Abstract
Previous studies have identified miR-182, miR-27a, FoxO1, and IL2RA as regulatory factors for Treg cell development and function. In order to investigate the association of miR-182, miR-27a, FoxO1, and IL2RA gene polymorphisms with Behçet’s disease (BD) and Vogt–Koyanagi–Harada (VKH) syndrome in a Chinese Han population, a two-stage association study was performed in 820 BD, 900 VKH patients, and 1,800 controls using polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) assay. In the first stage study, association analysis of 10 single nucleotide polymorphisms (SNPs) was performed in 400 BD, 400 VKH patients, and 600 controls. The results showed significantly decreased frequencies of the miR-182/rs76481776 CC genotype and C allele in BD (P = 3.36 × 10−4, OR = 0.55; P = 4.74 × 10−4, OR = 0.59) and VKH patients (P = 1.11 × 10−4, OR = 0.53; P = 1.26 × 10−4, OR = 0.56). No significant association of the other nine SNPs with BD or VKH was observed. In the second stage study, association analysis of miR-182/rs76481776 was performed in 420 BD, 500 VKH patients, and 1,200 controls. The second stage and combined studies confirmed the association of miR-182/rs76481776 with BD (CC genotype: P = 3.25 × 10−7, OR = 0.58; C allele: P = 1.81 × 10−7, OR = 0.60) and VKH (CC genotype: P = 7.89 × 10−8, OR = 0.57; C allele: P = 2.52 × 10−8, OR = 0.59). Real-time PCR analysis showed a significantly increased expression of miR-182 in TT/CT cases compared to CC cases in anti-CD3/CD28 antibodies-stimulated CD4+ T cells (P = 2.1 × 10−2). In conclusion, this study suggests that miR-182, but not miR-27a, FoxO1, and IL2RA, contributes to the genetic susceptibility of BD and VKH.
Key Message
-
MiR-182 contributes to genetic susceptibility of BD and VKH.
-
No significant association of miR-27a, FoxO1, and IL2RA with BD or VKH was observed.
-
Significantly increased expression of miR-182 in TT/CT cases compared to CC cases was observed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Behçet’s disease (BD) is a chronic inflammatory disease characterized by recurrent uveitis, oral aphthae, typical skin lesion, and genital ulcerations [1]. It is more prevalent in Chinese, Japanese, and Turkish ethnic populations as compared to Caucasian populations in the UK and USA [2]. Vogt–Koyanagi–Harada (VKH) syndrome is an autoimmune disorder characterized by bilateral granulomatous panuveitis frequently associated with alopecia, vitiligo, poliosis, and central nervous system symptoms [3]. It frequently affects Asians and Native Americans. Although the precise etiology and pathogenesis of both diseases remain unknown, the role of genetic factors in their pathogenesis has been extensively acknowledged [4]. Recent surveys have identified that IL23R, miR-146a, and EGF/ErbB signaling pathway genes confer risk of BD [5–7]. CTLA-4 and IL-17 polymorphisms have been reported to be associated with VKH disease [8, 9]. These reports suggest that genetic factors may play an important role in both diseases.
Regulatory T cells (Treg) are a subpopulation of T cells which modulate the immune system, maintain tolerance to self-antigens, thereby controlling the occurrence of overt autoimmune disease. Recent studies have shown that the transcription factor forkhead-box O1 (FoxO1) controls Treg cell development and function [10, 11]. MiR-27a, miR-96, and miR-182 can target the 3′ UTR of FoxO1, which is then degraded, resulting in a decrease in Treg cells and subsequent clonal expansion of helper T cells [12, 13]. Meanwhile, miR-182 can be induced by IL-2 receptor-alpha (IL2RA, also known as CD25) which regulates Treg specialization and stability [13–15]. Recently, miR-182 (rs76481776), miR-27a (rs895819), and IL2RA (rs706778, rs3118470, rs2104286, and rs7093069) have been considered as genetic predisposing factors for various diseases [16–21]. To the best of our knowledge, none of the SNPs of FoxO1 has been reported to be significantly associated with autoimmune-related diseases. In addition, SNPs for FoxO1 (rs2297626, rs17592236, rs9549241, and rs12585277) showed a lack of association with type 2 diabetes [22, 23]. As yet, no common SNPs (minor allele frequency >0.05) have been found in miR-96, which as mentioned above, is also a regulator of FoxO1 function.
In view of the fact that these genes play an important role in Treg cell development and function and since the level of Treg cells is associated with active uveitis in patients with VKH syndrome [24], we investigated whether polymorphisms of these genes were associated with uveitis. We chose to examine this question in two uveitis entities, BD and VKH, which are relatively common in China so as to obtain a sufficient sample size. Our results showed that miR-182, but not miR-27a, FoxO1, and IL2RA, contributes to the genetic susceptibility of BD and VKH.
Materials and methods
Study population
The study group comprised 1,720 unselected, consecutive patients (820 BD patients and 900 VKH disease patients) who were recruited from the First Affiliated Hospital of Chongqing Medical University (Chongqing, China) and the Zhongshan Ophthalmic Center of the Sun Yat-sen University (Guangzhou, China) between October 2006 and January 2013. A total of 1,800 unselected, consecutive control subjects were matched ethnically and geographically with the patients. The diagnosis for BD and VKH disease was strictly based on the criteria of International Study Group for BD and First International Workshop for VKH [25], respectively. The study received the approval of the Local Ethics Research Committee and all the investigated subjects provided informed consent before collection of blood. The tenets of the Declaration of Helsinki were conducted during all procedures of this study.
DNA extraction and genotyping
Genomic DNA samples of BD, VKH patients, and healthy controls were extracted by using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA). The target DNA sequence was amplified by the polymerase chain reaction (PCR) using proper primers [17–22] (Table S1). PCR products were digested with 3 U of PdiI, DraIII, AluI, Eco47I, BshNI, HindIII, TscAI, NdeI, BsmAI, BstNI (Fermentas, Shenzhen, China), and RsaI (Promega, Madison, WI, USA) restriction enzymes in a 10-μL reaction volume overnight. Digestion products were separated on 4 % agarose gels and stained with GoldView™ (SBS Genetech Beijing, China).
Cell isolation and culture
The peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood samples by Ficoll-Hypaque density-gradient centrifugation. Isolated PBMCs cells (2 × 106 cells per well) were seeded in 24-well plates and cultured in RPMI medium 1640 supplemented with 10 % fetal calf serum (FCS, Greiner, Wemmel, Belgium), 100 U/mL penicillin, and 100 μg/mL streptomycin. PBMCs were also cultured with 100 ng/mL LPS (100 ng/mL, Sigma, Missouri, USA) for 24 h. Magnetic beads (Miltenyi Biotec, Palo Alto, CA) were used to isolate CD4+ T cells according to the manufacturer’s protocol. Purified CD4+ T cells were treated with anti-CD3/CD28 antibodies (5:1) (Miltenyi Biotec, Palo Alto, CA) at 37 °C for 72 h.
Real-time PCR
Total RNA was extracted from PBMCs obtained from normal controls using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), followed by reverse transcription using a transcriptase kit (Applied Biosysterms, ABI, Foster City, CA). Real-time PCR was performed on the 7500 System (ABI) based on the SYBR-Green method. Bulge-loop™ qPCR primers for miR-182 and U6 were synthesized by Ribobio (Guangzhou, China). The expression of FoxO1 was examined using the following primers: forward 5′-AAGAGCGTGCCCTACTTCAA-3′ and reverse 5′-CTGTTGTTGTCCATGGAT GC-3′. β-Actin was chosen as the internal reference gene and its expression was detected by the following primers: forward 5′-GGATGCAGAAGGAGATCAC TG-3′ and reverse 5′-CGATCCACACGGAGTACTTG-3′. All reactions were conducted in triplicate. Relative expression levels were calculated using the 2−ΔΔCt method.
Statistical analysis
For single SNP analysis, genotype and allele frequencies were compared between patients and controls by the Chi-square test using SPSS version 17.0 (SPSS, Inc., Chicago, IL). The χ 2 test was also applied to analyze the Hardy–Weinberg equilibrium (HWE). Odds ratios (OR) and 95 % confidence intervals (95 % CI) were calculated using SPSS version 17.0 to estimate disease risk. Genotype test was performed by one genotype versus the other two pooled together. P values were corrected with the Bonferroni correction by multiplying with the number of analyses performed. The number of independent comparisons is 40. A P value less than 1.25 × 10−3 (0.05/40) was considered to be statistically significant. The independent samples t test or nonparametric Mann–Whitney U test was used to compare miR-182 and FoxO1 expression levels among three genotype groups.
Results
Clinical features of BD patients and VKH patients
The detailed demographic characteristics and clinical features of enrolled BD and VKH patients are shown in Table S2. The distribution of genotype frequencies of the 10 SNPs investigated did not deviate from the Hardy–Weinberg equilibrium in the controls.
Allele and genotype frequencies of tested SNPs in patients and controls in the first stage study
Ten SNPs were genotyped in 400 BD patients, 400 VKH patients, and 600 normal controls for the first stage study. Our results showed significantly decreased frequencies of the miR-182/rs76481776 CC genotype and C allele in BD (P = 3.36 × 10−4, OR = 0.55; P = 4.74 × 10−4, OR = 0.59) and VKH patients (P = 1.11 × 10−4, OR = 0.53; P = 1.26 × 10−4, OR = 0.56) (Tables 1 and 2). However, none of the other nine SNPs showed a significant association with BD and VKH disease (Table S3 and S4).
Allele and genotype frequencies of tested SNPs in patients and controls in the second stage and combined studies
In order to validate the significant association between miR-182/ rs76481776 and BD or VKH found in the first stage, another 420 BD patients, 500 VKH patients, and 1,200 controls were enrolled in the second stage study. The results again showed significantly decreased frequencies of the miR-182/rs76481776 CC genotype and C allele in BD (P = 8.00 × 10−4, OR = 0.61; P = 3.20 × 10−4, OR = 0.62) and VKH (P = 4.16 × 10−4, OR = 0.61; P = 1.28 × 10−4, OR = 0.61) (Tables 1 and 2). The combined data confirmed the association of rs76481776 with BD (CC genotype: P = 3.25 × 10−7, OR = 0.58; C allele: P = 1.81 × 10−7, OR = 0.60) and VKH (CC genotype: P = 7.89 × 10−8, OR = 0.57; C allele: P = 2.52 × 10−8, OR = 0.59) (Tables 1 and 2).
Stratified analysis for miR-182/ rs76481776 with main clinical features of BD and VKH disease
A stratified analysis was performed to examine the association of rs76481776 with the main clinical features of BD and VKH disease. The main clinical manifestations of BD included genital ulcer, skin lesions, arthritis, positive pathergy reaction, and hypopyon. The main clinical features of VKH consisted of headache, tinnitus, vitiligo, poliosis, and alopecia. We could not demonstrate a significant association between the rs76481776 genotype frequency and any clinical manifestation of BD or VKH (Table S5 and S6).
The influence of rs76481776 on miR-182 and FoxO1 expression
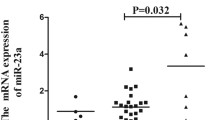
The aforementioned result showed a significant association of miR-182/ rs76481776 with BD and VKH. To investigate a possible function associated with this SNP, we performed real-time PCR analysis to evaluate its effect on the expression of miR-182 using PBMCs derived from 24 healthy individuals with known genotype. The results did not reveal an effect of the various rs76481776 genotypes on miR-182 expression (Fig. S1, P > 0.05). Furthermore, there was no significant association in the expression of miR-182 by LPS-stimulated PBMCs and the different genotypes (Fig. S1, P > 0.05). We subsequently examined whether the expression of pre-miR-182 and mature miR-182 was affected by the various rs76481776 genotypes in purified CD4+ T cells and anti-CD3/CD28 antibodies-stimulated CD4+ T cells. The results showed no significant differences of pre-miR-182 expression between rs76481776 CC cases and TT/CT cases in purified CD4+ T cells and anti-CD3/CD28 antibodies-stimulated CD4+ T cells (Fig. 1, P > 0.05). Our results also showed no significant difference in the miR-182 expression between rs76481776 CC cases and TT/CT cases by purified CD4+ T cells (Fig. 2, P > 0.05). However, the results showed a significantly increased expression of miR-182 in TT/CT cases compared to CC cases in anti-CD3/CD28 antibodies-stimulated CD4+ T cells (Fig. 2, P = 2.1 × 10−2). We also measured the expression of miR-182*, which is the less predominantly expressed mature form originating from the passenger strand of the pre-miR-182. Unfortunately, it was not detectable in both purified CD4+ T cells and anti-CD3/CD28 antibodies-stimulated CD4+ T cells. Moreover, we examined the expression of FoxO1 between miR-182/rs76481776 CC cases and TT/CT cases in purified CD4+ T cells and anti-CD3/CD28 antibodies-stimulated CD4+ T cells. Although the results showed lower FoxO1 expression in the miR-182/rs76481776 TT/CT genotypes than CC genotypes both in purified CD4+ T cells and anti-CD3/CD28 antibodies-stimulated CD4+ T cells, there were no statistically significant differences between the two groups (Fig. S2, P > 0.05).
Discussion
In the present study, we performed a two-stage case–control assay to investigate the association of 10 SNPs of miR-182, miR-27a, FoxO1, and IL2RA with BD and VKH diseases in a Chinese Han population. Our results showed that miR-182, but not FoxO1, miR-27a, and IL2RA, contributes to the genetic susceptibility of BD and VKH syndrome.
MiR-182 plays a dominant role in the regulation of adaptive immune responses and binds to a specific site of the 3′UTR of FoxO1 mRNA and reduces its expression [14, 26, 27]. Polymorphism analysis reported a significant association between the rs76481776 polymorphism in the pre-miR-182 and late insomnia in major depression patients [20]. To the best of our knowledge, the association between miR-182/rs76481776 polymorphisms and autoimmune-related diseases has not yet been reported, and there were no published genome-wide association studies (GWAS) for VKH. Although there were several published GWAS for BD [28–30], including Chinese datasets, no evidence of an association between miR-182/rs76481776 and BD was reported. Our result showed that the CC genotype of miR-182/rs76481776 had a significantly decreased frequency in both BD and VKH patients despite the fact that both diseases are quite different in pathogenesis. The fact that miR-182 is critical for Treg cell development may expand our knowledge concerning the role of these cells in the pathogenesis of BD and VKH and is in agreement with earlier findings showing a decreased level of Treg cells in VKH uveitis patients [24]. Moreover, we found a significantly increased expression of miR-182 in rs76481776 TT/CT cases compared to CC cases in anti-CD3/CD28 antibodies-stimulated CD4+ T cells, which confirmed recent data by Saus et al. [20]. However, the pre-miR-182 was not differentially expressed between rs76481776 CC cases and TT/CT cases either in purified CD4+ T cells or anti-CD3/CD28 antibodies-stimulated CD4+ T cells. Therefore, it implies that the processing of pre-miR-182 may be more efficient in the miR-182/rs76481776 TT/CT genotypes and that it does not affect the transcription of the miRNA. Although lower expression of FoxO1 was found in the miR-182/rs76481776 TT/CT cases than CC cases both in purified CD4+ T cells and anti-CD3/CD28 antibodies-stimulated CD4+ T cells, no significant association was observed. Further studies are needed to investigate whether polymorphisms of miR-182 can influence the expression of its target genes such as FoxO1. A recent study showed that miR-182 was identified as a sensory organ-specific microRNA and that it is highly expressed in the mouse retina [31]. Whether the association of miR-182/rs76481776 with BD and VKH syndrome disclosed by our study is linked to the miR-182 expression in the retina rather than an effect on the differentiation of regulatory T cells is an intriguing subject. More studies are needed to address this issue.
FoxO1, one of the four FoxO isoforms of Forkhead transcription factors, controls Treg cell development and function by binding to the promoter regions of Foxp3 and CTLA-4 genes [10, 11]. Previous studies suggested that SNPs of FoxO1 (rs2297626, rs17592236, rs9549241, and rs12585277) were not associated with type 2 diabetes [22, 23]. We also did not find a direct association between SNPs of FoxO1 with uveitis in either BD or VKH.
MiR-27a can target the 3′UTR of FoxO1, which is then degraded, resulting in a decrease in Treg cell numbers and helper T cell clonal expansion [12, 13]. Recent surveys have identified that miR-27a/ rs895819 is associated with gastric cancer, renal cell cancer, and breast cancer [17, 32]. In this study, we did not find an association between miR-27a/ rs895819 and BD or VKH disease.
IL2RA, also known as CD25, can induce miR-182 by constitutively activating STAT5. Previous studies showed that the IL2RA gene was associated with various autoimmune-related diseases, such as type 1 diabetes, rheumatoid arthritis, and multiple sclerosis [19, 33, 34]. In our study, SNPs for IL2RA (rs706778, rs3118470, rs2104286, and rs7093069) were not associated with neither BD nor VKH disease. This finding is in agreement with a recent study that showed that several SNPs of IL2RA (rs2104286, rs11594656, and rs12722495) were not associated with endogenous non-anterior uveitis [35].
Our study has a number of limitations. First of all, although we tried our best to match the controls for gender, it was not achieved in the BD group, which has an overrepresentation of male patients. Further studies should be validated in a gender-matched population. A limited number of SNPs of FoxO1 and IL2RA were tested in our study and it is possible that other as yet unknown SNPs might be involved. We limited our study to BD and VKH, and our data need to be confirmed in other ethnic populations but also in other uveitis entities.
In conclusion, our results identified that miR-182, but not miR-27a, FoxO1, and IL2RA, contributes to the genetic susceptibility of BD and VKH syndrome. Further research towards understanding the role of miR-182 and the biochemical pathways are needed to investigate during the development of BD and VKH.
References
Yang P, Fang W, Meng Q, Ren Y, Xing L, Kijlstra A (2008) Clinical features of chinese patients with Behcet's disease. Ophthalmology 115:312–318
Khairallah M, Accorinti M, Muccioli C, Kahloun R, Kempen JH (2012) Epidemiology of Behcet disease. Ocul Immunol Inflamm 20:324–335
Yang P, Ren Y, Li B, Fang W, Meng Q, Kijlstra A (2007) Clinical characteristics of Vogt–Koyanagi–Harada syndrome in Chinese patients. Ophthalmology 114:606–614
Gul A, Inanc M, Ocal L, Aral O, Konice M (2000) Familial aggregation of Behcet's disease in Turkey. Ann Rheum Dis 59:622–625
Jiang Z, Yang P, Hou S, Du L, Xie L, Zhou H, Kijlstra A (2010) IL-23R gene confers susceptibility to Behcet's disease in a Chinese Han population. Ann Rheum Dis 69:1325–1328
Zhou Q, Hou S, Liang L, Li X, Tan X, Wei L, Lei B, Kijlstra A, Yang P (2014) MicroRNA-146a and Ets-1 gene polymorphisms in ocular Behcet's disease and Vogt–Koyanagi–Harada syndrome. Ann Rheum Dis 73:170–176
Xavier JM, Krug T, Davatchi F, Shahram F, Fonseca BV, Jesus G, Barcelos F, Vedes J, Salgado M, Abdollahi BS et al (2013) Gene expression profiling and association studies implicate the neuregulin signaling pathway in Behcet's disease susceptibility. J Mol Med 91:1013–1023
Du L, Yang P, Hou S, Lin X, Zhou H, Huang X, Wang L, Kijlstra A (2008) Association of the CTLA-4 gene with Vogt–Koyanagi–Harada syndrome. Clin Immunol 127:43–48
Shu Q, Yang P, Hou S, Li F, Chen Y, Du L, Jiang Z (2010) Interleukin-17 gene polymorphism is associated with Vogt–Koyanagi–Harada syndrome but not with Behcet's disease in a Chinese Han population. Hum Immunol 71:988–991
Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch'en IL, Stockmann C, Katayama CD, Hedrick SM (2010) Foxo transcription factors control regulatory T cell development and function. Immunity 33:890–904
Ouyang W, Liao W, Luo CT, Yin N, Huse M, Kim MV, Peng M, Chan P, Ma Q, Mo Y et al (2012) Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature 491:554–559
Guttilla IK, White BA (2009) Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 284:23204–23216
O'Neill LA (2010) Outfoxing Foxo1 with miR-182. Nat Immunol 11:983–984
Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F et al (2010) The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol 11:1057–1062
Kelada S, Sethupathy P, Okoye IS, Kistasis E, Czieso S, White SD, Chou D, Martens C, Ricklefs SM, Virtaneva K et al (2013) miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS pathogens 9:e1003451
Mussig K, Staiger H, Machicao F, Stancakova A, Kuusisto J, Laakso M, Thamer C, Machann J, Schick F, Claussen CD et al (2009) Association of common genetic variation in the FOXO1 gene with beta-cell dysfunction, impaired glucose tolerance, and type 2 diabetes. J Clin Endocrinol Metab 94:1353–1360
Sun Q, Gu H, Zeng Y, Xia Y, Wang Y, Jing Y, Yang L, Wang B (2010) Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci 101:2241–2247
Klinker MW, Schiller JJ, Magnuson VL, Wang T, Basken J, Veth K, Pearce KI, Kinnunen L, Harjutsalo V, Wang X et al (2010) Single-nucleotide polymorphisms in the IL2RA gene are associated with age at diagnosis in late-onset Finnish type 1 diabetes subjects. Immunogenetics 62:101–107
Kawasaki E, Awata T, Ikegami H, Kobayashi T, Maruyama T, Nakanishi K, Shimada A, Uga M, Kurihara S, Kawabata Y et al (2009) Genetic association between the interleukin-2 receptor-alpha gene and mode of onset of type 1 diabetes in the Japanese population. J Clin Endocrinol Metab 94:947–952
Saus E, Soria V, Escaramis G, Vivarelli F, Crespo JM, Kagerbauer B, Menchon JM, Urretavizcaya M, Gratacos M, Estivill X (2010) Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet 19:4017–4025
Hinks A, Cobb J, Sudman M, Eyre S, Martin P, Flynn E, Packham J, Barton A, Worthington J, Langefeld CD et al (2012) Investigation of rheumatoid arthritis susceptibility loci in juvenile idiopathic arthritis confirms high degree of overlap. Ann Rheum Dis 71:1117–1121
Li T, Wu X, Zhu X, Li J, Pan L, Li P, Xin Z, Liu Y (2011) Association analyses between the genetic polymorphisms of HNF4A and FOXO1 genes and Chinese Han patients with type 2 diabetes. Mol Cell Biochem 353:259–265
Karim MA, Craig RL, Wang X, Hale TC, Elbein SC (2006) Analysis of FOXO1A as a candidate gene for type 2 diabetes. Mol Genet Metab 88:171–177
Chen L, Yang P, Zhou H, He H, Ren X, Chi W, Wang L, Kijlstra A (2008) Diminished frequency and function of CD4 + CD25high regulatory T cells associated with active uveitis in Vogt–Koyanagi–Harada syndrome. Invest Ophthalmol Vis Sci 49:3475–3482
Read RW, Holland GN, Rao NA, Tabbara KF, Ohno S, Arellanes-Garcia L, Pivetti-Pezzi P, Tessler HH, Usui M (2001) Revised diagnostic criteria for Vogt–Koyanagi–Harada disease: report of an international committee on nomenclature. Am J Ophthalmol 131:647–652
Ceribelli A, Satoh M, Chan EK (2012) MicroRNAs and autoimmunity. Curr Opin Immunol 24:686–691
Zhu S, Pan W, Qian Y (2013) MicroRNA in immunity and autoimmunity. J Mol Med 91:1039–1050
Remmers E, Cosan F, Kirino Y, Ombrello M, Abaci N, Satorius C, Julie M, Yang B, Korman B, Cakiris A et al (2010) Genome-wide association study indentifies variants in the MHC class I, IL 10, and IL23R/IL12RB2 regions associated with Behçet’s disease. Nat Genet 42:698–702
Hou S, Yang Z, Du L, Jiang Z, Shu Q, Chen Y, Li F, Zhou Q, Ohno S, Chen R et al (2012) Identification of a susceptibility locus in STAT4 for Behçet’s disease in Han Chinese in a genome-wide association study. Arthritis Rheum 64:4104–4113
Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, Ozyazgan Y, Sacli F, Erer B, Inoko H et al (2013) Genome-wide association study indentifies new susceptibility loci for Behçet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet 45:202–207
Xu S, Witmer P, Lumayag S, Kovacs B, Valle D (2007) MicroRNA (miRNA) transcriptome of mouse retina and indentification of a sensory organ-specific miRNA cluster. J Biol Chem 282:25053–25066
Shi D, Li P, Ma L, Zhong D, Chu H, Yan F, Lv Q, Qin C, Wang W, Wang M et al (2012) A genetic variant in pre-miR-27a is associated with a reduced renal cell cancer risk in a Chinese population. PLoS ONE 7:e46566
Schmied MC, Zehetmayer S, Reindl M, Ehling R, Bajer-Kornek B, Leutmezer F, Zebenholzer K, Hotzy C, Lichtner P, Meitinger T et al (2012) Replication study of multiple sclerosis (MS) susceptibility alleles and correlation of DNA-variants with disease features in a cohort of Austrian MS patients. Neurogenetics 13:181–187
Knevel R, de Rooy DP, Zhernakova A, Grondal G, Krabben A, Steinsson K, Wijmenga C, Cavet G, Toes RE, Huizinga TW et al (2013) Association of variants in IL2RA with progression of joint destruction in rheumatoid arthritis. Arthritis Rheum 65:1684–1693
Cenit MC, Marquez A, Cordero-Coma M, Fonollosa A, Adan A, Martinez-Berriotxoa A, Llorenc V, Diaz Valle D, Blanco R, Canal J et al (2013) Evaluation of the IL2/IL21, IL2RA and IL2RB genetic variants influence on the endogenous non-anterior uveitis genetic predisposition. BMC Med Gen 14:52
Acknowledgments
The authors would like to thank all donors enrolled in the present study. This work was supported by Natural Science Foundation Major International (Regional) Joint Research Project (81320108009), National Basic Research Program of China (973 Program) (2011CB510200), Key Project of Natural Science Foundation (81130019), National Natural Science Foundation Project (31370893, 81200678), Basic Research program of Chongqing (cstc2013jcyjC10001), Chongqing Key Laboratory of Ophthalmology (CSTC, 2008CA5003), National Clinical Key Department Construction Program, Key Project of Health Bureau of Chongqing (2012-1-003), and Fund for PAR-EU Scholars Program.
Conflict if interest
The authors declare no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Comparison of the relative expression levels of miR-182 variants. PBMCs and LPS-stimulated PBMCs samples of 12 CC genotype, 12 CT and TT genotype of rs76481776 were freshly obtained from healthy controls in the study (GIF 10 kb)
Fig. S2
The influence of various rs76481776 genotypes on expression of FoxO1. Purified CD4+ T cells and anti-CD3/CD28 antibodies-stimulated CD4+ T cells of 12 CC genotypes, 8 CT and TT genotypes of rs76481776 were freshly obtained from healthy controls in the study (GIF 19 kb)
ESM 1
(DOC 220 kb)
Rights and permissions
About this article
Cite this article
Yu, H., Liu, Y., Bai, L. et al. Predisposition to Behçet’s disease and VKH syndrome by genetic variants of miR-182. J Mol Med 92, 961–967 (2014). https://doi.org/10.1007/s00109-014-1159-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-014-1159-9