Abstract
Purpose of Review
The disease burden of inherited dilated cardiomyopathy (DCM) is large and likely underestimated. This population stands to benefit immensely from therapeutic approaches tailored to the underlying genetic causes. Here, we review recent advances in understanding novel genotype–phenotype relationships and how these can improve the care of patients with inherited DCM.
Recent Findings
In the last several years, discovery of novel DCM-associated genes, gene-specific DCM outcomes, and nuanced information about variant-environment interactions have advanced our understanding of inherited DCM. Specifically, novel associations of genes with specific clinical phenotypes can help to assess sudden cardiac death risk and guide counseling around behavioral and environmental exposures that may worsen disease.
Summary
Important expansions of the current genotype-phenotype profiling include the newly DCM-associated FLNC variant, prognostically significant LMNA, DSP inflammatory cardiomyopathy, and the highly penetrant features of RBM20 variants as well as the role of TTN variants in compounding the effects of environmental factors on toxin-mediated DCM. Future directions to improve diagnostic accuracy and prognostic improvement in DCM will center not just on identification of new genes, but also on understanding the interaction of known and novel variants in known DCM genes with patient genetic background and environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heart failure (HF) therapeutics have seen exponential growth in recent years with the advent of angiotensin receptor blocker plus neprilysin inhibitors (ARNIs) and the SGLT2 inhibitors dapagliflozin and empagliflozin [1]. However, the incidence of HF continues to grow worldwide, and significant heterogeneity in the clinical outcomes and presentation of HF patients indicates an outsized need for personalized approaches to treatment. Among patients with HF with reduced ejection fraction (HFrEF), a significant portion has dilated cardiomyopathy (DCM) with inherited DCM making up an estimated 50% of nonischemic CM cases [2]. This highlights a population that stands to benefit immensely from therapeutic approaches tailored to the underlying genetic cause of their disease.
DCM was last defined by the ESC and AHA in 2008 and 2006, respectively, as the left ventricular (LV) or biventricular dilatation and systolic dysfunction with normal wall thickness, and the absence of abnormal loading conditions or coronary artery disease sufficient to cause global systolic impairment [3, 4]. Since that time, several new HfrEF classification systems have been proposed in an effort to improve diagnosis and therapeutics, but none has captured the full heterogeneity of disease inherent to DCM [5, 6]. The presentation of DCM may include decline in LV contractile function and progressive LV dilation, clinical heart failure, ventricular and supraventricular arrhythmias, conduction system disease, thromboembolism, and/or sudden cardiac death; however, a number of patients present with symptoms preceding any structural changes [7]. DCM affects younger individuals with a 3:1 male to female predominance, and some studies demonstrate a threefold increased risk for DCM among Black individuals, who also have a twofold increased mortality risk [8]. Otherwise, risk predictors for DCM disease progression remain difficult to personalize for patients due both to this inherent heterogeneity of presentation and the lack of precise disease definitions.
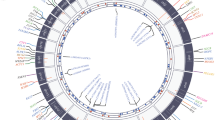
However, a substantial proportion of DCM patients may carry a diagnostic sign that enables precision or personalized therapy: an underlying genotype causative of their DCM (Fig. 1). Indeed, emerging evidence suggests that genetic diagnosis in DCM can confer differential outcomes, in particular, with respect to progression to transplant or early sudden cardiac death [9]. Further, emerging programs for gene-specific cardiomyopathy therapies are underway. However, the diagnostic yield of genetic testing in DCM lags behind that of other cardiovascular diseases [10]. This is not likely due to a low prevalence of genetic causes, but rather to our incomplete understanding of the genetic causes of DCM both at the level of specific variants in known DCM genes and at the level of novel genes themselves [11]. Further, prior models of strict Mendelian variants solely causative of disease versus truly polygenic risk alleles have evolved toward a more nuanced model in which variants of differential effect size are additive with each other (e.g., genetic background) as well as with environmental exposures to explain disease penetrance and expressivity [12••]. In the last several years, discovery of novel DCM-associated genes, gene-specific DCM outcomes, and nuanced information about variant-environment interactions has pushed this field forward (Fig. 2). Here, we will briefly review the current state of knowledge regarding the genetic architecture of DCM and then explore deeply gene-specific cases of recent advances in this field.
Current State of Knowledge: Genetic Architecture of DCM
While genetically heterogeneous, DCM is largely autosomal dominant in inheritance. A number of genes affecting a spectrum of cellular function and myocardial structures have been associated with inherited DCM. While the typical DCM clinical genetic testing panel sequences ~50 genes previously associated with DCM, marginal return of genetic diagnosis in expanding the number of genes tested is small and comes with increased identification of variants of uncertain significance and variants within genes of uncertain association with DCM [13].
In likely the most comprehensive gene burden analysis to date for inherited cardiomyopathies, Walsh et al. investigated 46 genes previously reported to be associated with DCM [14]. Among the most striking findings of this study was that only 15 of these genes displayed strong evidence for DCM association in the population (Table 1). In fact, in 2020, a reassessment of the Mendelian genetics of DCM found only 11 of these to be robustly disease-associated (Indicated by * in Table 1) and added BAG family molecular chaperone regulator 3 (BAG3) to this list [12••]. Critically, in both studies, particular classes of variants (truncating or missense) were differentially associated with DCM in these genes. Additionally, a recent comprehensive systematic review of 51 genes highlighted twelve genes categorized as definitively or strongly association with idiopathic DCM (BAG3, DES, DSP, FLNC, LMNA, MYH7, PLN, RBM20, SCN5A, TNNC1, TNNT2, TTN), whereas seven others showed only moderate evidence (ACTC1, ACTN2, JPH2, NEXN, TNNI3, TPM1, VCL) [15]. A 2022 Expert Consensus Statement by the European Heart Rhythm Association highlights the diagnostic yield of testing for these genes in patients with DCM, recommending that those 12 genes with the strongest evidence above be included and adding that moderate evidence genes may also be included at provider discretion [16].
These data suggest that while much of currently recognized inherited DCM is explained by these specific genetic diagnoses, familial disease may account for up to one half of cases of asymptomatic DCM [6, 8]. Given this, the low diagnostic yield of current testing methods belies significant missing heritability in DCM. Although these studies suggest that adding additional genes to genetic testing panels may not significantly increase the diagnostic power of genetic testing, the possibility remains that the majority of inherited DCM is due to small numbers of patients with rare variants in a large number of novel genes. It is also likely that variants of uncertain significance (VUS) in established DCM genes preclude diagnostic identification in a majority of patients. Additionally, the complexity of polygenic risk and environmental influences on DCM expression and penetrance remain to be fully understood.
Filamin C (FLNC): a Novel Gene Causing Arrhythmogenic DCM
The Filamin C (FLNC) gene was historically implicated in myofibrillar myopathy and hypertrophic cardiomyopathy due to missense variants in its ROD2 domain [17, 18]. However, new data has reported a significant association between truncating variants in FLNC and dilated and arrhythmogenic cardiomyopathies (ACM) with multiple cohort level studies confirming pathogenicity [19, 20, 21•]. The filamin C protein is responsible for actin cross-linkage between the sarcomeric Z-disc and the sarcolemma and therefore physiologically instructs mechanotransduction in cardiomyocytes [22]. This specific pathophysiology is highly associated with ACM and arrhythmogenic phenotypes [19].
The most recent longitudinal data available shows that FLNC truncating variants (FLNCtv) result in variable phenotypic presentations of DCM, generally characterized by high rates of malignant ventricular rhythms, even in the absence of significant ventricular remodeling, consistent with an arrhythmogenic cardiomyopathy phenotype [20]. These same reports estimate that FLNCtvs account for approximately 3–4% of DCM cases [20, 21•]. The presence of FLNC truncating variants has potential to inform indication for primary prevention ICD in DCM patients even in the absence of structural disease based on additional clinical risk stratification, which may include the presence of nonsustained ventricular tachycardia [21•]. Other cardiac phenotypes associated with FLNC variants include restrictive cardiomyopathy, congenital heart disease, ARVC, and noncompaction cardiomyopathy [17, 19]. In addition to FLNC being a gene newly associated with DCM, the arrhythmogenic clinical profile of FLNCtv DCM exemplifies a group of DCM genes that have recently revealed themselves to be highly arrhythmogenic. A 2019 Heart Rhythm Society Expert Consensus Statement indicates ICD implantation as a reasonable recommendation (class IIa) in FLNC variant ACM patients with EF < 45% [23]; though given recent data above from Gigli et al. [21•], it is clear than additional risk factors other than EF alone may be helpful in identifying those at high risk of SCD (e.g., nonsustained ventricular tachycardia).
Genotype-Specific Outcomes in DCM: Lamin A/C, Desmoplakin, and RBM20
The importance of expanding the current profiling of DCM genotype–phenotype relationships lies in the potential to enable risk prediction and disease-modifying strategies. While some DCM patients experience cardiac recovery with appropriate guideline directed medical therapy and advanced interventions, patients often present late in the disease course with high rates of morbidity and mortality, arrhythmic complications, and sudden cardiac death (SCD), and genetic diagnosis may be an important arbiter of these outcomes [24]. Additionally, understanding of genotype–phenotype relationships can guide the care of genotype-positive, phenotype-negative patients identified on cascade testing or in the general population [25, 26]. In recent years, more studies have embarked on the quest to confirm the genotype–phenotype relationship of various DCM presentations. Below, we outline growing evidence for the value of genetic diagnosis in risk prediction, as well as a complicated interface between genotype and environmental factors affecting prognosis [27].
Lamin A/C (LMNA) DCM: a Rapidly Progressive Cause of Arrhythmogenic DCM and Conduction System Disease
Lamin A/C (LMNA) variants are associated with both Hitchinson-Gilford progeria (accelerated aging) and DCM and account for approximately 5% of DCM cases [28]. LMNA is located on chromosome 1 and regulates mechanosensing, DNA replication, and transportation of genetic material from the nucleus to cellular cytoplasm for further processing and expression [29]. In mouse models, Lamin A/C-deficient mice developed rapidly progressive dilated cardiomyopathy characterized by LV dilation and systolic contractile dysfunction [30]. Both missense and nonmissense variants in LMNA have been associated with severe, progressive DCM [31, 32]. Initial reports described LMNA cardiomyopathy to be associated with LV dilation and less commonly by left ventricular noncompaction [28]. Recently, the first large, longitudinal cohort of LMNA DCM patients was published. This revealed an impressive prevalence of conduction system disease, atrial and ventricular arrhythmias, and rapid progression toward end-stage HF and advanced therapies [32].
In combination with prior descriptive reports and animal data, these observations led to the first ever guideline-based recommendation for genotype-specific ventricular arrhythmia risk stratification in patients with DCM: For DCM patients with pathogenic LMNA variants, risk stratification is now much more precise than the prior stratification based on LV ejection fraction alone; implantation of an implantable cardiac defibrillator (ICD) is recommended for those with any two of four risk factors specific to LMNA cardiomyopathy: male sex, truncating variant, nonsustained ventricular tachycardia by ambulatory monitoring, or LV ejection fraction ≤ 45%(class Iia) [33]. Additionally, early detection of LMNA variants with DCM may guide prognostic evaluation for early advanced heart failure therapies including mechanical circulatory support and transplantation [34].
Desmoplakin (DSP) Cardiomyopathy: Inflammatory and Surprisingly Specific to the Left Ventricle
Desmoplakin (DSP) plays a critical role in myocardial contraction, linking cardiac desmosomes to intermediate filaments and strengthening the cytoskeleton, driving the necessary force for myocardial contraction. In the early 2000s, DSP was the second gene discovered to cause ACM with autosomal recessive inheritance, initially thought to predominantly affect the right ventricle. A rapidly progressive form of DCM, characterized by heart failure and SCD in adolescence, was identified in Carvajal syndrome associated with homozygous and compound heterozygous mutations in DSP, suggesting a correlation between disease severity and loss of function of both DSP alleles [35]. In 2008, increased recognition of LV involvement with marked fibrosis resulted in expanded diagnostic considerations for ACM in the absence of typical RV involvement [36]. Truncating DSP variants have the most evidence of pathogenicity and currently account for ~ 15% of inherited ACM cases [15, 37, 38•].
Recent studies have continued to expand our understanding of DSPtv cardiomyopathy such that we now understand it to be a LV-dominant ACM more so than a cause of ARVC [38•]. Additional description of its underlying pathophysiology involves cyclical episodes of inflammation similar to myocarditis resulting in cardiomyocyte damage and fibrofatty replacement with a clinically significant proportion of patients in a multicenter cohort presenting with myocarditis-like symptoms and elevated troponins [38•]. This is corroborated by FDG-PET studies that reveal an association between left myocardial ventricular myocardial injury and myocardial inflammation, suggesting a potential immunogenic component of immune activation in response to desmosomal disruption. These episodes drive disease progression even in the absence of LV dysfunction. Further studies are needed to confirm whether variant type (truncating vs. missense) or location can predict disease severity.
RBM20: a Particularly Penetrant and Arrhythmogenic Cardiomyopathy
RNA-binding motif protein 20 (RBM20) is a post-transcriptional splice-regulator of transcripts critical to myocardial and calcium homeostasis, with missense variants associated with 3–6% of genetic DCM cases [39]. Alternative splicing results in significant heterogeneity in protein expression, structure, regulation, and function of a number of genes implicated in LV function and arrhythmia including TTN, LDB3, CAMK2D, and RYR2 [40, 41].
RBM20 was first associated with inherited DCM in 2009 and 2010 in several kindred studies showing autosomal dominant inheritance [42, 43]. A recent report including an international cohort of RBM20 patients showed that RBM20 cardiomyopathy is a highly arrhythmogenic and penetrant form of DCM, with prevalence of sustained ventricular arrhythmias and atrial fibrillation similar to LMNA cardiomyopathy and significant increased compared to TTNtv DCM [44]. Growing clinical and pre-clinical evidence corroborates these findings, with some suggestion that missense variants in RBM20 act to sequester the protein in stress and processing bodies [39, 45,46,47]. Clinically, a genetic diagnosis of RBM20 cardiomyopathy therefore portends a relatively high risk of sudden cardiac arrest even with normal or mildly reduced LV ejection fraction. Future longitudinal studies will be required to understand RBM20-DCM-specific risk stratification for life-threatening arrhythmias.
Titin Truncating Variants (TTNtv): Exemplar of Genotype Interactions with Genetic Background and the Environment
The Titin protein is an integral part of the cardiomyocyte sarcomere, binding the thin and thick filaments of striated muscle, and modulating myocardial contraction and stiffness. Most pathogenic variants in TTN are thought to be truncating variants (TTNtvs) with data indicating the benign nature of missense variants [48]. In general, TTNtvs located in exons within the A-band experience high inclusion probability (high percent spliced in, PSI) and are more likely to be disease causing [49, 50]. TTNtvs found in the I-band generally have low PSI, are prevalent in healthy individuals, and therefore are thought much less likely to be pathogenic [49].
TTNtvs are estimated to account for 15–25% of inherited DCM cases with physiologic effects thought related to faulty cardiomyocyte- and tissue-level mechanotransduction [13, 51]. Compared to DCM patients without other genetic diagnoses, those with TTNtv variants have worse LV dysfunction and LV dilation [52]. However, among genetic causes of DCM, TTNtv patients are thought to have somewhat milder disease [53]. While TTNtv was one of the first and broad-reaching genes connected to DCM and many other cardiomyopathies, there continues to be growth in our understanding of the pathogenicity of the numerous variants. The historical focus on culprit loss of function mutations is now expanded to include rare splice-altering TTN variants that were previously categorized as unknown significance [54]. Recent population data also reveals that TTNtvs are associated with DCM previously attributed to toxins alone (e.g., alcohol and chemotherapy), indicating that these variants may compound the effects of environmental exposure on DCM phenotypes, and vice versa. The effect of TTNtvs on LV morphology in an unselected population is also affected by genetic background. Polygenic risk for change in LV morphology modulates these effects as imaged by cardiac magnetic resonance [55, 56].
Toxin-Mediated TTNtv DCM
A number of toxins are known to have direct deleterious effects on the myocardium, but the heterogeneity of dose–response relationships in DCM has raised the possibility of genetic susceptibility. Only in the past 5 years has data corroborated this suspicion, specifically with respect to TTNtvs. For example, in alcohol-related DCM, one large cohort study evaluated nine DCM genes and found an almost 10% incidence of TTNtv among alcohol-related CM patients compared to unaffected subjects [57]. Additionally, among DCM patients with TTNtv, moderate alcohol intake predicts worse biventricular function and negative remodeling [57].
A similarly heterogeneous clinical presentation pattern is seen among chemotherapy-associated CM (excluding immune-checkpoint inhibitors). Genetic data in this population is limited; however, one moderate-sized study identified 7.5% (n = 16/213) TTNtv prevalence among women with hematologic, breast, and other solid-tumor cancers and confirmed chemotherapy-associated cardiomyopathy, compared to healthy volunteers (0.7%, n = 3/445) [58]. These patients were also found to have more heart failure hospitalizations and incidence of atrial fibrillation but no significant correlation with disease severity or recovery. Further evaluation is needed in a larger and more diverse patient population to improve our understanding of the relationship and risk implications for patients with TTNtvs. Beyond counseling to avoid cardiotoxic substance use (e.g., excessive alcohol, amphetamines), for example, data-driven chemotherapy selection or cardiac monitoring may be enabled by a genome-first approach.
TTNtvs in Peripartum Cardiomyopathy
Peripartum cardiomyopathy (PPCM) is a rare yet highly morbid disease with significant contribution to a persistently elevated maternal mortality rate in the USA particularly among Black women [59, 60]. It is phenotypically similar to DCM and has reported significant familial clustering raising concern for a genetic contributions, which were confirmed in a large population in 2016 [61]. Recently, a large effort to sequence women with PPCM led to recapitulation of these findings: Not only did ~ 10% of these women have TTNtvs, but also overrepresentation of pathogenic variants in FLNC, DSP, and BAG3 was also identified [62]. These data suggest that a significant portion of PPCM may be heritable in the form of DCM elsewhere in the family, and therefore, that approaches to genetic testing in PPCM should mirror those for DCM. Further, future development of gene-specific therapies for DCM may also be applicable to PPCM.
Future Directions to Realize the Potential of Clinical Genetic Testing for DCM
With increasing access to next-generation sequencing and rapidly growing population-based biobanks, our ability to identify the genes that cause and modify DCM phenotypes has been greatly accelerated. However, genetic diagnosis is still hindered by a lack of robust precision genotype–phenotype data at the level of variants. This is largely driven by incomplete knowledge of variants of uncertain significance (VUS) even within known DCM genes. Additionally, identification of VUS that disrupt splicing is imperfect, resulting in hampered discovery of pathogenic variants [63]. Concerted efforts to prospectively classify these variants with respect to their pathogenicity are ongoing. As our ability to sequence and examine large datasets expands, advances in transcriptomic and proteomic technologies may offer a route to better understand the molecular endophenotypes resulting from pathogenic variants in DCM genes as well.
More precise genotype–phenotype profiling will also guide clinical applications ranging from arrhythmia monitoring to early initiation of goal-directed medical therapy and expedited evaluation for advanced heart failure interventions. Early studies of polygenic risk scores using cardiac-MRI-derived markers have yielded promising results in correlating with incident DCM while uncovering that the penetrance of high-impact rare variants may be influenced by carriers’ polygenic backgrounds [55]. These, in combination with clear evidence of genetic susceptibility to environmental exposures, have highlighted the need for further studies to evaluate the interplay between common variants, rare variants, and environmental factors in developing DCM. These approaches have the potential to bridge the diagnostic and therapeutic gaps for populations currently unresponsive to the standard of care for DCM. Ultimately, a combination of scaled and low throughput approaches will be necessary to achieve the mission of precision therapy for DCM. Recent progress is heartening, and the enthusiasm of the cardiovascular genetics community for collaboration will be critical to our progress in this vein.
References
Papers of particular interest, published recently, have been highlighted as:• Of importance •• Of major importance
Committee W, Maddox TM, Januzzi JL Jr, et al. 2021 Update to the 2017 ACC expert consensus decision pathway for optimization of heart failure treatment: answers to 10 pivotal issues about heart failure with reduced ejection fraction: a report of the American College of Cardiology Solution Set Oversight Committee. J Am Coll Cardiol. 2021;77:772–810.
McNally EM, Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res. 2017;121:731–48.
Katritsis DG, Katritsis D, Gersh BJ, John Camm A. Clinical cardiology: current practice guidelines. Oxford University Press; 2013.
Maron BJ, Towbin JA, Thiene G, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–16.
Pinto YM, Elliott PM, Arbustini E, et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 2016;37:1850–8.
Bozkurt B, Colvin M, Cook J, et al. Current diagnostic and treatment strategies for specific dilated cardiomyopathies: a scientific statement from the American Heart Association. Circulation. 2016;134:e579–646.
Schultheiss H-P, Fairweather D, Caforio ALP, et al. Dilated cardiomyopathy. Nat Rev Dis Primers. 2019;5:32.
Huggins GS, Kinnamon DD, Haas GJ, et al. Prevalence and cumulative risk of familial idiopathic dilated cardiomyopathy. JAMA. 2022;327:454–63.
Escobar-Lopez L, Ochoa JP, Mirelis JG, et al. Association of genetic variants with outcomes in patients with nonischemic dilated cardiomyopathy. J Am Coll Cardiol. 2021;78:1682–99.
Sturm AC, Hershberger RE. Genetic testing in cardiovascular medicine: current landscape and future horizons. Curr Opin Cardiol. 2013;28:317–25.
Verdonschot JAJ, Hazebroek MR, Krapels IPC, et al. Implications of genetic testing in dilated cardiomyopathy. Circ Genom Precis Med. 2020;13:476–87.
•• Mazzarotto F, Tayal U, Buchan RJ, et al. Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation. 2020;141:387–398. This large DCM cohort sequencing study of over 2500 probands identified genes and variants most relevant for DCM genetic testing.
Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–8.
Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2016;19:192–203.
Jordan E, Peterson L, Ai T, et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation. 2021;144:7–19.
Wilde AAM, Semsarian C, Márquez MF, et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the state of genetic testing for cardiac diseases. Europace. 2022. https://doi.org/10.1093/europace/euac030.
Verdonschot JAJ, Vanhoutte EK, Claes GRF, et al. A mutation update for the FLNC gene in myopathies and cardiomyopathies. Hum Mutat. 2020;41:1091.
Brodehl A, Gaertner-Rommel A, Milting H. (Filamin-C): a new(er) player in the field of genetic cardiomyopathies. Circ Cardiovasc Genet. 2017. https://doi.org/10.1161/CIRCGENETICS.117.001959.
Brun F, Gigli M, Graw SL, et al. FLNC truncations cause arrhythmogenic right ventricular cardiomyopathy. J Med Genet. 2020. https://doi.org/10.1136/jmedgenet-2019-106394.
Begay RL, Graw SL, Sinagra G, et al. Filamin C truncation mutations are associated with arrhythmogenic dilated cardiomyopathy and changes in the cell-cell adhesion structures. JACC Clin Electrophysiol. 2018;4:504–14.
• Gigli M, Stolfo D, Graw SL, et al. Phenotypic expression, natural history, and risk stratification of cardiomyopathy caused by filamin C truncating variants. Circulation. 2021. https://doi.org/10.1161/CIRCULATIONAHA.121.053521. Gigli et al. detected the arrhythmogenic phenotype associated with FLNC truncating variants independent of left ventricular dysfunction, an example of high risk cardiomyopathy with heterogenous clinical presentations.
Agarwal R, Paulo JA, Toepfer CN, et al. Filamin C cardiomyopathy variants cause protein and lysosome accumulation. Circ Res. 2021;129:751–66.
Towbin JA, McKenna WJ, Abrams DJ, et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm. 2019;16:e301–72.
Merlo M, Pivetta A, Pinamonti B, Stolfo D, Zecchin M, Barbati G, Di Lenarda A, Sinagra G. Long-term prognostic impact of therapeutic strategies in patients with idiopathic dilated cardiomyopathy: changing mortality over the last 30 years. Eur J Heart Fail. 2014;16:317–24.
James CA, Calkins H. Arrhythmogenic right ventricular cardiomyopathy: progress toward personalized management. Annu Rev Med. 2019;70:1–18.
Ho CY, Day SM, Axelsson A, et al. Valsartan in early-stage hypertrophic cardiomyopathy: a randomized phase 2 trial. Nat Med. 2021;27:1818–24.
Hazebroek MR, Moors S, Dennert R, et al. Prognostic relevance of gene-environment interactions in patients with dilated cardiomyopathy: applying the MOGE(S) classification. J Am Coll Cardiol. 2015;66:1313–23.
Crasto S, My I, Di Pasquale E. The broad spectrum of LMNA cardiac diseases: from molecular mechanisms to clinical phenotype. Front Physiol. 2020. https://doi.org/10.3389/fphys.2020.00761.
Captur G, Arbustini E, Bonne G, et al. Lamin and the heart. Heart. 2018;104:468–79.
Nikolova V, Leimena C, McMahon AC, et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113:357–69.
Sinagra G, Dal Ferro M, Merlo M. Lamin A/C cardiomyopathy: cutting edge to personalized medicine. Circ Cardiovasc Genet. 2017. https://doi.org/10.1161/CIRCGENETICS.117.002004.
Kumar S, Baldinger SH, Gandjbakhch E, et al. Long-term arrhythmic and nonarrhythmic outcomes of lamin A/C mutation carriers. J Am Coll Cardiol. 2016;68:2299–307.
Al-Khatib SM, Stevenson WG, Ackerman MJ, et al. 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Heart Rhythm. 2018;15:e190–252.
Hasselberg NE, Haland TF, Saberniak J, Brekke PH, Berge KE, Leren TP, Edvardsen T, Haugaa KH. Lamin A/C cardiomyopathy: young onset, high penetrance, and frequent need for heart transplantation. Eur Heart J. 2018;39:853–60.
Yermakovich D, Sivitskaya L, Vaikhanskaya T, Danilenko N, Motuk I. Novel desmoplakin mutations in familial Carvajal syndrome. Acta Myol. 2018;37:263.
Corrado D, van Tintelen PJ, McKenna WJ, et al. Arrhythmogenic right ventricular cardiomyopathy: evaluation of the current diagnostic criteria and differential diagnosis. Eur Heart J. 2020;41:1414–29.
Wang W, Murray B, Tichnell C, Gilotra NA, Zimmerman SL, Gasperetti A, Scheel P, Tandri H, Calkins H, James CA. Clinical characteristics and risk stratification of desmoplakin cardiomyopathy. Europace. 2022;24:268–77.
•Smith ED, Lakdawala NK, Papoutsidakis N, et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation. 2020. https://doi.org/10.1161/CIRCULATIONAHA.119.044934. DSP, previously associated with predominantly right ventricular arrhythmogenic cardiomyopathy, is described in association with a unique left ventricular fibrotic and inflammatory cardiomyopathy in this study.
Hey TM, Rasmussen TB, Madsen T, Aagaard MM, Harbo M, Mølgaard H, Møller JE, Eiskjær H, Mogensen J. Pathogenic RBM20-variants are associated with a severe disease expression in male patients with dilated cardiomyopathy. Circ Heart Fail. 2019;12: e005700.
Guo W, Schafer S, Greaser ML, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–73.
Maatz H, Jens M, Liss M, et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–30.
Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV, Olson TM. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–41.
Li D, Morales A, Gonzalez-Quintana J, Norton N, Siegfried JD, Hofmeyer M, Hershberger RE. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3:90–7.
Parikh VN, Caleshu C, Reuter C, et al. Regional variation in RBM20 causes a highly penetrant arrhythmogenic cardiomyopathy. Circ Heart Fail. 2019. https://doi.org/10.1161/CIRCHEARTFAILURE.118.005371.
van den Hoogenhof MMG, Beqqali A, Amin AS, et al. RBM20 mutations induce an arrhythmogenic dilated cardiomyopathy related to disturbed calcium handling. Circulation. 2018;138:1330–42.
Schneider JW, Oommen S, Qureshi MY, et al. Dysregulated ribonucleoprotein granules promote cardiomyopathy in RBM20 gene-edited pigs. Nat Med. 2020;26:1788–800.
Fenix AM, Miyaoka Y, Bertero A, et al. Gain-of-function cardiomyopathic mutations in RBM20 rewire splicing regulation and re-distribute ribonucleoprotein granules within processing bodies. Nat Commun. 2021;12:6324.
Akinrinade O, Heliö T, Lekanne Deprez RH, et al. Relevance of titin missense and non-frameshifting insertions/deletions variants in dilated cardiomyopathy. Sci Rep. 2019;9:4093.
Deo RC. Alternative splicing, internal promoter, nonsense-mediated decay, or all three: explaining the distribution of truncation variants in titin. Circ Cardiovasc Genet. 2016;9:419–25.
Romano R, Ghahremani S, Zimmerman T, Legere N, Thakar K, Ladha FA, Pettinato AM, Hinson JT. Reading frame repair of truncation variants restores titin quantity and functions. Circulation. 2022;145:194–205.
Akhtar MM, Lorenzini M, Cicerchia M, et al. Clinical phenotypes and prognosis of dilated cardiomyopathy caused by truncating variants in the TTN gene. Circ Heart Fail. 2020. https://doi.org/10.1161/CIRCHEARTFAILURE.119.006832.
Haggerty CM, Damrauer SM, Levin MG, et al. Genomics-first evaluation of heart disease associated with titin-truncating variants. Circulation. 2019;140:42–54.
Tayal U, Newsome S, Buchan R, et al. Phenotype and clinical outcomes of titin cardiomyopathy. J Am Coll Cardiol. 2017;70:2264–74.
Patel PN, Ito K, Willcox JAL, et al. Contribution of noncanonical splice variants to truncating variant cardiomyopathy. Circ Genom Precis Med. 2021;14: e003389.
Pirruccello JP, Bick A, Wang M, et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat Commun. 2020. https://doi.org/10.1038/s41467-020-15823-7.
Pirruccello JP, Bick A, Chaffin M, et al. Titin truncating variants in adults without known congestive heart failure. J Am Coll Cardiol. 2020;75:1239–41.
Ware JS, Amor-Salamanca A, Tayal U, et al. Genetic etiology for alcohol-induced cardiac toxicity. J Am Coll Cardiol. 2018. https://doi.org/10.1016/j.jacc.2018.03.462.
Garcia-Pavia P, Kim Y, Restrepo-Cordoba MA, et al. Genetic variants associated with cancer therapy-induced cardiomyopathy. Circulation. 2019;140:31–41.
McNamara DM, Elkayam U, Alharethi R, et al. Clinical outcomes for peripartum cardiomyopathy in North America: results of the IPAC study (investigations of pregnancy-associated cardiomyopathy). J Am Coll Cardiol. 2015;66:905–14.
Gad MM, Elgendy IY, Mahmoud AN, Saad AM, Isogai T, Sande Mathias I, Misbah Rameez R, Chahine J, Jneid H, Kapadia SR. Disparities in cardiovascular disease outcomes among pregnant and post-partum women. J Am Heart Assoc. 2021;10: e017832.
Ware JS, Li J, Mazaika E, et al. Shared genetic predisposition in peripartum and dilated cardiomyopathies. N Engl J Med. 2016;374:233–41.
Goli R, Li J, Brandimarto J, et al. Genetic and phenotypic landscape of peripartum cardiomyopathy. Circulation. 2021. https://doi.org/10.1161/CIRCULATIONAHA.120.052395.
Orphanou N, Papatheodorou E, Anastasakis A. Dilated cardiomyopathy in the era of precision medicine: latest concepts and developments. Heart Fail Rev. 2021. https://doi.org/10.1007/s10741-021-10139-0.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
VNP is on the SAB of and receives research support from BioMarin, Inc., and has minor consulting relationships with Viz.ai, and Constantiam Biosciences. VNP also reports support from NHLBI K08HL14318503, as well as payment for UCSD genomic medicine grand rounds. The other authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Myocardial Disease
Rights and permissions
About this article
Cite this article
Njoroge, J.N., Mangena, J.C., Aribeana, C. et al. Emerging Genotype–Phenotype Associations in Dilated Cardiomyopathy. Curr Cardiol Rep 24, 1077–1084 (2022). https://doi.org/10.1007/s11886-022-01727-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11886-022-01727-z