to coordinate one zinc atom (Collet et al. 2003). Although the two E. coli thioredoxins are equivalent for most of their in vivo functions, the transcriptional regulation of trxA and trxC is different (Ritz et al. 2000). While both trx genes are not essential for viability of E. coli [both genes can be deleted from the genome (Ritz et al. 2000)], Trx 1 is required for viability of a number of other bacteria, e.g., Rhodobacter sphaeroides (Pasternak et al. 1997), Bacillus subtilis (Scharf et al. 1998), Anacystis nidulans (Muller and Buchanan 1989), Synechocystis sp. PCC 6803 (Navarro and Florencio 1996).
Several reviews have addressed the general structure and function of thioredoxins (e.g., Aslund and Beckwith 1999; Arner and Holmgren 2000; Carmel-Harel and Storz 2000; Ritz and Beckwith 2001). The fact that cancer cells have high levels of thioredoxin and that reduced cellular thioredoxin levels can cause cancer-prone disease (reviewed in Kontou et al. 2004) emphasizes the importance of this protein in humans. In plants, the thioredoxin system is particularly complex because at least 20 thioredoxin isoforms have been found (reviewed in, e.g., Gelhaye et al. 2005). Different pathways allowing thioredoxin reduction coexist in plants involving ferredoxin–thioredoxin reductase and thioredoxin reductases (Gelhaye et al. 2005). There are far too many reports on thioredoxins and their function to give a single comprehensive overview. Therefore, this review is restricted to bacterial thioredoxins and focuses on the role of thioredoxins in gene regulation and on the regulation of thioredoxin genes.
Functions of bacterial thioredoxins
Many important functions are fulfilled by bacterial thioredoxins, which are summarized in Fig. 1. Thioredoxins are involved in the reduction of a number of enzymes. As hydrogen donor to ribonucleotide reductase (Orr and Vitols 1966) and methionine sulfoxide reductase (Gonzalez Porqué et al. 1970; Boschi-Muller et al. 2000), thioredoxin fulfills an important role in DNA synthesis and protein repair, respectively. As hydrogen donor for phosphoadenosine–phosphosulfate reductase (Lillig et al. 1999), it is implicated in sulfur assimilation (Gonzalez Porqué et al. 1970; Russel et al. 1990).
Functions of thioredoxin. Rectangles include those functions for which the mechanisms of thioredoxin action are known and which involve direct interaction with target molecules. Arrows indicate functions that include the interaction of thioredoxin with other proteins (the interaction partners are circled). ROS reactive oxygen species; TCC tricarboxylic acid cycle; PPC pentose phosphate cycle; AhpC alkyl hydroperoxide reductase; FtsZ cell division protein, tubulin homolog; MreB cell division protein, actin homolog
Thioredoxins are not only involved in reducing cytoplasmic proteins but can directly reduce hydrogen peroxide, H2O2 (Spector et al. 1988; Kang et al. 1998). Kang et al. (1998) were able to show that the thioredoxin system provides reducing equivalents to peroxiredoxins that in turn reduce H2O2. Furthermore, thioredoxins function as singlet oxygen quencher and hydroxyl radical scavenger (Das and Das 2000) and act as hydrogen donor for peroxidases (Chae et al. 1994). These features imply an important function of thioredoxin in the oxidative stress response. Oxidative stress is defined as a disturbance of the pro-oxidant–antioxidant balance in favor of pro-oxidants (Sies 1985). It is caused by reactive oxygen species (ROS) that are generated by auto-oxidation of components of the respiratory chain and other cellular compounds (Gonzalez-Flecha and Demple 1995; Imlay and Fridovich 1991; Messner and Imlay 1999; Seaver and Imlay 2004) or by exposure of aerobically grown cells to metals, redox-active chemicals [such as Butyl-hydroperoxide (t-BOOH) or diamide], or by radiation. Oxidative stress conditions promote disulfide bond formation of redox-sensitive proteins resulting in the functional modulation of these proteins (reviewed in Imlay 2003). The oxidative stress response is aimed to prevent, to counteract, and to repair damages caused by ROS. A role of thioredoxins in the oxidative stress response has been shown for several bacterial species. An E. coli double mutant, lacking both Trx 1 and Trx 2, was shown to be more sensitive to the disulfide bond-inducing agent diamide, suggesting an active role of thioredoxins in dealing with the accumulation of nonnative disulfide bonds. Surprisingly the same mutant was found to be more resistant to high levels of H2O2 (Ritz et al. 2000). This was explained by the fact that the cytoplasmic redox potential of this mutant is more oxidized, which in turn results in the activation of the oxidative stress response (e.g., induction of catalase). This activation of the stress response results in a higher resistance toward H2O2. The deletion of the trxC gene alone results in a more sensitive phenotype in response to H2O2 in E. coli implicating a role of Trx 2 in the oxidative stress response (Ritz et al. 2000). A number of proteins that participate in the oxidative stress response (superoxide dismutase, hydrogen peroxidase I, alkyl hydroperoxide reductase) or have key regulatory functions in the oxidative stress response (ferric uptake regulator, aconitase) were found to be associated with thioredoxin 1 in E. coli (Kumar et al. 2004).
Beside the functions for redox regulation and oxidative stress defense, thioredoxin is an essential subunit of the bacteriophage T7 DNA polymerase (Huber et al. 1987; Mark and Richardson 1976) and is essential for the assembly of several filamentous phages (Russel and Model 1985). The Trx 1 of Synechocystis sp. PCC 6803 is required for both photoautotrophic and heterotrophic growth (Navarro and Florencio 1996). An interaction analysis of the Synechocystis thioredoxin indicated numerous thioredoxin-linked processes in Cyanobacteria, such as glycogen synthesis, sugar-nucleotide metabolism, oxidative stress response, and light harvesting (Lindahl and Florencio 2003). In E. coli, a proteome analysis resulted in the identification of many thioredoxin-targeted proteins (Kumar et al. 2004). A total of 80 proteins was found to be associated with the E. coli Trx 1, implicating the involvement of thioredoxin in at least 26 distinct cellular processes that include cell division, transcriptional regulation, energy transduction, protein folding and degradation, and several biosynthetic pathways.
Recent studies addressed the role of thioredoxins in facultatively photosynthetic bacteria, which provide an excellent model system to study the oxidative stress response of free-living bacteria. Bacteria of the facultatively phototrophic genus Rhodobacter are metabolically highly versatile and can rapidly adapt to changes in their environment. Rhodobacter species are found in aquatic environments, where the oxygen concentration may rapidly change due to external conditions or the metabolic activities of other organisms. While Rhodobacter capsulatus, like E. coli, contains Trx 1 and Trx 2, R. sphaeroides lacks Trx 2. Although trxA is essential in Rhodobacter (Pasternak et al. 1999; Li and Klug, unpublished results), strains that produce lower amounts of trxA could be constructed and analyzed (Pasternak et al. 1999). An R. sphaeroides strain with decreased Trx 1 level shows higher sensitivity to diamide and H2O2 than the wild type; however, it shows higher resistance to the superoxide anion-generating agent paraquat and to the glutathione depleting and oxidizing organic peroxide t-BOOH (Li et al. 2003b). The trxC deletion mutant of R. capsulatus is more sensitive to all oxidative-stress generating agents tested (H2O2, paraquat, t-BOOH, and diamide) than the isogenic wild-type strain (Li et al. 2003a). These results implicate that in Rhodobacter, TrxA and TrxC have a role in the oxidative stress response.
In Lactococcus lactis, the thioredoxin system was believed to be essential because this organism does not produce glutathione. However, cells lacking thioredoxin reductase were viable, even under aerobic conditions (Vido et al. 2005). This strongly suggests that other molecules besides thioredoxin and glutathione can maintain the cytoplasmic redox potential in L. lactis.
Reactive oxygen and nitrogen molecules are generated by mammalian cells and plant cells as a defense strategy against bacterial infections. Therefore, thioredoxins are not only important proteins for the oxidative stress response in nonpathogenic bacteria, but they may also influence the survival of pathogens in host cells. Mycobacterium leprae harbors a thioredoxin–thioredoxin reductase hybrid gene that increases intracellular survival of Mycobacterium smegmatis (Wieles et al. 1997). Helicobacter pylori trxA or trxA2 (for Trx 2) mutants show increased sensitivity to agents generating oxidative stress (Comtois et al. 2003; McGee et al. 2006). In other bacteria, a role of thioredoxins in the oxidative stress response has not been demonstrated by mutant analysis, but an increased expression of trx genes in the presence of ROS implies their role in oxidative stress defense [e.g., B. subtilis (Scharf et al. 1998); Oenococcus oeni (Jobin et al. 1999)].
Regulation of trx gene expression in response to external stimuli is important in all bacteria, but different strategies have evolved to properly adjust the level of thioredoxins. An overview about the different strategies used by bacteria to regulate expression of trx gene is addressed below.
Regulation of bacterial thioredoxin gene expression
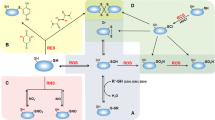
Despite the importance of thioredoxins in many cellular functions, our knowledge on the regulation of trx genes is still limited and restricted to few species. As in many other respects, enteric bacteria served as the first bacterial systems to study the oxidative stress response. Two key regulons of the adaptive responses to oxidative stress were defined by the analysis of a number of genes with known or predicted functions in the oxidative stress response. The OxyR regulon comprises genes that respond to H2O2 including genes encoding thioredoxin 2 (trxC), catalase (katG), alkyl hydroperoxidase (ahpCF), a small RNA (oxyS), glutaredoxin 1 (grxA), and the glutathione reductase gene, gorA (Storz et al. 1990; Zheng et al. 2001). OxyR, a transcriptional regulator of the LysR family, binds to its target sites (Toledano et al. 1994) in its oxidized form and, in most cases, activates gene expression by contacting the alpha subunit of DNA polymerase (Tao et al. 1993). In some cases, however, repression of gene expression by OxyR was observed (Zheng et al. 2001). Oxidized OxyR is reduced by glutaredoxin 1 accompanied by the consumption of glutathione, resulting in a feedback loop for OxyR-regulated genes (Zheng et al. 1998; Storz and Zheng 2000, Fig. 2a). In addition, the Stamler group was able to show that OxyR not only responds to oxidative stress but can also be activated by nitrosative events by S-nitrosylation (Hausladen et al. 1996; Kim et al. 2002). The second regulon for the oxidative stress response in E. coli is the SoxRS regulon. In this system, SoxR and SoxS serve as regulators of the response to superoxide in enteric bacteria (reviewed in, e.g., Nunoshiba 1996; Demple 1996; Storz and Zheng 2000). Interestingly, thioredoxins seem to contribute to SoxR regulation by affecting the disassembly and reassembly of the [2Fe-2S] clusters (Ding and Demple 1998).
Regulation of thioredoxin genes in a Escherichia coli and b Streptomyces coelicolor by oxidative stress. a Oxidized OxyR regulates expression of the OxyR regulon in response to oxidative and nitrosative stress, thereby activating expression of trxC, grxA, and gorA. Oxidized OxyR is reduced by glutaredoxin 1 accompanied by the consumption of glutathione, resulting in a feedback loop for OxyR-regulated genes. b The activity of σR is controlled by the anti-sigma factor RsrA. Oxidative stress induces intramolecular disulfide bond formation in RsrA. In this form, RsrA releases σR that activates expression of trxBA. Reduced thioredoxin A oxidizes RsrA, allowing the formation of the σR–RsrA complex and thereby establishing a feedback loop of regulation. ROS reactive oxygen species, RNS reactive nitrogen species, GrxA glutaredoxin A, GorA glutathione reductase, GSSH/GSH oxidized/reduced glutathione, TrxA thioredoxin A, TrxB thioredoxin reductase
As described above, E. coli thioredoxins are involved in the response to oxidative stress. Therefore, it is of no surprise that the expression of the trxC gene is induced by H2O2 and that trxC is a member of the OxyR regulon (Ritz et al. 2000; Fig. 2a). In contrast, trxA expression is not increased by H2O2 in E. coli and is not under control of OxyR (Michan et al. 1999; Garrido and Grant 2002). It was described that the thioredoxin 1 gene of E. coli (trxA) is under the control of guanosine 3′, 5′-bispyrophosphate (ppGpp), is expressed in the stationary phase (Lim et al. 2000) and is negatively regulated by cyclic AMP (Sa et al. 1997).
Oxygen tension and ROS also affect the expression of thioredoxin genes in the related facultatively photosynthetic bacteria R. sphaeroides and R. capsulatus. Both Rhodobacter strains have OxyR homologues, but no SoxRS homologues. The trxA genes of both R. sphaeroides and R. capsulatus are induced by an increase of oxygen, while the trxC gene of R. capsulatus is slightly repressed (Pasternak et al. 1996, Li et al. 2003a). All Rhodobacter thioredoxin genes also respond to oxidative stress. Expression of trxC in R. capsulatus is strongly induced in response to diamide (20-fold, 1.5 mM final concentration), moderately induced by paraquat (1 mM final concentration), and shows little response to t-BOOH (0.6 mM final concentration) and H2O2 [1 mM final concentration (Zeller, Li, and Klug, unpublished results)]. The response of the R. capsulatus trxA gene to oxidative stress is quite different from that of the trxC gene. It most remarkably shows very little response to diamide (twofold). The addition of t-BOOH results in threefold increase of trxA expression, while the response to paraquat and H2O2 is similar to the trxC response (Zeller, Li, and Klug, unpublished results). Similar to the trxC gene of R. capsulatus the trxA gene of R. sphaeroides shows a strong response to diamide. It increases about sixfold after addition of t-BOOH and two- to threefold after exposure to paraquat or H2O2 (Li et al. 2003b). Apparently, glutathione depletion (induced by t-BOOH) is a stronger stimulus for trxA expression in R. sphaeroides than reactive oxygen species. Expression studies of trx genes in oxyR mutants of Rhodobacter indicate an involvement of OxyR in the regulation of the trxC gene (Zeller, Li, and Klug, unpublished results). The exact mechanisms of this regulation in Rhodobacter are currently under study.
Although many gram-positive bacteria encode OxyR homologues, they use other regulators to control trx gene expression under oxidative stress. The essential trxA gene of B. subtilis is not only under control of the vegetative sigma factor σA but is also transcribed by the general stress sigma factor σB (Scharf et al. 1998). Transcription initiating at the σA-dependent promoter is induced by H2O2 (Scharf et al. 1998). The induction of the B. subtilis trxA and trxB (encoding the thioredoxin reductase) genes by disulfide stress (induced by diamide, Leichert et al. 2003) involves the Spx protein that also represses activator-stimulated transcription by interacting with the C-terminal domain of RNA polymerase alpha subunit (Nakano et al. 2003a,b). A B. subtilis Spx mutant is hypersensitive to diamide (Nakano et al. 2003b). It was proposed that Spx, on one hand, functions as an activator that mobilizes the operations necessary to reverse oxidative stress, but on the other hand, serves as a negative regulator that causes the postponement of developmental programs and energy-consuming functions while the cells cope with stress (Nakano et al. 2003b). Disulfide stress causes an increase of Spx level, possibly due to posttranscriptional regulation (Nakano et al. 2003b). The transcriptional activation by Spx requires formation of an intramolecular disulfide bond within a highly conserved Cys-X1-X2-Cys motif (Nakano et al. 2005). A similar motif is present at the C-terminal end of the transcriptional repressor PerR, another regulator of the oxidative stress response in Bacillus (Bsat et al. 1998; Herbig and Helmann 2001).
In Streptomyces coelicolor, trxB and trxA constitute an operon that is under direct control of the alternative sigma factor σR (Paget et al. 1998; Li et al. 2002; Li et al. 2003c). The trxC gene was also found to be a member of the σR regulon (Paget et al. 2001; Li et al. 2002). The activity of σR is controlled by the anti-sigma factor RsrA. Oxidative stress induces intramolecular disulfide bond formation in RsrA, which causes it to lose affinity for σR, thereby releasing σR to activate transcription of trxBA (Kang et al. 1999; Li et al. 2002; Bae et al. 2004). Interestingly, oxidized RsrA is a direct substrate for reduced thioredoxin, which allows the formation of the σR–RsrA complex, thereby establishing a feedback loop of regulation (Kang et al. 1999; Li et al. 2002; Li et al. 2003c) (Fig. 2b). While OxyR is a positive regulator that is active in its oxidized form, RsrA is a negative regulator and the reduced form of the protein is active (Fig. 2).
An alternative sigma factor, SigH, is involved in the regulation of the trxC and trxB2 genes in the intracellular pathogen Mycobacterium tuberculosis (Raman et al. 2001; Manganelli et al. 2002). SigH regulates the expression of the stress-responsive (heat and oxidative stress) sigma factors SigE and SigB, suggesting a central role of SigH in a network regulating heat and oxidative stress responses (Raman et al. 2001; Manganelli et al. 2002). In Staphylococcus aureus, several oxidative stress compounds (diamide, t-BOOH and the redox cycling agent menadione) induce the trxA and trxB genes, while no effect of H2O2 was observed (Uziel et al. 2004). This induction is independent of the stress sigma factor σB, but the regulators involved in this response remain to be identified.
Thioredoxins can influence the expression of genes
A direct effect of thioredoxins in the oxidative stress response is expected because of their capability to reduce oxidized proteins. However, thioredoxins can also participate in the oxidative stress response by affecting the expression of other genes involved in this response. As outlined in the previous paragraph and shown in Fig. 2, thioredoxins are part of regulatory feedback loops including the sigmaR/RsrA proteins in S. coelicolor. Therefore, thioredoxins affect the regulation of other genes that are under the control of sigmaR/RsrA.
Significantly increased expression of the genes grxA, fpg (DNA repair glycosylase Fpg), nrdA, and nrdB (ribonucleotide reductase) were observed in E. coli strains lacking both thioredoxin 1 and glutathione reductase or thioredoxin 1 and glutaredoxin 1 (Gallardo-Madueno et al. 1998; Prieto-Alamo et al. 2000). The trxC mutant of R. capsulatus shows much stronger, H2O2-induced expression of acnA (aconitase A), fur (ferric uptake regulator), gorA, katG, and stronger paraquat-induced expression of acnA, fpr (ferredoxin/flavodoxin reductase), fur, gorA, and katG than the wild type (Li et al. 2004a). The induction of acnA by superoxide in E. coli results in the synthesis of higher levels of aconitase A, which is resistant to superoxide and can therefore keep the tricarboxylic acid cycle functional (Varghese et al. 2003). The fur gene encodes a regulatory protein, which represses genes required for iron uptake. Upon oxidative stress a stronger repression of iron uptake can prevent the formation of hydroxyl radicals by the Fenton reaction. gorA encodes glutathione reductase, an important component of the glutathione/glutaredoxin system. katG encodes catalase, an important enzyme for the detoxification of H2O2. These findings confirm an interplay of different defense systems.
Smits et al. (2005) reported the effects of thioredoxin depletion on global transcription levels in B. subtilis. The results of this study indicate that changes in thioredoxin A level cause transcriptional changes in B. subtilis. Because thioredoxins have so far not been reported to act as transcriptional regulators, the authors suggest that these transcriptional changes are likely to represent indirect effects of thioredoxin A (e.g., interaction or influence on transcription factors or other proteins).
In Rhodobacter, thioredoxins have been demonstrated to be involved in the redox-dependent regulation of photosynthesis genes (Clement-Metral 1979; Pasternak et al. 1999; Li et al. 2003b). Oxygen tension is the major factor that determines the regulation of photosynthesis genes and, consequently, the formation of photosynthetic complexes in Rhodobacter. Decreased levels of Trx 1 lead to lower increase of puf and puc mRNA levels after a drop of oxygen tension compared to wild-type strains in R. sphaeroides and R. capsulatus (Pasternak et al. 1999; Li et al. 2004b). The puf and the puc operon encode pigment-binding proteins and other proteins required for the formation of photosynthetic complexes. Surprisingly, a trxC deletion mutant of R. capsulatus showed a stronger increase of puf and puc mRNA levels after drop of oxygen tension (Li et al. 2003a). This finding of a signal from thioredoxin to transcription of photosynthesis genes resulted in the discovery of a new signaling pathway.
In a search for proteins interacting with Rhodobacter thioredoxins, the gyrase B subunit was identified by a yeast-two hybrid screening (Li et al. 2004b). A model in which thioredoxin affects gene expression by modifying gyrase activity was experimentally confirmed (Li et al. 2004b). TrxA mutants of Rhodobacter exhibit lower supercoiling activity than the wild type; in contrast, the TrxC mutant exhibits higher supercoiling activity. In vitro experiments supported the modulation of gyrase supercoiling activity by thioredoxin. Because the expression of many genes is influenced by the supercoiling status of the DNA (Dorman et al. 1988, Franco and Drlica 1989; Schneider et al. 2000), this implies an important function of thioredoxins on the expression of many genes. A model for the action of thioredoxins on gene expression is shown in Fig. 3. Reduced, but not oxidized, Trx 1 interacts with the gyrase B subunit and increases its supercoiling activity. In contrast, oxidized, but not reduced, Trx 2 interacts with gyrase B and decreases its supercoiling activity. Because a reduced supercoiling leads to decreased puf and puc transcription (Zhu and Hearst 1988), a reduction of oxygen tension results in increased gyrase activity and, consequently, in increased puf and puc transcription. The same opposite effect of Trx 1 and Trx 2 on gyrase activity was observed in E. coli (Li et al. 2004b). This strongly suggests that the gyrase-mediated effect of thioredoxins on gene expression is a common redox-dependent signalling pathway in bacterial adaptation. Based on the above-mentioned model, one can also speculate that by regulating photosynthesis genes via gyrase, thioredoxins may also be involved in the regulation of ROS generation in Rhodobacter. Because the simultaneous presence of pigments, light, and oxygen results in the formation of toxic ROS, the effect of thioredoxins on the gyrase activity decreases the expression of photosynthesis genes under high oxygen tension and therefore limits the generation of ROS.
Model for redox regulation on gene expression through thioredoxins as established for the thioredoxins of R. sphaeroides and R. capsulatus. The redox status of the cell determines the ratio of reduced to oxidized thioredoxin. The redox switch of thioredoxins alters the supercoiling activity of gyrase, which further affects gene expression. Reduced but not oxidized thioredoxin A binds to gyrase and increases its supercoiling activity. Oxidized but not reduced thioredoxin C binds to gyrase and decreases its supercoiling activity. TrxA thioredoxin A, TrxC thioredoxin C, GyrB subunit B of gyrase. S2 and (SH)2 indicate oxidized or reduced redox state of thioredoxins, respectively
Concluding remarks
The elucidation of the regulation of bacterial thioredoxin genes and the effects of thioredoxin on gene regulation is still in an early phase. Nevertheless, the available data demonstrate that thioredoxins are parts of complex regulatory networks that control the bacterial oxidative stress response and, most likely also, many additional physiological functions. The intensified studies on the functions of bacterial thioredoxins will most likely reveal further strategies to build up such regulatory networks to maintain many important cellular functions under changing environmental conditions.
References
Arner ES, Holmgren A (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267:6102–6109
Aslund F, Beckwith J (1999) The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. j Bacteriol 181:1375–1379
Aslund F, Berndt KD, Holmgren A (1997) Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein–protein redox equilibria. J Biol Chem 272:30780–30786
Bae JB, Park JH, Hahn MY, Kim MS, Roe JH (2004) Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide band formation. J Mol Biol 335:425–435
Boschi-Muller S, Azza S, Sanglier-Cianferani S, Talfournier F, van Dorsselear A, Branlant G (2000) A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli. J Biol Chem 275:35908–35913
Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD (1998) Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol 29:189–198
Capitani G, Markovic-Housley Z, DelVal G, Morris M, Jansonius JN, Schurmann P (2000) Crystal structures of two functionally different thioredoxins in spinach chloroplasts. J Mol Biol 302:135–154
Carmel-Harel O, Storz G (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol 54:439–461
Chae HZ, Chung SJ, Rhee SG (1994) Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269:27670–27678
Clement-Metral JD (1979) Activation of ALA synthetase by reduced thioredoxin in Rhodopseudomonas sphaeroides Y. FEBS Lett 101:116–120
Collet JF, D Souza JC, Jakob U, Bardwell JC (2003) Thioredoxin 2, an oxidative stress-induced protein, contains a high affinity zinc binding site. J Biol Chem 14:45325–45332
Comtois SL, Gidley MD, Kelly DJ (2003) Role of thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology 149:121–129
Das KC, Das CK (2000) Thioredoxin, a singlet oxygen quencher and hydroxyl radical scavenger: redox independent functions. Biochem Biophys Res Commun 277:443–447
Demple B (1996) Redox signalling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53–57
Ding H, Demple B (1998) Thiol-mediated disassembly and reassembly of [2Fe-2S] clusters in the redox-regulated transcription factor SoxR. Biochemistry 49:17280–17286
Dorman CJ, Barr GC, Bhriain NN, Higgins CF (1988) DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J Bacteriol 170:2816–2826
Eklund H, Gleason FK, Holmgren A (1991) Structural and functional relations among thioredoxins of different species. Proteins 11:13–28
Franco RJ, Drlica K (1989) Gyrase inhibitors can increase gyrA expression and DNA supercoiling. J Bacteriol 171:6573–6579
Gallardo-Madueno R, Leal JF, Dorado G, Holmgren, Lopez-Barea J, Pueyo C (1998) In vivo transcription of nrdAB operon and of grxA and fpg genes is triggered in Escherichia coli lacking both thioredoxin and glutaredoxin 1 or thioredoxin and glutathione, respectively. J Biol Chem 273:18382–18388
Garrido EO, Grant CM (2002) Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol Microbiol 43:993–1003
Gelhaye E, Rouhier N, Navrot N, Jacquot JP (2005) The plant thioredoxin system. Cell Mol Life Sci 62:24–35
Gonzalez-Flecha B, Demple B (1995) Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem 270:13681–13687
Gonzalez Porqué P, Baldesten A, Reichard P (1970) The involvement of the thioredoxin system in the reduction of methionine sulfoxide and sulphate. J Biol Chem 245:2371–2374
Hausladen A, Privalle CT, Keng T, DeAngelo J, Stamler JS (1996) Nitrosative stress: activation of the transcriptional factor OxyR. Cell 86:719–729
Herbig AF, Helmann JD (2001) Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol 44:849–859
Holmgren A (1985) Thioredoxin. Annu Rev Biochem 54:237–271
Holmgren A (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264:13963–13966
Holmgren A (1995a) Thioredoxin and glutaredoxin systems. J Biol Chem 3:233–316
Holmgren A (1995b) Thioredoxin structure and mechanism: conformational changes on oxidation of the active-site sulfhydryls to a disulfide. Structure 3:239–243
Huber HE, Tabor S, Richardson CC (1987) Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem 262:16224–16232
Imlay JA (2003) Pathways of oxidative stress damage. Annu Rev Microbiol 57:395–418
Imlay JA, Fridovich I (1991) Assay of metabolic superoxide production in Escherichia coli. J Biol Chem 266:6957–6965
Jobin MP, Garmyn D, Divies C, Guzzo J (1999) Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiology 145:1245–1251
Kang SW, Chae, HZ, Seo, MS, Kim K, Baines IC, Rhee SG (1998) Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-alpha. J Biol Chem 273:6297–6302
Kang JG, Paget MSB, Seok YJ, Hahn MY, Bae JB, Hahn JS, Leanthous C, Buttner MJ, Roe JH (1999) RsrA, an anti-sigma factor regulated by redox change. EMBO J 18:4292–4298
Kim SO, Merchant K, Nudelman R, Beyer WF Jr, Keng T, DeAngelo J, Hausladen A, Stamler JS (2002) OxyR: a molecular cod for redox-related signaling. Cell 109:383–396
Kontou M, Will RD, Adelfalk C, Wittig R, Poustka A, Hirsch-Kauffmann M, Schweiger M (2004) Thioredoxin, a regulator of gene expression. Oncogene 23:2146–2152
Krause G, Lundstrom J, Barea JL, Pueyo de la Cuesta C, Holmgren A (1991) Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J Biol Chem 266:9494–9500
Kumar JK, Tabor S, Richardson CC (2004) Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc Natl Acad Sci U S A 101:3759–3764
Laurent TC, Moore EC, Reichard P (1964) Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli. J Biol Chem 239:3436–3444
Leichert LI, Scharf C, Hecker M (2003) Global characterization of disulfide stress in Bacillus subtilis. J Bacteriol 185:1967–1975
Li W, Stevenson CE, Burton N, Jakimowicz P, Paget MS, Buttner MJ, Lawson DM, Kleanthous C (2002) Identification and structure of the anti-sigma factor-binding domain of the disulphide-stress regulated sigma factor σR from Streptomyces coelicolor. J Mol Biol 323:225–236
Li K, Haertig E, Klug G (2003a) Thioredoxin 2 is involved in oxidative stress defense and redox-dependent expression of photosynthesis genes in Rhodobacter capsulatus. Microbiology 149:419–430
Li K, Pasternak C, Klug G (2003b) Expression of the trxA gene for thioredoxin 1 in Rhodobacter sphaeroides during oxidative stress. Arch Microbiol 180:484–489
Li W, Bottrill AR, Bibb MJ, Buttner MJ, Paget MS, Kleanthous C (2003c) The role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J Mol Biol 333:461–472
Li K, Hein S, Zou W, Klug G (2004a) The glutathione–glutaredoxin system in Rhodobacter capsulatus: part of a complex regulatory network controlling defense against oxidative stress. j Bacteriol 186:6800–6808
Li K, Pasternak C, Hartig E, Haberzettl K, Maxwell A, Klug G (2004b) Thioredoxin can influence gene expression by affecting gyrase activity. Nucleic Acids Res 32:4563–4575
Lillig CH, Prior A, Schwenn JD, Aslund F, Ritz D, Vlamis-Gardikas A, Holmgren A (1999) New thioredoxins and glutaredoxins as electron donors of 3′-phosphoadenylylsulfate reductase. J Biol Chem 274:7695–7698
Lim CJ, Daws T, Gerami-Nejad M, Fuchs JA (2000) Growth-phase regulation of the Escherichia coli thioredoxin gene. Biochim Biophys Acta 25:1–6
Lindahl M, Florencio FJ (2003) Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc Natl Acad Sci U S A 100:16107–16112
Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I (2002) Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tubercolosis global gene expression. Mol Microbiol 45:365–374
Mark DF, Richardson CC (1976) Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A 73:780–784
Martin JL (1995) Thioredoxin—a fold for all reasons. Structure 3:245–250
McGee DJ, Kumar S, Viator RJ, Bolland JR, Ruiz J, Spadafora D, Testerman TL, Kelly DJ, Pannell LK, Windle HJ (2006) Helicobacter pylori thioredoxin is an arginase chaperone and guardian against oxidative and nitrosative stresses. J Biol Chem 281:3290–3296
Messner KR, Imlay JA (1999) The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem 274:10119–10128
Michan C, Manchado M, Dorado G, Pueyo C (1999) In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. j Bacteriol 181:2759–2764
Miranda-Vizuete A, Damdimopoulos AE, Gustafsson J, Spyrou G (1997) Cloning, expression, and characterization of a novel Escherichia coli thioredoxin. J Biol Chem 272:30841–30847
Muller EG, Buchanan BB (1989) Thioredoxin is essential for photosynthetic growth. The thioredoxin m gene of Anacystis nidulans. J Biol Chem 264:4008–4014
Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P (2003a) A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc Natl Acad Sci U S A 100:4233–4238
Nakano S, Kuster-Schock E, Grossman AD, Zuber P (2003b) Spx-dependent global transcriptional control is induced by thiol-specific stress in Bacillus subtilis. Proc Natl Acad Sci USA 100:13603–13608
Nakano S, Erwin KN, Ralle M, Zuber P (2005) Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol 55:498–510
Navarro F, Florencio FJ (1996) The cyanobacterial thioredoxin gene is required for both photoautotrophic and heterotrophic growth. Plant Physiol 111:1067–1075
Nunoshiba T (1996) Two-stage gene regulation of the superoxide stress response soxRS system in Escherichia coli. Crit Rev Eukaryot Gene Expr 6:377–389
Orr MD, Vitols E (1966) Thioredoxin from Lactobacillus leichmannii and its role as hydrogen donor for ribonucleoside triphosphate reductase. Biochem Biophys Res Commun 25:109–115
Paget MS, Buttner MJ (2003) Thiol-based regulatory switches. Annu Rev Genet 37:91–121
Paget MS, Kang J-G, Roe J-H, Buttner MJ (1998) SigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor. EMBO J 17:5776–57782
Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ (2001) Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigma R regulon. Mol Microbiol 42:1007–1020
Pasternak C, Assemat K, Breton AM, Clement-Metral JD, Klug G (1996) Expression of the thioredoxin gene (trxA) in Rhodobacter sphaeroides Y is regulated by oxygen. Mol Gen Genet 250:189–196
Pasternak C, Assemat K, Clement-Metral JD, Klug G (1997) Thioredoxin is essential for Rhodobacter sphaeroides growth by aerobic and anaerobic respiration. Microbiology 143:83–91
Pasternak C, Haberzettl K, Klug G (1999) Thioredoxin is involved in oxygen-regulated formation of the photosynthetic apparatus of Rhodobacter sphaeroides. j Bacteriol 181:100–106
Prieto-Alamo MJ, Jurado J, Gallardo-Madueno R, Monje-Casas F, Holmgren A, Pueyo C (2000) Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J Biol Chem 275:13398–13405
Raman S, Song T, Puyang X, Bardarov S, Jacobs WR, Husson RN (2001) The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. j Bacteriol 183:6119–6125
Ritz D, Patel H, Doan B, Zheng M, Aslund F, Storz G, Beckwith J (2000) Thioredoxin 2 is involved in the oxidative stress response in E. coli. J Biol Chem 275:2502–2512
Ritz D, Beckwith J (2001) Roles of thiol-redox pathways in bacteria. Annu Rev Microbiol 55:21–48
Russel M, Model P (1985) Thioredoxin is required for filamentous phage assembly. Proc Natl Acad Sci U S A 82:29–33
Russel M, Model P, Holmgren A (1990) Thioredoxin or glutaredoxin in Escherichia coli is essential for sulfate reduction but not for deoxyribonucleotide synthesis. j Bacteriol 172:1923–1929
Sa JH, Namgung MA, Lim CJ, Fuchs JA (1997) Expression of the Escherichia coli thioredoxin gene is negatively regulated by cyclic AMP. Biochem Biophys Res Commun 234:564–567
Scharf C, Riethdorf S, Ernst H, Engelmann S, Volker U, Hecker M (1998) Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. j Bacteriol 180:1869–1877
Schneider R, Travers A, Muskhelishvili G (2000) The expression of the Escherichia coli fis gene is strongly dependent on the superhelical density of DNA. Mol Microbiol 38:167–175
Seaver LC, Imlay JA (2004) Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J Biol Chem 279:48742–48750
Sies H (1985) Oxidative stress. Academic, London
Smits WK, Dubois JY, Bron S, van Dijl JM, Kuipers OP (2005) Tricksy business: transcriptome analysis reveals the involvement of thioredoxin A in redox homeostasis, oxidative stress, sulfur metabolism, and cellular differentiation in Bacillus subtilis. j Bacteriol 187:3921–3930
Spector A, Yan GZ, Huang RR, McDermott MJ, Gascoyne PR, Pigiet V (1988) The effect of H2O2 upon thioredoxin-enriched lens epithelial cells. J Biol Chem 263:4984–4990
Storz G, Zheng M (2000) Oxidative stress. In: Bacterial stress responses. ASM Press, Washington, DC, pp 47–59
Storz G, Tartaglia LA, Ames BN (1990) Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189–194
Tao K, Fujita N, Ishihama A (1993) Involvement of the RNA polymerase alpha subunit C-terminal region in co-operative interaction and transcriptional activation with OxyR protein. Mol Microbiol 7:859–864
Toledano MB, Kullik I, Trinh F, Baird PT, Schneider T, Storz G (1994) Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897–909
Uziel O, Borovok I, Schreiber R, Cohen G, Aharonowitz Y (2004) Transcriptional regulation of the Staphylococcus areus thioredoxin and thioredoxin genes in response to oxygen and disulfide stress. j Bacteriol 186:326–334
Varghese S, Tang Y, Imlay JA (2003) Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. j Bacteriol 185:221–230
Vido K, Diemer H, Van Dorsselaer A, Leize E, Juillard V, Gruss A, Gaudu P (2005) Roles of thioredoxin reductase during the aerobic life of Lactococcus lactis. j Bacteriol 187:601–610
Wieles B, Ottenhoff TH, Steenwijk TM, Franken KL, de Vries RR, Langermans JA (1997) Increased intracellular survival of Mycobacterium smegmatis containing the Mycobacterium leprae thioredoxin–thioredoxin reductase gene. Infect Immunol 65:2537–2541
Zheng M, Aslund F, Storz G (1998) Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718–1721
Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. j Bacteriol 183:4562–4570
Zhu YS, Hearst JE (1988) Transcription of oxygen-regulated photosynthetic genes requires DNA gyrase in Rhodobacter capsulatus. Proc Natl Acad Sci U S A 85:4209–4213
Acknowledgements
The work from the authors was supported by the Deutsche Forschungsgemeinschaft (Kl563/16), the Fonds der chemischen Industrie, the BMBF, and the DAAD.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zeller, T., Klug, G. Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften 93, 259–266 (2006). https://doi.org/10.1007/s00114-006-0106-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00114-006-0106-1