Abstract
Dilated cardiomyopathy (DCM) is a heart muscle disease characterized by impaired contractility and dilation of the ventricles. In a subset of DCM patients, classical inheritance patterns occur (familial DCM), which have led to the identification of specific genomic loci and gene defects causing monogenic DCM subtypes. In the majority of DCM patients, however, there is no evidence for a monogenic etiology of the disorder (sporadic DCM), and in the absence of other recognizable etiological factors, these cases were classified as “idiopathic”. Recent research suggests that cardiotropic viruses are important environmental factors in the pathogenesis of “idiopathic” cases and that DCM commonly results from interactions between genetic and environmental factors, whereas “pure” genetic forms are rather rare. Regarding genetics, the clinical cardiomyopathic phenotype associated with single gene defects may be highly variable for unknown reasons. Furthermore, a novel class of genetic defects was identified recently which provide a molecular basis for abnormal reactions of cardiomyocytes to environmental stress. These defects are paradigms of specific molecular links between genome and environment during the pathogenesis of DCM. Regarding environmental factors, a recent molecular virological study based on myocardial biopsies in a large series of sporadic DCM patients has detected cardiac viral infections in the majority of patients, with a broad spectrum of virus species being involved. Apparently, DCM does not only occur as a late sequela of acute viral myocarditis, but also in patients without clinical history of cardiac viral disease. Cardiotropic viruses thus emerge as prevalent environmental factors which may cause or influence the course of DCM in a large fraction of cases. Synopsis of current data suggests that a comprehensive picture of DCM pathogenesis can only be drawn if both genetic and environmental pathogenetic factors are considered. The course of cardiac viral infections depends strongly on genetic host factors and may range from rapid and complete virus elimination or silencing without clinical symptoms, to rapidly progressive or fatal disease. Viruses interact not only with genetically heterogenous host systems of virus uptake, migration, and antiviral immunity, but, due to their prevalence in DCM hearts, are also likely to encounter multiple structural proteins of cardiac cells known to be defective in familial DCM. The combined knowledge on DCM-associated gene defects and viruses therefore suggests in-depth studies on genome–environment interactions in DCM pathogenesis which may underlie the high clinical variability observed both in monogenic and virus-associated DCM and have implications for the clinical management of DCM patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
DCM as a complex disorder
The term “dilated cardiomyopathy” (DCM) is used for the clinical phenotypic description of a heart muscle disease characterized by impaired contractility and dilation of the ventricles. This phenotype encompasses a variety of pathogenetically distinct subtypes. Regarding the genetic basis of the disease, specific genomic loci and gene defects have been identified which cause DCM in the sense of classic monogenic disorders (familial DCM) in a subset of DCM patients. Disease genes have been identified by linkage analysis in DCM families or by mutation screening of candidate genes in DCM populations. Regarding environmental factors which may cause the disease, recent molecular virological investigations of large DCM populations based on myocardial biopsies have revealed chronic cardiac viral infections in the majority of cases, suggesting that viruses are important environmental factors in the pathogenesis of DCM. In the absence of any recognizable etiological factor (neither genetic nor environmental), the disorder is designated as “idiopathic,” but these cases become rarer as new genetic and environmental causes of DCM are being identified. We review current knowledge on monogenic DCM subtypes and cardiotropic viral infections associated with DCM. From a synopsis of the available genetic and virological data, we derive the need to pay close attention to genome–environment interactions in order to explain the highly variable clinical courses observed both in monogenic and virus-associated DCM.
Importantly, the full scale of variability cannot be appreciated during the initial studies of selected patient groups, but only after systematic evaluation of disease phenotype and course in large population-based studies. In other diseases, such stepwise revelation of the phenotype spectrum resulting from single gene defects has often led to the identification of genome–environment interactions of paramount importance for proper clinical management of mutation carriers. α1-Antitrypsin deficiency (ATD) may serve as a paradigm for this stepwise process. ATD was initially identified in patients with chronic obstructive lung disease (COPD), and the underlying gene defects were thought to cause this disease per se [1]. However, when mutation screening of patients with other diseases and normal control populations was conducted, it became apparent that ATD is associated with a very broad clinical spectrum ranging from normal lung function and morphology to rapidly destructive lung disease and early death [2, 3]. Cigarette smoking was identified as the key environmental factor determining both symptomatology and survival in mutation carriers [2]. Individuals with ATD may have normal life expectancy and minimal symptoms if they never smoke which has the important clinical implication that many mutations carriers need not be treated at all [2, 4]. Liver cirrhosis is another possible consequence of ATD due to a completely distinct pathogenic process [5] but occurs only very rarely if another independent gene defect is present in the same individual [6, 7].
In complex disorders such as COPD or DCM, a comprehensive picture of the pathogenesis can only be drawn if both genetic and environmental pathogenetic factors are considered. Broadened understanding of the pathogenesis may then decisively alter the clinical risk assessment and treatment of patients. One current challenge in DCM research is the identification of all environmental pathogenetic factors which act on the host genetic background. Cardiotropic viruses appear to be important in this respect.
Monogenic subtypes of DCM
About one third of DCM cases are currently estimated to have a genetic basis [8]. Numerous genomic loci have been mapped in different DCM families, and so far, 20 individual disease genes have been identified and screened for mutations in affected individuals. The genes involved in DCM encode a broad variety of proteins including sarcomeric, Z-disk-associated, sarcolemma cytoskeleton, intermediate filament, and calcium cycle proteins (Table 1). The identification of numerous monogenic subtypes of DCM was a major achievement of the past decade. On the other hand, the extensive heterogeneity of disease phenotypes, which are associated with mutations in a single gene and even in the same codon of that gene, is currently unexplained. Such heterogeneity has been observed in genetic animal models of DCM, but far more pronounced in DCM patients. Basically, it may be due to independent genetic factors in as yet unknown modifier genes or to environmental factors not investigated or not yet recognized in the affected individuals.
Variable phenotypic expression of single gene defects
Table 1 summarizes data documenting the very high phenotypic variability in single gene defects associated with DCM. Not included are the numerous complex genetic syndromes which include “pure” DCM as only one component since the vast majority of clinical DCM patients display cardiac dilation and dysfunction as an isolated phenotype. From this table on “pure” DCM, it is obvious that genetic and/or environmental cofactors need to be taken into account to obtain a comprehensive perspective on the complex pathogenesis of this disease. The full scale of phenotypic variability in single gene defects cannot be appreciated in the initial studies which are mostly genetic linkage analyses in DCM families recognized on the basis of a severe early onset phenotype in several members, with little variation on similar genetic backgrounds. Once the disease gene is mapped and sequenced, however, mutation scanning of that gene in large sporadic and familial DCM populations and also in healthy controls is required to assess the quantitative clinical relevance of that gene within the population and also the full phenotype spectrum associated with that gene. First steps towards broadened assessment have recently been made, e.g. for the β- and δ-sarcoglycan gene [9] and the lamin A/C gene [10]. In these studies, mutations of the respective genes were found to be very rare in population-based sporadic DCM groups. A recent survey of troponin C and T mutations in a major DCM series with 43% familial cases revealed 5% mutation carriers in the latter subgroup [11].
Investigation of large DCM collectives for mutations in a candidate gene may already give some impression of variability but still be severely biased by genetic founder effects and differences between geographically distant populations. In a recent study addressing these issues, we identified a mutation (W4R) of the MLP gene in ten out of 516 DCM patients from central Europe. W4R was not found among 320 European controls or among DCM patients or 400 controls from Japan. Haplotyping of the MLP mutation carriers confirmed a founder effect for central Europe. Thus, the MLP defect initially discovered by a candidate gene approach in our local patients is quantitatively relevant in parts of Europe but not elsewhere. A survey of lamin A/C mutations showed a similar founder effect for Finland [12]. Experimental investigations showed that the W4R mutation results in dysfunction of a cardiomyocyte stress sensor protein complex including MLP. This work led to the delineation of a novel DCM pathomechanism, but the clinical phenotype in mutation carriers was rather variable with age at diagnosis ranging from 32 to 70 years and NYHA class from I to III. This is consistent with W4R being a strong predisposing genetic factor whose actual sequelae, however, are dependent on environmental cofactors challenging the cellular stress sensor.
Similarly, extensive investigation of troponin gene mutations in cardiomyopathy patients has revealed that mutations in the same gene may be associated with either dilated or hypertrophic cardiomyopathy (HCM) (for review, see [13]). Moreover, mutations in the same codon (R145) of the troponin I gene were associated with either HCM or the completely distinct phenotype of restrictive cardiomyopathy (RCM) [14]. Another troponin I mutation (A2V) was neither associated with autosomal dominant HCM nor RCM, but with DCM and an autosomal recessive mode of inheritance [15]. These differences are currently unexplained at the molecular level, and data on possible environmental confounding factors are sparse.
In most reported DCM families, the inheritance pattern was autosomal dominant, but recessive, X-linked, and mitochondrial inheritance are also encountered. Distinct inheritance patterns may become indiscernible if the phenotypic penetrance of the inherited defect is low or the families are small, leading to classification of the index case as “sporadic” DCM. Even within clearly discernible cardiomyopathy families with their rather confined genetic background, phenotypic expression may be highly variable [16, 17]. Clinical experience further indicates the existence of genetic DCM predispositions which gain relevance only, however, when specific environmental noxious factors are encountered. Among cancer patients receiving similar doses of potentially cardiotoxic drugs (e.g. anthracyclines, herceptin), only a small fraction will develop DCM. Similarly, alcohol abuse is frequent but leads to the DCM phenotype in only a few cases. In the absence of any other explanation, a genetic DCM predisposition is obvious in these cases.
The complexity of DCM genetics renders routine molecular screening for monogenic DCM subtypes impracticable. This explains the lack of genetic data on DCM patients investigated for other reasons (e.g. virological diagnostics as outlined below) and impedes the study of genome–environment interactions. Conversely, data on environmental factors are sparse in molecular genetic studies of DCM populations. In clinical practice, the recognition of familial DCM predisposition by the clinician is pivotal since it may lead to in-depth genetic studies. These are of high scientific interest but may also gain relevance for early recognition and treatment of relatives at risk [18]. In summary, the molecular genetics of DCM is particularly complex, and unequivocal genotype–phenotype correlations are lacking in many cases.
Viruses as environmental agents in DCM
An unexpected finding of recent virological surveys was the high prevalence of cardiotropic viruses in DCM hearts. Initially, only enteroviruses (EVs) were detected in endomyocardial biopsies (EMBs) of DCM patients [19, 20] and were shown to be important prognostic factors [21–23]. The incidence of EV infections in DCM hearts was rather low, however. The recent detection of non-EV genomes (e.g. Parvovirus B19) in patients presenting with a sudden onset of cardiac symptoms mimicking acute myocardial infarction [24, 25] raised the question if the total prevalence of cardiotropic viruses in DCM may have been underestimated. Indeed, a high prevalence (67.4%) of viral genomes was found in the heart of 245 consecutive DCM patients in a recent survey investigating a broad panel of viruses [26]. EV and adenoviral (AdV) genomes were found in 9.4 and 2.0%, respectively, parvovirus B19 (PVB19) in 51.4%, and human herpes virus-6 (HHV6) in 21.6% of cases. Dual or multiple infections occurred in 27.3% of patients. The clinical course in patients carrying both PVB19 and HHV6 was particularly severe, and interferon-β treatment [21] was less effective in these cases. In only 32.6% no viral genome was detected. This high prevalence of viral genomes in the heart of adult DCM patients was unexpected, although possible viral etiologies have long been discussed in “idiopathic” DCM [27–29]. PVB19 and HHV6 genomes have previously been reported in childhood cardiomyopathies [30–33], but detected only in rare cases in adults [24, 25, 34]. Apparently, a broad panel of cardiotropic viral agents needs to be investigated during the diagnostic workup of DCM patients since otherwise possible viral etiologies are likely to be missed.
Virus–host interactions in DCM pathogenesis
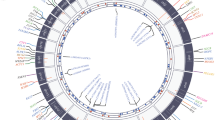
The frequent detection of viral genomes in human DCM hearts strongly suggests that viral infections play a major role in the pathogenesis of DCM. Even a small cardiac virus load during chronic latent infections is able to sustain significant viral transcriptional activity over long periods of time. This may result in slow but progressive deterioration of cardiac function, either by direct cytopathic effects of virus-encoded proteins or indirectly by virus-induced alterations of expression of host-encoded genes (e.g. chronic cardiac inflammation [35], local cytokine release [36, 37]). Cardiac PVB19 was recently found to be associated with diastolic dysfunction in a major series of patients [38]. At all stages from the initial virus entry to late chronic sequelae, the host’s reactions are determined by its genetic constitution. Key interaction sites of viruses with their hosts are depicted in Fig. 1.
Interaction sites of cardiotropic viruses with their target cells. Panel A shows the frequency at which various cardiotropic viruses were detected in a series of 245 DCM patients. In order of decreasing frequency: PVB19 (51.4%), HHV6 (21.6%), EV (9.4%), EBV (2.0%), AdV (1.6%), HCV (0.8%). Also screened but not detected were HSV1/2, InfA/B, and CMV. Panel B shows the cellular entry mechanisms for most of these viruses via cell surface receptors and interacting coreceptors, followed by receptor-mediated endocytosis of the virus particles, release of the viral RNA (CVB3) or DNA genomes (other viruses), and their nuclear import via the nuclear pore complex (NPC). Toll-like receptors (TLRs) 3, 7, 8, 9 are located in the membrane of the endosomes and may trigger signaling cascades of innate antiviral immunity (INFβ-dependent and -independent pathways). Cardiac virus receptors form a first structural level at which genetic variants may influence an individual’s susceptibility to viral infections, by mutations in the receptors or coreceptors or in their regulatory pathways. CAR shows high interindividual variability and strong induction in human DCM [39, 40], but little is known on the regulation and genetics of other receptors. Endosomes, TLRs, and TLR-dependent signaling pathways form a second level at which mutations may alter the course of viral disease [43–45]. A third, genetically polymorphic level is formed by the nuclear envelope (nuclear membranes+NPCs+nuclear lamina). Lamins A and C are intermediate filaments encoded by the LMNA gene mutated in a subset of DCM patients [10, 12]. After the viral genomes are transcribed in the nucleus and translated in the cytosol, their products may act upon and damage the multiple structural proteins of the cardiomyocyte known to carry mutations in familial DCM (Table 1). Whereas for most cardiotropic virus-encoded proteins no data on interactions with mutated structural proteins vs their normal counterparts are available, dystrophin deficiency has already been shown to markedly increase CVB3-induced cardiac damage via enteroviral protease 2A [47]
Most known cardiotropic viruses are taken up into the cell by receptor-mediated endocytosis and then further transported into the nucleus. Virus entry and migration within the host are determined by the expression of virus receptors on target cells. The viral entry pathways into the heart have not been fully delineated, but induction or inhibition of these pathways would decisively alter the individual susceptibility for cardiac viral infections. Induction of a common receptor (CAR) for coxsackieviruses and adenoviruses was observed in human DCM but not in further cardiomyopathies [39, 40]. Receptor induction may open a path for infections with receptor-dependent viruses (EV/AdV) and contribute to dual infections by the respective viruses which are further influenced by distinct coreceptors. Another common receptor (CD46) shared by otherwise unrelated viruses (HHV6 and CAR-independent group B AdV) has been identified [41]. It is currently unknown what genetic or environmental factors cause the dysregulation of CAR in DCM, but its induction may critically influence pathogenesis. Since virus genomes are prevalent in DCM hearts, genetic variants of virus receptors or their regulatory pathways could provide a molecular basis for important genome–environment interactions influencing the disease process.
During their transport through the endosomal pathway, the RNA or DNA viral genome may engage certain Toll-like receptors (TLR 3, 7, 8, 9) linked to key signaling cascades of innate immunity [42]. Genetic alterations of immune pathways decisively alter the disease course in viral infections. Thus, the mortality after CVB3 infection is very low in mice lacking MyD88, a key component of TLR-dependent innate immune signaling, as compared to wild-type animals. Mice are protected from CVB3 myocarditis by gene-targeted knockout of p56Lck, the Src family kinase essential for T-cell activation [43]. In contrast, CVB3 infection of INF-β deficient mice results in excessive mortality as compared to controls [44]. Extracellular signal-regulated kinase 1 and 2 (ERK-1/2) is intense in the heart of myocarditis-susceptible A/J mice, in contrast to myocarditis-resistant C57BL/6 mice. The ERK-1/2 response to CVB3 may thus contribute also to differential host susceptibility [45].
Beyond the proteins involved in cellular virus entry and antiviral defense, several structural proteins of the cardiomyocyte building the sarcomere, sarcolemma cytoskeleton, Z-bands, and intermediate filaments are known to be mutated in cases of familial DCM. No population-wide comprehensive mutation scanning has so far been published for these genes, and thus, the full extent of their genetic variability is unknown. However, one pioneering experimental study has already demonstrated the extent to which such mutations may aggravate the cardiac damage caused by a virus, when CoxB3 infection led to grossly aggravated disease in dystrophin-deficient mice via enteroviral protease 2A-mediated cleavage of dystrophin [46, 47]. Identification of further cellular sites vulnerable to attack by the newly identified cardiotropic virus will be facilitated by the availability of a broad spectrum of genetic animal models for known DCM-associated mutations.
Towards a synopsis of genetic and environmental factors in DCM
Various approaches are available to obtain further insight into genome–environment interactions. First, appreciation of the full phenotype spectrum associated with a single gene defect requires mutation screening of that gene not only in large DCM groups, but also in healthy controls representative of the general population. The latter are required to recognize if mutation carriers may remain disease-free unless they encounter certain environmental factors (viruses, drugs) which then precipitate the disorder. The paradigm of genetic ATD illustrates that environmental agents may indeed play such a decisive role from the clinical point of view. Several groups have begun to investigate the quantitative clinical relevance of DCM-associated mutations on a population-wide scale [9, 10, 17] as opposed to the initial studies mostly focused on highly selected subgroups of DCM patients.
Second, patients suffering from known familial DCM subtypes should be examined for possible cardiac viral infections which were frequent in a recent survey of DCM hearts [26]. In contrast to obvious environmental DCM risk factors (e.g. cardiotoxic drugs), cardiac viruses will remain hidden unless specifically searched for. On the other hand, viruses are, due to their prevalence in DCM hearts, likely to encounter multiple structural proteins of cardiac cells known to be defective in familial DCM. Mutation carriers may react far more sensitively to these viruses than normal hearts [47], and virus detection in familial DCM could gain therapeutic relevance if antiviral therapy in mutation carriers was shown to favourably influence the course of the disease. Conversely, the screening of DCM genes may provide additional prognostic information in patients with viral cardiomyopathy, in particular, if experimental data suggest that defects in a certain gene may enhance the cardiac damage induced by a specific virus. In addition to genes known to cause DCM in humans, numerous other risk genes identified in animal models [48, 49] may confer high susceptibility. Interestingly, environmental factors other than viruses are required for certain mutations to become pathogenetically relevant, e.g. cardiac dysfunction is triggered by hemodynamic stress in vinculin [50] and KDEL receptor mutations [51].
Third, we need to learn more also about the effects of the known cardiotropic viruses on cardiomyocyte function and morphology and on possible specific interactions between these viruses and cardiac cells. Interaction of any virus-encoded protein with a particularly sensitive cardiac cellular protein may have relevance for a large number of DCM patients due to the high prevalence of those viruses in DCM hearts. This may be studied in vitro and in genetic DCM animal models. In addition to defects in cardiac structural genes, genetic variants in cellular signaling pathways involved in antiviral immunity may aggravate or ameliorate the effects of cardiotropic viruses. Genetic dissection of virus–cardiomyocyte interactions may thus lead to the identification of new therapeutic targets. Functional or interaction proteomics may reveal specific interactions between virus- and host-encoded proteins and lead to the identification of novel proteins which are genetically variable or defective in a subset of the human population.
In summary, future investigations into the molecular pathogenesis of human DCM should encompass detailed clinical phenotyping, in conjunction with both molecular genetic data and data on possible noxious factors from the environment, in particular, cardiotropic viruses. This should help to better understand the interactions between host genetics and environmental agents and to further improve the clinical management of DCM patients who still carry an adverse prognosis.
Abbreviations
- AdV:
-
adenovirus
- ATD:
-
α1-antitrypsin deficiency
- CAR:
-
coxsackievirus–adenovirus receptor
- CMV:
-
cytomegalovirus
- COPD:
-
chronic obstructive pulmonary disease
- CVB3:
-
coxsackievirus B3
- DAF:
-
decay accelerating factor
- DCM:
-
dilated cardiomyopathy
- EBV:
-
Epstein–Barr virus
- EV:
-
enteroviruses (coxsackieviruses and echoviruses)
- HCM:
-
hypertrophic cardiomyopathy
- HCV:
-
hepatitis C virus
- HHV6:
-
human herpes virus 6
- InfA/B:
-
Influenza virus A/B
- INFβ:
-
interferon-β
- MLP:
-
muscle LIM protein
- NBC:
-
nuclear pore complex
- PVB19:
-
parvovirus B19
- RCM:
-
restrictive cardiomyopathy
- TLR:
-
toll-like receptor
References
Eriksson S (1964) Pulmonary emphysema and α1-antitrypsin deficiency. Acta Med Scand 175:197–205
Crystal R (1990) α1-antitrypsin deficiency, emphysema, and liver disease. J Clin Invest 85:1343–1352
Poller W, Faber J-P, Olek K (1990) Highly variable clinical course in severe α1-antitrypsin deficiency—use of polymerase chain reaction for the detection of rare deficiency alleles. Klin Wochenschr 68:857–863
Stecenko A, Brigham K (2003) Gene therapy progress and prospects: α1 antitrypsin. Gene Ther 10:95–99
Eriksson S, Carlson J, Veley R (1986) Risk of cirrhosis and primary liver cancer in α1-antitrypsin deficiency. N Engl J Med 314:736–739
Teckman J, Qu D, Perlmutter D (1996) Molecular pathogenesis of liver disease in α1-antitrypsin deficiency. Hepatology 24:1504–1516
Wu Y, Whitman I, Molmenti E, Moore K, Hippenmeyer P, Perlmutter D (1994) A lag in intracellular degradation of mutant α1-antitrypsin correlates with the liver disease phenotype in homozygous PI ZZ α1-antitrypsin deficiency. Proc Natl Acad Sci U S A 91:9014–9018
Franz W, Muller O, Katus H (2001) Cardiomyopathies: from genetics to the prospect of treatment. Lancet 358:1627–1637
Sylvius N, Duboscq-Bidot L, Bouchier C et al (2003) Mutational analysis of the β- and δ-sarcoglycan genes in a large number of patients with familial and sporadic dilated cardiomyopathy. Am J Med Genet 120A:8–12
Sebillon P, Bouchier C, Bidot L et al (2003) Expanding the phenotype of LMNA mutations in dilated cardiomyopathy and functional consequences of these mutations. J Med Genet 40:560–567
Mogensen J, Murphy R, Shaw T et al (2004) Severe disease expression of cardiac troponin C and T mutations in patients with dilated cardiomyopathy. J Am Coll Cardiol 44:2033–2040
Kärkkäinen S, Heliö T, Miettinen R et al (2004) A novel mutation, Ser143Pro, in the lamin A/C gene is common in Finnish patients with familial dilated cardiomyopathy. Eur Heart J 25:885–893
Gomes A, Potter J (2004) Molecular and cellular aspects of troponin cardiomyopathies. Ann N Y Acad Sci 1015:214–224
Mogensen J, Kubo T, Duque M et al (2003) Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest 111:209–216
Murphy R, Mogensen J, Shaw A, Kubo T, Hughes S, McKenna W (2004) Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet 363:371–372
Brito D, Richard P, Isnard R, Pipa J, Komajda M, Madeira H (2003) Familial hypertrophic cardiomyopathy: the same mutation, different prognosis. Comparison of two families with a long follow-up. Rev Port Cardiol 22:1445–1461
Knöll R, Hoshijima M, Hoffman H et al (2002) The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111:943–955
Mahon N, Madden B, Caforio A et al (2002) Immunohistologic evidence of myocardial disease in apparently healthy relatives of patients with dilated cardiomyopathy. J Am Coll Cardiol 39:455–462
Bowles N, Richardson P, Olsen E, Archard L (1986) Detection of coxsackie-B virus-specific RNA sequences in myocardial biopsy samples from patients with myocarditis and dilated cardiomyopathy. Lancet 1:1120–1123
Pauschinger M, Doerner A, Kuehl U et al (1999) Enteroviral RNA replication in the myocardium of patients with left ventricular dysfunction and clinically suspected myocarditis. Circulation 99:889–895
Kühl U, Pauschinger M, Schwimmbeck P et al (2003) Interferon-β treatment eliminates cardiotropic viruses and improves left ventricular function in patients with myocardial persistence of viral genomes and left ventricular dysfunction. Circulation 107:2793–2798
Frustaci A, Chimenti C, Calabrese F, Pieroni M, Thiene G, Maseri A (2003) Immunosuppressive therapy for active lymphocytic myocarditis: virologic and immunologic profile of responders versus non-responders. Circulation 107:857–863
Bowles N, Towbin J (2000) Molecular aspects of myocarditis. Curr Infect Dis Rep 2:308–314
Kühl U, Pauschinger M, Bock T et al (2003) Parvovirus B19 infection mimicking acute myocardial infarction. Circulation 108:945–950
Bültmann B, Klingel K, Sotlar K et al (2003) Fatal parvovirus B19-associated myocarditis clinically mimicking ischemic heart disease: an endothelial cell-mediated disease. Human Pathol 23:92–95
Kühl U, Pauschinger M, Noutsias M et al. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation, in press
Richardson P, McKenna W, Bristow M et al (1996) Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the definition and classification of cardiomyopathies. Circulation 93:841–842
Dec G, Palacios I, Fallon J et al (1985) Active myocarditis in the spectrum of acute dilated cardiomyopathies. Clinical features, histological correlates and clinical outcome. N Engl J Med 312:885–890
Feldman A, McNamara D (2000) Medical progress: myocarditis. N Engl J Med 343:1388–1398
Rohayem J, Dinger J, Fischer R, Klingel K, Kandolf R, Rethwilm A (2001) Fatal myocarditis associated with acute parvovirus B19 and herpesvirus 6 coinfection. J Clin Microbiol 39:4585–4587
Nigro G, Bastianon V, Colloridi V et al (2000) Human parvovirus B19 infection in infancy associated with acute and chronic lymphocytic myocarditis and high cytokine levels: report of 3 cases and review. Clin Infect Dis 31:65–69
Murry C, Jerome K, Reichenbach D (2001) Fatal parvovirus myocarditis in a 5-year-old girl. Human Pathol 32:342–345
Yoshikawa T, Ihira M, Suzuki K et al (2001) Fatal acute myocarditis in an infant with human herpesvirus 6 infection. J Clin Pathol 54:792–795
Pankuweit S, Moll R, Baandrup U, Portig I, Hufnagel G, Maisch B (2003) Prevalence of the parvovirus B19 genome in endomyocardial biopsy specimens. Human Pathol 94:497–500
Klingel K, Kandolf R (1993) The role of enterovirus replication in the development of acute and chronic heart muscle disease in different immunocompetent mouse strains. Scand J Infect Dis Suppl 88:79–85
Shioi T, Matsumori A, Sasayama S (1996) Persistent expression of cytokine in the chronic stage of viral myocarditis in mice. Circulation 94:2930–2937
Horwitz M, LaCava A, Fine C, Rodriguez E, Ilic A, Sarvetnick N (2000) Pancreatic expression of interferon-γ protects mice from lethal coxsackievirus B3 infection and subsequent myocarditis. Nat Med 6:693–697
Tschöpe C, Bock C, Kasner M et al. High prevalence of cardiac parvovirus B19 infection in patients with isolated left ventricular diastolic dysfunction. Circulation, in press
Noutsias M, Fechner H, Jonge H et al (2001) Human coxsackie-adenovirus-receptor is co-localized with Iintegrins αvβ3 and αvβ5 on the cardiomyocyte sarcolemma and upregulated in dilated cardiomyopathy—implications for cardiotropic viral infections. Circulation 104:275–280
Fechner H, Noutsias M, Tschoepe C et al (2003) Induction of coxsackievirus–adenovirus-receptor expression during myocardial tissue formation and remodeling—identification of a cell–cell contact dependent regulatory mechanism. Circulation 107:876–882
Gaggar A, Shayakhmetov D, Lieber A (2003) CD46 is a cellular receptor for group B adenoviruses. Nat Med 9:1408–1412
Ulevitch R (2004) Therapeutics targeting the innate immune system. Nat Rev Immunol 4:512–520
Liu P, Aitken K, Kong YY et al (2000) The tyrosine kinase p56lck is essential in coxsackievirus B3-mediated heart disease. Nat Med 6:429–434
Deonarain R, Cerullo D, Fuse K, Liu P, Fish E (2004) Protective role for interferon-β in coxsackievirus B3 infection. Circulation 110 (in press)
Opavsky M, Martino T, Rabinovitch M et al (2002) Enhanced ERK-1/2 activation in mice susceptible to coxsackievirus-induced myocarditis. J Clin Invest 109:1561–1569
Badorff C, Lee G, Iamphear B et al (1999) Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nat Med 5:320–326
Xiong D, Lee G-H, Badorff C et al (2002) Dystrophin deficiency markedly increases enterovirus-induced cardiomyopathy: a genetic predisposition to viral heart disease. Nat Med 8:782–877
Eigenthaler M, Engelhardt S, Schinke B et al (2003) Disruption of cardiac Ena-VASP protein localization in intercalated disks causes dilated cardiomyopathy. Am J Physiol Heart Circ Physiol 285:H2471–H2481
Arimura T, Hayashi T, Terada H et al (2004) A Cypher/ZASP mutation associated with dilated cardiomyopathy alters the binding affinity to protein kinase C. J Biol Chem 279:6746–6752
Zemljic-Harpf AE, Ponrartana S, Avalos RT et al (2004) Heterozygous inactivation of the vinculin gene predisposes to stress-induced cardiomyopathy. Am J Pathol 165:1033–1044
Hamada H, Suzuki M, Yuasa S et al (2004) Dilated cardiomyopathy caused by aberrant endoplasmic reticulum quality control in mutant KDEL receptor transgenic mice. Mol Cell Biol 24:8007–8017
Kamisago M, Solomon S, Sharma P et al (2000) Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med 343:1688–1696
Bonne G, Carrier L, Richard P, Hainque B, Schwartz K (1998) Familial hypertrophic cardiomyopathy: from mutations to functional defects. Circ Res 83:580–593
Olson T, Michels V, Thibodeau S, Tai Y-S, Keating M (1998) Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 280:750–752
Mogensen J, Klausen IC, Pedersen AK et al (1999) α-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest 103:R39–R43
Li D, Czernuszewicz GZ, Gonzalez O et al (2001) Novel cardiac troponin T mutation as a cause of familial dilated cardiomyopathy. Circulation 104:2188–2193
Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Gessner R (2001) First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Human Mutat 17:524
Olson T, Kishimoto N, Whitby F, Michels V (2001) Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J Mol Cell Cardiol 33:723–732
Thierfelder L, Watkins H, MacRae C et al (1994) α-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 77:701–712
Gerull B, Gramlich M, Atherton J et al (2002) Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet 30:201–204
Itoh-Satoh M, Hayashi T, Nishi H et al (2002) Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem Biophys Res Commun 291:385–393
Mohapatra B, Jimenez S, Lin JH et al (2003) Mutations in the muscle LIM protein and α-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol Genet Metab 80:207–215
Tsubata S, Bowles KR, Vatta M et al (2000) Mutations in the human d-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J Clin Invest 106:655–662
Maeda M, Holder E, Lowes B, Valent S, Bies R (1997) Dilated cardiomyopathy associated with deficiency of the cytoskeletal protein metavinculin. Circulation 95:17–20
Li D, Tapscoft T, Gonzalez O et al (1999) Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation 100:461–464
Goldfarb LG, Park KY, Cervenakova L et al (1998) Missense mutation in desmin associated with familial cardiac and skeletal myopathy. Nat Genet 19:402–403
Fatkin D, MacRae C, Sasaki T et al (1999) Missense mutations in the rod domain of the lamin A/C gene as caused of dilated cardiomyopathy and conduction-system disease. N Engl J Med 341:1715–1724
Schmitt J, Kamisago M, Asahi M et al (2003) Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 299:1410–1413
Haghighi K, Kolokathis F, Pater L et al (2003) Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest 111:869–876
Forleo C, Resta N, Sorrentino S et al (2004) Association of β-adrenergic receptor polymorphisms and progression to heart failure in patients with idiopathic dilated cardiomyopathy. Am J Med 117:451–458
Mizon-Gerard F, de Groote P, Lamblin N et al (2004) Prognostic impact of matrix metalloproteinase gene polymorphisms in patients with heart failure according to the aetiology of left ventricular systolic dysfunction. Eur Heart J 25:688–693
Acknowledgements
This publication has been supported by the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich/Transregio 19.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Poller, W., Kühl, U., Tschoepe, C. et al. Genome–environment interactions in the molecular pathogenesis of dilated cardiomyopathy. J Mol Med 83, 579–586 (2005). https://doi.org/10.1007/s00109-005-0664-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-005-0664-2