Abstract
Pyrimidine ring and its fused derivatives including pyrazolo[3,4-d]pyrimidine, pyrido[2,3-d]pyrimidine, quinazoline, and furo[2,3-d]pyrimidine compounds had received much interest due to their diverse biological potential, in addition fused pyrimidine are considered as bioisosteres with purines and consequently many pyrimidine and fused pyrimidine derivatives as pyrazolo[3,4-d]pyrimidine, pyrido[2,3-d]pyrimidine, quinazoline, and furo[2,3-d]pyrimidine possessed promising anticancer activity. These pyrimidine derivatives exerted their anticancer potential through different action mechanisms; one of these mechanisms is inhibiting protein kinases that are considered as essential enzymes for controlling cell growth, differentiation, migration, and metabolism. The present review sheds the light on the anticancer significance of some privileged pyrimidine and fused pyrimidine derivatives via selective inhibition of protein kinases, revealing structure-activity relationships and some synthetic pathways used for constructing these scaffolds in an attempt to assist medicinal chemists to construct novel pyrimidines with higher selectivity as anticancer agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is still one of the most fatal diseases, affecting nearly 7 million persons per year all over the world. It is characterized by loss of cell growth control causing a cellular mass called cancer [1,2,3,4,5,6]. However, death is most often accompanies cancer because of metastasis which is responsible for spreading cancer to another part in the body to establish secondary cancerous growths [7,8,9,10,11,12]. It was found that strategies for cancer treatment such as surgery and radiation cannot control the spread of tumor; so many scientific trials to treat cancer had been depending on conventional chemotherapy [13,14,15,16]. Unfortunately, the conventional chemotherapy did not differentiate between the normal human cells and affected cells, causing many drawbacks [17, 18]. Consequently, to circumvent these drawbacks a new strategy for cancer treatment compromising the use of selective tumor drugs called molecular targeted therapies which inhibits certain receptors and signaling pathways which stimulate tumor cell growth had been developed [19,20,21,22]. Protein kinases (PKs) are enzymes that stimulate phosphate transfer from ATP to amino acids tyrosine, serine and/ or threonine residues in protein substrates [23,24,25,26]. Also, PKs are important enzymes responsible for cellular signaling processes such as cell growth regulation, differentiation, migration, and metabolism [27,28,29]. Mutation or overexpression of many PKs had been reported in multiple human cancers [30,31,32,33]. Consequently, inhibiting protein kinase has been used as a selective way for targeting cancer cells [34,35,36,37,38]. The literature survey explained that the binding site of kinase inhibitor consists of five essential regions; adenine-binding site, sugar region, phosphate-binding region (hydrophilic channel), hydrophobic region I and hydrophobic region II [39, 40]. Most discovered kinase inhibitors should be small molecules and ATP-competitive inhibitors that exhibited up to three hydrogen bonding interactions with the amino acids present in the target kinase [41,42,43].
Pyrimidine ring attracted much attention due to its diverse array of biological and pharmacological activities especially anticancer activity [44,45,46,47,48,49]. Many fused pyrimidine derivatives exerted their anticancer activity through inhibiting many types of PKs as they are considered as bioisosteres to purine scaffold from which ATP is formed [12, 50,51,52,53]. Herein, this review reveals the new approaches and medicinal chemistry efforts in search for new pyrimidine and fused pyrimidine scaffolds as PKs inhibitors.
Pyrimidine derivatives
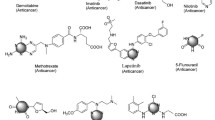
At the beginning of the twenty-first century, scientist’s efforts discovered more potent and selective tyrosine kinase inhibitors, imatinib (GleevecTM) (1) [54, 55], nilotinib (TasignaTM) (2) [56, 57], and dasatinib (SpryclTM) (3) [58, 59] (Fig. 1). FDA approved these pyrimidine derivatives for treating chronic myelogenous leukemia via inhibiting Bcr-Abl tyrosine kinases [60,61,62].
In 2013, a group of investigators designed and synthesized a new series of pyrimidine derivatives as anaplastic lymphoma kinase (ALK) inhibitors with antiproliferative potential against H3122 [63]. From this series, compound 9 (IC50 = 0.004 µM, IC50 = 0.034 µM in cell based assay) exhibited 2.5-fold better ALK inhibitory activity and antiproliferative potential than the reference compound crizotinib (IC50 = 0.01 µM in ALK assay, IC50 = 0.092 µM in cell based assay) (10). SAR study revealing the effect of various substituents is illustrated in Fig. 2.
The target compound 9 was obtained in 75% yield by reacting compound 4 with 4-(N-Boc-piperazin-1-yl)-2-methoxyaniline 5 in the presence of diisopropylethylamine, followed by separating the obtained regioisomers (6&7) and reacting compound 6 with trimethoxy aniline (Scheme 1).
In 2020, Xu et al. [64] designed and constructed novel 2,4-disubstitutedpyrimidines as Aurora kinase inhibitors. In addition, the prepared derivatives were evaluated for their activity toward A549, HCT-116, and MCF-7 cell lines and SAR of the designed derivatives are depicted in Fig. 3. From these constructed derivatives, compound 11 demonstrated similar antitumor potential (IC50 equal to 12.05, 1.31, and 20.53 µM toward A549, HCT-116, and MCF-7, respectively) to that recorded by the standard VX680 (IC50 = 3.9, 1.49, and 17.39 µM, respectively). Regarding Aurora suppression activity, this derivative demonstrated suppression potential toward Aurora A (IC50 = 79.4 µM) and Aurora B (IC50 = 66.3 µM). SAR studies showed that cyclohexyl moiety at the tail exhibited better antiproliferative than aromatic ring toward HCT-116 cell lines. Replacing NH on the urea group with CH2 did not affect the anticancer activity on the tested three cell lines.
Furthermore, the N-(4-(2-(4-(morpholinophenyl)amino)pyrimidin-4-yl)phenyl)acrylamide derivative 12 (Fig. 4) was newly synthesized and showed high JAK3 suppression activity with IC50 = 1.7 nM with higher selectivity than JAK2 and JAK1 [65]. This selectivity was attributed to the formation of hydrophobic interaction between the aromatic amine moiety and Leu828 and Gly908 amino acids. Moreover, this candidate 12 demonstrated better suppression of T-cell proliferation potential (IC50 = 0.83 µM) than exhibited by tofacitinib (IC50 = 1.38 µM). SAR study revealing the effect of various substituents is illustrated in Fig. 4. In addition, the benzoxazolopyrimidine derivative 13 (Fig. 4) showed excellent anticancer potential toward leukemia cell line (RPMI-8226), prostate cancer cell line (PC-3), and renal cancer cell line (A498) with IC50 = 0.72, 0.7932, and 0.8511 µM, respectively. The mechanism of anticancer potential of this candidate was suggested to be PTK inhibition on MDA-MB-468 cell lines with IC50 = 0.07 µM [66].
VX680 (14) (Fig. 5) is a small pyrimidine heterocycle reported to suppress Aurora kinases A, B, and C with ki equal to 0.6, 18, and 4.6 nµ, sequentially [67]. This compound 14 has the ability to inhibit many cancer types as ovarian cancer, colon cancer, and leukemia [68]. While, ENMD-2076 (15) suppress Aurora A selectively with IC50 = 14 nM [69]. Luo et al. [70] designed and prepared new C-2, C-4, C-6 trisubstitutedpyrimidines as Aurora A suppressor. The design of these pyrimidine targets was based upon some modifications of both VX680 and ENMD-2076. These modifications include introducing N-substituted aniline at C-2 instead of S-substituted moiety in VX680. In addition, modification included replacement of styrene side chain in ENMD-2076 with N-benzylamine. SAR explaining effect of substituent at C-6 on the inhibitory potential on Aurora A is appeared in Fig. 5. From the constructed pyrimidines compound 16 recorded potential inhibitory effect on aurora A with IC50 = 25 nM through hydrophobic interaction and three hydrogen bondings with K162, A213, and L139. Moreover, the designed derivatives were assayed for anticancer potential in vitro by the use of VX680 as a standard toward breast cancer cell line as MCF-7, SK-BR-3, MDA-MD-231, and leukemia cell lines as K-562, U937, HL-60, MOLT-4. Results of compounds 16 and 17 are explained in Fig. 5.
Long et al. [71] in 2018 revealed that the disubstituted anilino moiety at C-2 of pyrimidine nucleus showed higher Aurora A inhibitory potential than the monosubstituted anilino fragment. This is obvious upon comparing compound 18 (IC50 = 7.7 nM) with 4-aminobenzoate 17 (IC50 = 46 nM).
Recently, in 2019, novel series of 4,6-diaryl-2-amine derivatives were evaluated for their anticancer potential toward colon cancer cell line (HCT-116) [72]. All the new compounds showed anticancer activity with half maximal cell growth inhibitory concentrations equal to 1.45–40.82 µM. The candidate 19—the most active anticancer agent (GI50 = 1.45)—was subjected to kinase assay using 25 cancer-associated kinases applying EMD Millipore Kinase Profiler service assay protocol. This tested compound 19 recorded high selectivity toward Aurora A kinase than other tested kinases. In silico docking of this derivative demonstrated that this derivative performed two hydrogen bondings with Ala213 and Pro214, in addition to seven hydrophobic interactions (Leu263, Arg220, Gly216, Tyr212, Glu211, Leu139, and Arg137) as seen in Fig. 6.
Pyrazolo[3,4-d]pyrimidine derivatives
Pyrazolopyrimidine derivatives 1NA-PP1 (20) and 1NM-PP1 (21) (Fig. 7) were documented to inhibit Src kinase and widely reported as standards for design of many pyrazolo[3,4-d]pyrimidine derivatives [73,74,75].
Moreover, novel derivatives of 1,4,6-trisubstituted pyrazolo[3,4-d]pyrimidines were designed based upon some chemical modifications of olomoucine 26 and roscovitine 27. These chemical modifications include replacing the imidazole ring in olomoucine with a pyrazole ring in addition to introduction different substituents at C-4, C-6, and N-1 of pyrazolopyrimidine. The newly synthesized derivatives were evaluated for their inhibitory activity against CDK2 and for their antiproliferative potential against A431, SNU638, and HCT-116 cell lines. In general, structure–activity relationship (SAR) recorded that compounds having anilino group at C-4 showed higher CDK2 and cell division than 4-benzyl derivatives. Compounds 25a, b having a 3-fluoroaniline group at C-4 were prepared and revealed comparable or superior cyclin-dependent kinase 2 (CDK2) (IC50 = 0.5 µM for 33a, 0.7 µM for 33b), inhibitory activity to those of olomoucine 26 (IC50 = 7 µM) and roscovitine 27 (IC50 = 0.5 µM) as reference compounds [76]. Moreover, unsubstitution at N-1 recorded higher potency for CDK2 inhibitory effect than substituted analogs. Regarding substitution at C-6, it was found that no difference between monoethanolamine and diethanolamine toward CDK2 activity (Fig. 8). The pyrazolopyrimidine derivatives (25a,b) were prepared as illustrated in Scheme 2. Accordingly, the reaction of 4-chloro-6-methylmercaptopyrazolo[3,4-d]pyrimidine (22) with 3-fluoroaniline in the presence of Hunig’s base in n-butanol yielded compound 23 which upon oxidation with m-CPBA followed by nucleophilic displacement of the resulting activated methylsulfonyl groups with hydroxylamines gave the target 1,4,6-trisubstituted pyrazolo[3,4-d]pyrimidines 25a,b.
Some 4-aminopyrazolo[3,4-d]pyrimidines substituted at position 1 and 6 were prepared and exhibited anticancer activity toward A431 cells, inhibited Src phosphorylation and induced apoptotic cell death. SAR studies of these derivatives are explained in Fig. 9. From the prepared compounds, derivative 26 incorporating a methylthio moiety at position 6 inhibited Src phosphorylation than the reference compound 27 (Fig. 9), so, the size of the alkyl group at position 6 was essential for activity [77].
Recently, novel derivatives of 4,6-disubstituted pyrazolopyrimidines incorporating various anilines at C-4 and thiophenethyl or thiopentane moieties at C-6 had been synthesized [78]. All the synthesized compounds were evaluated for in vitro CDK2/cyclin E and Abl kinase inhibitory activity as well as antiproliferative activity against K-562 (chronic myelogeneous leukemia), and MCF-7 (breast adenocarcinoma) cell lines. The SAR studies are explained in Fig. 10. From SAR, it is clear that derivative with thiophenethyl at C-6 with monosubstituted aniline showed better CDK2 inhibitory potential than thiopentane at C-6 and disubstituted anilines. From the prepared derivatives, compound 28 was the most CDK inhibitor with IC50 = 5.1 µM with antiproliferative activity toward K-562 (chronic myelogeneous leukemia), and MCF-7 (breast adenocarcinoma) cell lines with IC50 = 24.6 and 24.3 µM, respectively.
In 2008, new derivatives of 4-aminosubstitutedpyrazolo[3,4-d]pyrimidines were prepared and exhibited inhibitory activity against Src and proto-oncogene tyrosine protein kinase Ab1 kinases [79]. From this series, compound 29 was docked within adenine triphosphate (ATP)-binding sites of the two target enzymes (Fig. 11). This study recorded that in both enzymes compound 29 located pyrazolopyrimidine nucleus into the adenine region of the ATP-binding site, introducing the side chains at C-4 and N-1 toward two hydrophobic regions and the alkylthio substituent toward the external region of the binding pocket. Furthermore, this target compound performed two hydrogen bonding interactions with Src enzyme and one hydrogen bonding interaction with Abl as shown below (Fig. 11).
Ducray et al. [80] reported the preparation and in vitro evaluation of a new group containing anilinopyrazolo[3,4-d]pyrimidine derivatives 30a–c (Fig. 12) as receptor tyrosine protein kinase erb-2 (erbB2) and EGFR kinase inhibitors. Encouraging results with compound 30c provided potent, orally active erbB2 kinase inhibitor in rats and dogs with IC50 = 0.001 µM toward erbB2. Furthermore, novel derivatives of pyrazolo[3,4-d]pyrimdine-3,6-diamines (Fig. 12) were synthesized as potent and selective non-receptor tyrosine kinase, activated Cdc42Hs-asociated Kinase1 (ACK1) inhibitors. Compounds 31a and 31b showed promising ACK1 suppression with IC50 = 0.02 µM and 0.04 µM. A brief description SAR is illustrated in Fig. 12 [81].
Moreover, compound 32 (Fig. 13) was identified to inhibit EGFR enzyme at low-micromolar concentration with antiproliferative effect against cancer cells [82]. The in silico docking study of compound 32 demonstrated that the amino H and N-5 of pyrimidine ring make hydrogen bonds with backbone atoms of M769. The interaction of the ligand with T766 is mediated by a water molecule, and the 3-methylphenyl moiety substituted at N(1) is buried within the binding pocket (Fig. 13).
Recently in 2016, a new series of pyrazolo[3,4-d]pyrimidines hybridized with (4-substitutedbenzylidene)acetohydrazide derivatives (33a–g) and (Fig. 14) were synthesized and evaluated for their inhibitory activity toward epidermal growth factor receptor tyrosine kinase (EGFR-TK) [83]. Among these targets 33a–g, 33g was the most potent EGFR inhibitor (IC50 = 4.18 μM) followed by compound 33c (IC50 = 4.72 μM). Pyrazolo[3,4-d]pyrimidine combined with pyrazole moiety (35) revealed antiproliferative activity with IC50 = 5.00–32.52 μM on breast (MCF-7), colon (HCT-116), and liver (HEPG2) cancer cell lines [84]. Furthermore, this studied candidate inhibited fibroblast growth factor receptor (FGFR) with IC50 = 5.18 μM.
The [4-(1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-ylamino)phenyl]methanone derivative (35) was synthesized by heating 4-(1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-ylamino)benzohydrazide (34) with acetylacetone in acetic acid for 10 h under reflux conditions, as shown in Scheme 3.
Pyrido[2,3-d]pyrimidine derivatives
Two decades ago, PD 089828 (36) was synthesized as a novel 6-aryl-pyrido[2,3-d]pyrimidine derivative (Fig. 15), which was discovered as inhibitor for many PKs as human full-length fibroblast growth factor receptor-1 (FGFR-1), platelet-derived growth factor (PDGF) receptor b subunit (PDGFR-b), Src nonreceptor tyrosine kinase (c-Src), and epidermal growth factor receptor (EGFR) [85]. In addition, another group of pyrido[2,3-d]pyrimidines (37a–d) (Fig. 15) had been prepared and assessed for their potential antitumor agents. These compounds showed ATP-competitive inhibition of c-Src kinase with IC50 values 10 nM and from 6 to 100-folds selectivity for c-Src tyrosine kinase relative to the basic fibroblast growth factor receptor (bFGFr) tyrosine kinase, platelet-derived growth factor receptor (PDGFr) tyrosine kinase, and epidermal growth factor receptor (EGFr) tyrosine kinase [86, 87]. Moreover, the pyridopyrimidine derivative (PD 180970, 38) (Fig. 15) was prepared by Kraker et al. [87] and subjected to biological evaluation, which revealed that they inhibit Gab2 tyrosine phosphorylation in K-562 cells.
A group of coworkers [88] synthesized a novel series of pyrido[2,3-d]pyrimidine derivatives 39a–l (Fig. 16) as tyrosine kinase inhibitors. From this series, PD 089828 (39b) exhibited highly inhibitory activity against PDGFr, FGFr, and c-Src tyrosine kinase with IC50 values of 1.25, 0.14, and 0.22 µM, respectively. A SAR study was performed to reveal the effect of phenyl substitution at the 6-position on the potency of these three kinases. This study ascertained the importance of the presence of phenyl moieties in the 6-position substituted with either halogen or methyl groups at 2- and/or 6-position of phenyl ring on the inhibitory activity against PDGFr, FGFr, and c-Src tyrosine kinase as in the target compounds (39c, 39d, and 39f) which showed inhibitory activity than the unsubstituted parent compound 39a. Moreover, this study recorded that the introduction of ethyl (39i), or methoxy (39j) in the 2-position of the phenyl ring resulted in a decreased the inhibitory activity against both PDGFr-TK and FGFr-TK but c-Src inhibitory activity is not affected. In addition, this work explained that compounds which substituted at 4-position of the phenyl ring such as 39e, 39k, and 39l exhibited marked decrease or loss in their inhibitory activity against all tested kinases. Furthermore, the 2,4,6-trimethyl derivative 39h exhibited the same inhibitory activity as the 2,6-dimethyl derivative 39f, even though it contained a methyl substituent in the 4-position. Furthermore, the 3,5-dimethyl derivative 39g displayed selectivity for the FGFr-TK relative to the PDGFr-TK and the c-Src tyrosine kinase.
In an attempt to design potent kinase inhibitors, Reddy et al. [89] tested more than 150 cyanopyrido[2,3- d]pyrimidine derivatives. They found pyrido[2,3-d]pyrimidine-6-carbonitrile derivative (40) (Fig. 17) revealed the most potent activity and the most apoptosis inducer in tumor cells at a concentration of 30–100 nM. Moreover, this target compound inhibited many kinases such as CDK4/CYCLIN D1 and ARK5 kinases. Furthermore, Edupugantiet al. [90] reported the design and synthesis of a novel series of pyrido[2,3-d]pyrimidine-2,4-dione derivatives, which were evaluated for the eukaryotic elongation factor-2 kinase (eEF-2K) inhibitory activity. This study reported that compounds 43 (IC50 = 420 nM) and 44 (IC50 = 930 nM) (Fig. 17) were found to be the most active compounds among all the tested compounds.
The in silico docking studies of derivatives 43 within Eef-2k is shown in Fig. 18. The binding of inhibitor 6 involves hydrogen bonds to residues K170, I232, and G234. The hydrophobic cyclopropyl and ethyl groups are buried deep inside the adenine-binding pocket and underneath the Gly-rich-loop, respectively.
Synthesis of pyrido[2,3-d]pyrimidine-2,4-dione derivatives 43 and 44 were demonstrated in Scheme 4. The uracil derivatives 41a,b was subjected to Vilsmeierreactionto give the intermediate derivative 42, which then was treated with triethylamine and cyanoacetamide in ethanol, to afford the target compounds 43 and 44.
A novel 4-aminopyrido[2,3-d]pyrimidine derivative 45 (Fig. 19) was recorded to inhibit tyrosine kinases in the B-cell receptor and demonstrated antiproliferative activity toward 20 non-Hodgkin’s lymphomas (NHLs) cell lines with GI50 ranging from 1.3 to 6.9 μM at 24 h, and 1.4 to 7.2 μM at 48 h [83]. Also, a novel series of oxopyrido[2,3-d]pyrimidines was prepared and it was found that they inhibit gefitinib-resistant EGFRL858R, T790M with 100-fold selectivity over wild type. From this group, compound 46 (Fig. 19) showed strong antiproliferative activity against H1975 nonsmall cell lung cancer cell line, the first line mutant HCC827 cell line, and showed also promising antitumor activity in an EGFRL858R,T790M driven H1975 xenograft model sparing the side effects associated with the inhibition of wild-type EGFR [84].
Quinazoline derivatives
A new series of quinazoline derivatives was synthesized as EGFR kinase inhibitors and evaluated in cancer clinical trials. The anilinoquinazoline-containing compounds, Erlotinib (TarcevaTM) (47) [91, 92] and gefitinib (IressaTM) (48) [93, 94], had been used for the treatment of patients with advanced nonsmall lung cancer. In addition, lapatinib (TykerbTM) (49) was used for the treatment of human epidermal growth factor receptor 2 (HER2) positive advanced or metastatic breast cancer [95,96,97] (Fig. 20).
Another series of 4-anilinoquinazoline derivatives was prepared and tested for their inhibitory activity toward EGFR tyrosine kinase. SAR performed on this series recorded that substitution on the three position of aniline moiety with small lipophilic electron withdrawing group led to an increase in the potency of quinazoline derivative (50) (Fig. 21), which showed EGFR inhibitory activity with IC50 = 0.029 nM [98]. Moreover, compound 51 (Fig. 21) was synthesized and exhibited inhibitory activity toward both c-Src and Abl kinases [99].
A new series of 5-aminopyrimidinylquinazolines 52a–e (Fig. 22) had been designed as specific Aurora kinase inhibitors by Heron et al. [100]. The SAR study showed that the potency was dependent on the substitution on the phenyl ring of the benzamido group. The activity was increased by adding small hydrophobic groups such as 3-chloro (52a) and the 3-chloro, 4-fluoro (52b) analogs. On the other side a drastic negative effect on potency was revealed on the introduction of larger groups such as 3-bromo-4-methylbenzamido derivative (52c). Furthermore, solubility could be improved by adding heterocyclic group instead of phenyl ring (52d) or an alkyl substituent (52e) but a drop in the potency was observed. SAR of these derivatives 52a-e is explained in Fig. 22.
In addition, compound 53 (GW2016) (Fig. 23) was discovered to treat cancer cells selectively without affecting normal cells. This compound exerted its effect by inhibiting EGFR and ErbB-2 kinases and inducing apoptosis [101].
Furo[2,3-d]pyrimidine derivatives
Miyazaki et al. [102] recorded the synthesis and kinases inhibitory activity of a new series of 5,6-diaryl-furo-4-amino[2,3-d]pyrimidines. This work displayed that methoxyphenylfuro[2,3-d]pyrimidine (54) (Fig. 24) was the most active derivative with IC50 < 3 nM on both VEGFR2 and Tie-2 TK receptors. The activity of this compound was explained based on the X-ray crystal structure, which showed that the urea moiety formed two interactions with amino acids Asp1044 and Glu883. In addition, the amino group and nitrogen atom of the aminopyrimidine form interactions with Glu915 and Cys917.
In 2008, the same group modified the chemical structure of the previous furo[2,3-d]pyrimidine target compound 54 to change the activity of this compound away from Tie-2/VEGFR2 to target GSK-3 [103]. The structure modifications included the incorporation of the 3-pyridinyl moiety at the 6-position and different sulfonamides and amides at the para position of the 5-phenyl ring. From this series of the modified structure, compound 61 (Fig. 25) exhibited potent GSK-3b inhibitory activity with IC50 = 30 nM. The docking study of compound 61 displayed the aryl ring of the sulfonamide at 5-position appeared to clash with residues Met101, Leu112, and Leu130 of GSK-3. In addition, N3 nitrogen and NH2 of aminopyrimidine are anchored with the carbonyl moiety and NH of Val135, respectively, via hydrogen bond interactions (Fig. 26). Moreover, the 3-pyridine moiety at 6-position is close to Lys85 of the conserved salt bridge (Lys85/Glu97).
The target furopyrimidine derivative (61) was prepared as illustrated in Scheme 5. Accordingly, sequential acetylation, bromination, and hydroxylation of 4-aminoacetophenone (55) resulted in the α-hydroxyketone (56) which upon treatment with malononitrile in the presence of diethylamine provided 2-amino-3-cyano-furan 57. Reaction of 57 with triethylorthoformate, followed by amination and cyclization in the presence of sodium ethoxide yielded 4-amino-furo[2,3-d]pyrimidine 58. Bromination of 58 at 6-position with NBS followed by Suzuki coupling with 3-pyridineboronic acid pinacol ester gave furopyrimidine 60 which upon removal of the acetyl group and reaction of the resulting amine with benzenesulfonyl chloride provided the target compound (61).
In addition, a group of 2,4-diaminofuro[2,3-d]pyrimidines were prepared as a novel class of in an attempt to inhibit both dihydrofolate reductase and receptor tyrosine kinases [104]. Compound 62 (Fig. 27) serves as a template for rationally designed VEGFR2 and PDGFR-b inhibitory activity combined with DHFR inhibitory activity in one molecule. Moreover, the candidate compound (63) (Fig. 27) exhibited anticancer and anti-angiogenic activity in mouse HT-29 xenograft model via inhibition of both VEGFR2 and Tie-2 enzymes [105].
Furthermore, 3-pyridylfuro[2,3-d]pyrimidine derivative 64 (Fig. 28) was synthesized and evaluated for its inhibitory activity against a panel of many kinases [106]. This study demonstrated that compound 64 exhibited selective and potent inhibitory activity against GSK-3b over other tested kinases. Moreover, the furo[2,3-d]pyrimidine derivative 65 (Fig. 28) was found to possess high inhibitory activity against aurora kinase A with IC50 = 159 nM [107].
Conclusion
Pyrimidine ring and its fused derivatives including pyrazolo[3,4-d]pyrimidine, pyrido[2,3-d]pyrimidine, quinazoline, and furo[2,3-d]pyrimidine derivatives have proved as great target molecules in medicinal chemistry and drug development. These scaffolds exerted their effect through inhibiting PKs which are considered as essential enzymes to regulate cell growth, differentiation, migration, and metabolism. This review has highlighted the most promising compounds among these scaffolds based on primary literature. It is anticipated that information given in this review would give rise to design of better molecules with better anticancer activity and increased specificity, and finally will lead to the development of novel synthetic strategies.
References
Hill RP. Identifying cancer stem cells in solid tumors: case not proven. Cancer Res. 2006;66:1891–6.
Haddow S, Fowlis D, Parkinson K, Akhurst R, Balmain A. Loss of growth control by TGF-beta occurs at a late stage of mouse skin carcinogenesis and is independent of ras gene activation. Oncogene. 1991;6:1465–70.
Sozzi G, Pastorino U, Moiraghi L, Tagliabue E, Pezzella F, Ghirelli C, et al. Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res. 1998;58:5032–37.
Abdellatif KR, Abdelall EK, Abdelgawad MA, Ahmed RR, Bakr RB. Synthesis, docking study and antitumor evaluation of certain newly synthesized pyrazolo [3, 4-d] pyrimidine derivatives. Org Chem.: An Indian Journal 2014;10:157–67.
Abdelgawad MA, Bakr RB, Omar HA. Design, synthesis and biological evaluation of some novel benzothiazole/benzoxazole and/or benzimidazole derivatives incorporating a pyrazole scaffold as antiproliferative agents. Bioorg Chem. 2017;74:82–90.
Bakr RB, Abdelall EK, Abdel-Hamid MK, Kandeel MM. Design and synthesis of new EGFR-tyrosine kinase inhibitors containing pyrazolo [3, 4-d] pyrimidine cores as anticancer agents. Bull Pharm Sci Assiut Univ. 2012;35:1–16.
Klagsbrun M, Knighton D, Folkman J. Tumor angiogenesis activity in cells grown in tissue culture. Cancer Res. 1976;36:110–4.
Thurston DE. Chemistry and pharmacology of anticancer drugs. United States: CRC Press; 2006.
Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64.
Mundy GR. Mechanisms of bone metastasis. Cancer. 1997;80:1546–56.
Brownstein MH, Helwig EB. Spread of tumors to the skin. Arch Dermatol. 1973;107:80–6.
B Bakr R, BM Mehany A, RA Abdellatif K. Synthesis, EGFR inhibition and anti-cancer activity of new 3, 6-dimethyl-1-phenyl-4-(substituted-methoxy) pyrazolo [3, 4-d] pyrimidine derivatives. Anti-Cancer Agents Med Chem. 2017;17:1389–400.
Wu H-C, Chang D-K, Huang C-T. Targeted therapy for cancer. J Cancer Mol. 2006;2:57–66.
Hall JJ, Loggie BW, Shen P, Beamer S, Case LD, McQuellon R, et al. Cytoreductive surgery with intraperitoneal hyperthermic chemotherapy for advanced gastric cancer. J Gastrointest Surg. 2004;8:454–63.
Chabner BA, Roberts TG. Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5:65–72.
Abdellatif KR, Abdelall EK, Abdelgawad MA, Ahmed RR, Bakr RB. Synthesis and anticancer activity of some new pyrazolo [3, 4-d] pyrimidin-4-one derivatives. Molecules. 2014;19:3297–309.
Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579.
Klement G, Baruchel S, Rak J, Man S, Clark K, Hicklin DJ, et al. Continuous low-dose therapy with vinblastine and VEGF receptor-2 antibody induces sustained tumor regression without overt toxicity. J Clin Investig. 2006;116:2827–27.
Dachs GU, Dougherty GJ, Stratford IJ, Chaplin DJ. Targeting gene therapy to cancer: a review. Oncol Res. 1997;9:313–25.
Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 1998;279:377–80.
Escudier B, Michaelson M, Motzer R, Hutson T, Clark J, Lim H, et al. Axitinib versus sorafenib in advanced renal cell carcinoma: subanalyses by prior therapy from a randomised phase III trial. Br J Cancer. 2014;110:2821.
Sawyers C. Targeted cancer therapy. Nature. 2004;432:294.
Lavogina D, Enkvist E, Uri A. Bisubstrate inhibitors of protein kinases: from principle to practical applications. ChemMedChem. 2010;5:23–34.
Cheng Y, Zhang Y, McCammon JA. How does the cAMP-dependent protein kinase catalyze the phosphorylation reaction: an ab initio QM/MM study. J Am Chem Soc. 2005;127:1553–62.
Luković E, González-Vera JA, Imperiali B. Recognition-domain focused chemosensors: versatile and efficient reporters of protein kinase activity. J Am Chem Soc. 2008;130:12821–27.
Dissmeyer N, Schnittger A. The age of protein kinases. In: Plant Kinases. Clifton, NJ: Springer; 2011. p. 7–52. https://doi.org/10.1007/978-1-61779-264-9_2.
Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12:349.
Smith DS, Tsai L-H. Cdk5 behind the wheel: a role in trafficking and transport? Trends Cell Biol. 2002;12:28–36.
Weishaupt J, Neusch C, Bähr M. Cyclin-dependent kinase 5 (CDK5) and neuronal cell death. Cell Tissue Res. 2003;312:1–8.
Garcia-Echeverria C, Sellers W. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511.
Xu K, Shu H-KG. EGFR activation results in enhanced cyclooxygenase-2 expression through p38 mitogen-activated protein kinase–dependent activation of the SP1/SP3 transcription factors in human gliomas. Cancer Res. 2007;67:6121–9.
Meulenbeld HJ, Mathijssen RH, Verweij J, de Wit R, de Jonge MJ. Danusertib, an aurora kinase inhibitor. Expert Opin Investig Drugs. 2012;21:383–93.
Naula C, Parsons M, Mottram JC. Protein kinases as drug targets in trypanosomes and Leishmania. Biochim Biophys Acta. 2005;1754:151–9.
Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28.
Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291.
Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin cancer Res. 2007;13:1576–83.
Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010;16:205.
Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130.
Traxler P, Furet P. Strategies toward the design of novel and selective protein tyrosine kinase inhibitors. Pharmacol Ther. 1999;82:195–206.
Paul MK, Mukhopadhyay AK. Tyrosine kinase–role and significance in cancer. Int J Med Sci. 2004;1:101.
Yu K, Toral-Barza L, Shi C, Zhang W-G, Lucas J, Shor B, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69:6232–40.
Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, et al. Developing irreversible inhibitors of the protein kinase cysteinome. Chem Biol. 2013;20:146–59.
Akritopoulou-Zanze I, Hajduk PJ. Kinase-targeted libraries: the design and synthesis of novel, potent, and selective kinase inhibitors. Drug Discov Today. 2009;14:291–7.
Cocco MT, Congiu C, Lilliu V, Onnis V. Synthesis and in vitro antitumoral activity of new hydrazinopyrimidine-5-carbonitrile derivatives. Bioorg Med Chem. 2006;14:366–72.
Kassab AE, Gedawy EM. Synthesis and anticancer activity of novel 2-pyridyl hexahyrocyclooctathieno [2, 3-d] pyrimidine derivatives. Eur J Med Chem. 2013;63:224–30.
Hafez HN, El-Gazzar A-RB. Synthesis and antitumor activity of substituted triazolo [4, 3-a] pyrimidin-6-sulfonamide with an incorporated thiazolidinone moiety. Bioorg Med Chem Lett. 2009;19:4143–7.
Lauria A, Patella C, Dattolo G, Almerico AM. Design and synthesis of 4-substituted indolo [3, 2-e][1, 2, 3] triazolo [1, 5-a] pyrimidine derivatives with antitumor activity. J Med Chem. 2008;51:2037–46.
Zhang N, Ayral-Kaloustian S, Nguyen T, Hernandez R, Beyer C. 2-Cyanoaminopyrimidines as a class of antitumor agents that promote tubulin polymerization. Bioorg Med Chem Lett. 2007;17:3003–05.
Abdelgawad MA, Bakr RB, Azouz AA. Novel pyrimidine-pyridine hybrids: Synthesis, cyclooxygenase inhibition, anti-inflammatory activity and ulcerogenic liability. Bioorg Chem. 2018;77:339–48.
Lacbay CM, Mancuso J, Lin Y-S, Bennett N, Götte M, Tsantrizos YS. Modular assembly of purine-like bisphosphonates as inhibitors of HIV-1 reverse transcriptase. J Med Chem. 2014;57:7435–49.
Chauhan M, Kumar R. Medicinal attributes of pyrazolo [3, 4-d] pyrimidines: a review. Bioorg Med Chem. 2013;21:5657–68.
Ismail NS, Ali GM, Ibrahim DA, Elmetwali AM. Medicinal attributes of pyrazolo [1, 5-a] pyrimidine based scaffold derivatives targeting kinases as anticancer agents. Future J Pharm Sci. 2016;2:60–70.
Elrazaz EZ, Serya RA, Ismail NS, El Ella DAA, Abouzid KA. Thieno [2, 3-d] pyrimidine based derivatives as kinase inhibitors and anticancer agents. Future J Pharm Sci. 2015;1:33–41.
Joo YH, Min SU, Lee DH, Suh DH. A case of drug eruption with periorbital edema and exfoliative dermatitis induced by imatinib mesylate (STI571, Gleevec (TM)). Korean. J Dermatol. 2007;45:194–6.
Cozzi P, Mongelli N, Suarato A. Recent anticancer cytotoxic agents. Curr Med Chem-Anti-Cancer Agents. 2004;4:93–121.
Kantarjian HM, Giles F, Gattermann N, Bhalla K, Alimena G, Palandri F, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome–positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood. 2007;110:3540–6.
White DL, Saunders VA, Dang P, Engler J, Zannettino AC, Cambareri AC, et al. OCT-1–mediated influx is a key determinant of the intracellular uptake of imatinib but not nilotinib (AMN107): reduced OCT-1 activity is the cause of low in vitro sensitivity to imatinib. Blood. 2006;108:697–704.
Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362:2260–70.
Copland M, Hamilton A, Elrick LJ, Baird JW, Allan EK, Jordanides N, et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107:4532–9.
Rix U, Hantschel O, Dürnberger G, Rix LLR, Planyavsky M, Fernbach NV, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–63.
Tokarski JS, Newitt JA, Chang CYJ, Cheng JD, Wittekind M, Kiefer SE, et al. The structure of Dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66:5790–7.
Mariaule G, Belmont P. Cyclin-dependent kinase inhibitors as marketed anticancer drugs: where are we now? A short survey. Molecules. 2014;19:14366–82.
Yang EH, Yun JI, Latif M, Lee HJ, Yun C-S, Lee K, et al. New pyrimidine derivatives possessing ALK inhibitory activities. Bull Korean Chem Soc. 2013;34:3129–32.
Xu Y, Hao S-Y, Zhang X-J, Li W-B, Qiao X-P, Wang Z-X, et al. Discovery of novel 2, 4-disubstituted pyrimidines as Aurora kinase inhibitors. Bioorg Med Chem Lett. 2020;30:126885.
Shu L, Chen C, Huan X, Huang H, Wang M, Zhang J, et al. Design, synthesis, and pharmacological evaluation of 4-or 6-phenyl-pyrimidine derivatives as novel and selective Janus kinase 3 inhibitors. Eur J Med Chem. 2020;191:112148.
Chikhale R, Thorat S, Choudhary RK, Gadewal N, Khedekar P. Design, synthesis and anticancer studies of novel aminobenzazolyl pyrimidines as tyrosine kinase inhibitors. Bioorg Chem. 2018;77:84–100.
Bennett MJ, Zehnder LR, Ninkovic S, Kung P-P, Meng JJ, Buwen H. 2-amino pyrimidine compounds. Google Patents; 2010.
Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–7.
Katsha A, Belkhiri A, Goff L, El-Rifai W. Aurora kinase A in gastrointestinal cancers: time to target. Mol Cancer. 2015;14:106.
Luo Y, Deng Y-Q, Wang J, Long Z-J, Tu Z-C, Peng W, et al. Design, synthesis and bioevaluation of N-trisubstituted pyrimidine derivatives as potent aurora A kinase inhibitors. Eur J Med Chem. 2014;78:65–71.
Long L, Luo Y, Hou Z-J, Ma H-J, Long Z-J, Tu Z-C, et al. Synthesis and biological evaluation of aurora kinases inhibitors based on N-trisubstituted pyrimidine scaffold. Eur J Med Chem. 2018;145:805–12.
Lee YH, Park J, Ahn S, Lee Y, Lee J, Shin SY, et al. Design, synthesis, and biological evaluation of polyphenols with 4, 6-diphenylpyrimidin-2-amine derivatives for inhibition of Aurora kinase A. DARU. 2019;27:265–81.
Oh H, Ozkirimli E, Shah K, Harrison ML, Geahlen RL. Generation of an analog-sensitive Syk tyrosine kinase for the study of signaling dynamics from the B cell antigen receptor. J Biol Chem. 2007;282:33760–8.
Blethrow J, Zhang C, Shokat KM, Weiss EL. Design and use of analog‐sensitive protein kinases. Current Protoc Mol Biol. 2004;66:18.11.1–18.11.19.
Lopez MS, Choy JW, Peters U, Sos ML, Morgan DO, Shokat KM. Staurosporine-derived inhibitors broaden the scope of analog-sensitive kinase technology. J Am Chem Soc. 2013;135:18153–9.
Kim DC, Lee YR, Yang B-S, Shin KJ, Kim DJ, Chung BY, et al. Synthesis and biological evaluations of pyrazolo [3, 4-d] pyrimidines as cyclin-dependent kinase 2 inhibitors. Eur J Med Chem. 2003;38:525–32.
Schenone S, Bruno O, Ranise A, Bondavalli F, Brullo C, Fossa P, et al. New pyrazolo [3, 4-d] pyrimidines endowed with A431 antiproliferative activity and inhibitory properties of Src phosphorylation. Bioorg Med Chem Lett. 2004;14:2511–7.
Cherukupalli S, Chandrasekaran B, Kryštof V, Aleti RR, Sayyad N, Merugu SR, et al. Synthesis, anticancer evaluation, and molecular docking studies of some novel 4, 6-disubstituted pyrazolo [3, 4-d] pyrimidines as cyclin-dependent kinase 2 (CDK2) inhibitors. Bioorg Chem. 2018;79:46–59.
Schenone S, Brullo C, Bruno O, Bondavalli F, Mosti L, Maga G, et al. Synthesis, biological evaluation and docking studies of 4-amino substituted 1H-pyrazolo [3, 4-d] pyrimidines. Eur J Med Chem. 2008;43:2665–76.
Ducray R, Ballard P, Barlaam BC, Hickinson MD, Kettle JG, Ogilvie DJ, et al. Novel 3-alkoxy-1H-pyrazolo [3, 4-d] pyrimidines as EGFR and erbB2 receptor tyrosine kinase inhibitors. Bioorg Med Chem Lett. 2008;18:959–62.
Kopecky DJ, Hao X, Chen Y, Fu J, Jiao X, Jaen JC, et al. Identification and optimization of N 3, N 6-diaryl-1H-pyrazolo [3, 4-d] pyrimidine-3, 6-diamines as a novel class of ACK1 inhibitors. Bioorg Med Chem Lett. 2008;18:6352–6.
Cavasotto CN, Ortiz MA, Abagyan RA, Piedrafita FJ. In silico identification of novel EGFR inhibitors with antiproliferative activity against cancer cells. Bioorg Med Chem Lett. 2006;16:1969–74.
Abdelgawad MA, Bakr RB, Alkhoja OA, Mohamed WR. Design, synthesis and antitumor activity of novel pyrazolo [3, 4-d] pyrimidine derivatives as EGFR-TK inhibitors. Bioorg Chem. 2016;66:88–96.
Bakr RB, Mehany A. (3, 5-Dimethylpyrazol-1-yl)-[4-(1-phenyl-1H-pyrazolo [3, 4-d] pyrimidin-4-ylamino) phenyl] methanone. Molbank. 2016;2016:M915.
Dahring TK, Lu GH, Hamby JM, Batley BL, Kraker AJ, Panek RL. Inhibition of growth factor-mediated tyrosine phosphorylation in vascular smooth muscle by PD 089828, a new synthetic protein tyrosine kinase inhibitor. J Pharmacol Exp Ther. 1997;281:1446–56.
Boschelli DH, Wu Z, Klutchko SR, Showalter HH, Hamby JM, Lu GH, et al. Synthesis and tyrosine kinase inhibitory activity of a series of 2-amino-8 H-pyrido [2, 3-d] pyrimidines: identification of potent, selective platelet-derived growth factor receptor tyrosine kinase inhibitors. J Med Chem. 1998;41:4365–77.
Kraker AJ, Hartl BG, Amar AM, Barvian MR, Showalter HH, Moore CW. Biochemical and cellular effects of c-Src kinase-selective pyrido [2, 3-d] pyrimidine tyrosine kinase inhibitors. Biochem Pharmacol. 2000;60:885–98.
Connolly CJ, Hamby JM, Schroeder MC, Barvian M, Lu GH, Panek RL, et al. Discovery and structure-activity studies of a novel series of pyrido [2, 3-d] pyrimidine tyrosine kinase inhibitors. Bioorg Medicinal Chem Lett. 1997;7:2415–20.
Reddy MR, Akula B, Cosenza SC, Athuluridivakar S, Mallireddigari MR, Pallela VR. et al. Discovery of 8-cyclopentyl-2-[4-(4-methyl-piperazin-1-yl)-phenylamino]-7-oxo-7, 8-dihydro-pyrido [2, 3-d] pyrimidine-6-carbonitrile (7x) as a potent inhibitor of cyclin-dependent kinase 4 (CDK4) and AMPK-related kinase 5 (ARK5). J Med Chem. 2014;57:578–99.
Edupuganti R, Wang Q, Tavares CD, Chitjian CA, Bachman JL, Ren P, et al. Synthesis and biological evaluation of pyrido [2, 3-d] pyrimidine-2, 4-dione derivatives as eEF-2K inhibitors. Bioorg Med Chem. 2014;22:4910–6.
Huang S, Armstrong EA, Benavente S, Chinnaiyan P, Harari PM. Dual-agent molecular targeting of the epidermal growth factor receptor (EGFR): combining anti-EGFR antibody with tyrosine kinase inhibitor. Cancer Res. 2004;64:5355–62.
Gordon A, Finkler N, Edwards R, Garcia A, Crozier M, Irwin D, et al. Efficacy and safety of erlotinib HCl, an epidermal growth factor receptor (HER1/EGFR) tyrosine kinase inhibitor, in patients with advanced ovarian carcinoma: results from a phase II multicenter study. Int J Gynecol Cancer. 2005;15:785–92.
Kim ES, Hirsh V, Mok T, Socinski MA, Gervais R, Wu Y-L, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet. 2008;372:1809–18.
Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 2004;305:1163–7.
Higa HM. Breast cancer: beyond the cutting edge. Expert Opin Pharmacother. 2009;10:2479–98.
Ren H, Yang B, Rainov NG. Receptor tyrosine kinases as therapeutic targets in malignant glioma. Rev Recent Clin Trials. 2007;2:87–101.
Wiedmann W, Mossner M. J. Molecular targeted therapy of hepatocellular carcinoma-results of the first clinical studies. Curr Cancer Drug Targets. 2011;11:714–33.
Rewcastle GW, Denny WA, Bridges AJ, Zhou H, Cody DR, McMichael A, et al. Tyrosine kinase inhibitors. 5. Synthesis and structure-activity relationships for 4-[(phenylmethyl) amino]-and 4-(phenylamino) quinazolines as potent adenosine 5'-triphosphate binding site inhibitors of the tyrosine kinase domain of the epidermal growth factor receptor. J Med Chem. 1995;38:3482–87.
Hennequin LF, Allen J, Breed J, Curwen J, Fennell M, Green TP. et al. N-(5-Chloro-1, 3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl) ethoxy]-5-(tetrahydro-2 H-pyran-4-yloxy) quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor. J Med Chem. 2006;49:6465–88.
Heron NM, Anderson M, Blowers DP, Breed J, Eden JM, Green S, et al. SAR and inhibitor complex structure determination of a novel class of potent and specific Aurora kinase inhibitors. Bioorg Med Chem Lett. 2006;16:1320–3.
Spector N. Elucidating the role of truncated ErB2 receptor (p95) in breast cancer. United states: DTIC; 2011.
Miyazaki Y, Matsunaga S, Tang J, Maeda Y, Nakano M, Philippe RJ, et al. Novel 4-amino-furo [2, 3-d] pyrimidines as Tie-2 and VEGFR2 dual inhibitors. Bioorg Med Chem Lett. 2005;15:2203–7.
Miyazaki Y, Maeda Y, Sato H, Nakano M, Mellor GW. Rational design of 4-amino-5, 6-diaryl-furo [2, 3-d] pyrimidines as potent glycogen synthase kinase-3 inhibitors. Bioorg Med Chem Lett. 2008;18:1967–71.
Gangjee A, Zeng Y, Ihnat M, Warnke LA, Green DW, Kisliuk RL, et al. Novel 5-substituted, 2, 4-diaminofuro [2, 3-d] pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors with antiangiogenic and antitumor activity. Bioorg Med Chem. 2005;13:5475–91.
Miyazaki Y, Tang J, Maeda Y, Nakano M, Wang L, Nolte RT, et al. Orally active 4-amino-5-diarylurea-furo [2, 3-d] pyrimidine derivatives as anti-angiogenic agent inhibiting VEGFR2 and Tie-2. Bioorg Med Chem Lett. 2007;17:1773–8.
Maeda Y, Nakano M, Sato H, Miyazaki Y, Schweiker SL, Smith JL, et al. 4-Acylamino-6-arylfuro [2, 3-d] pyrimidines: potent and selective glycogen synthase kinase-3 inhibitors. Bioorg Med Chem Lett. 2004;14:3907–11.
Coumar MS, Chu C-Y, Lin C-W, Shiao H-Y, Ho Y-L, Reddy R, et al. Fast-forwarding hit to lead: aurora and epidermal growth factor receptor kinase inhibitor lead identification. J Med Chem. 2010;53:4980–8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abdellatif, K.R.A., Bakr, R.B. Pyrimidine and fused pyrimidine derivatives as promising protein kinase inhibitors for cancer treatment. Med Chem Res 30, 31–49 (2021). https://doi.org/10.1007/s00044-020-02656-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-020-02656-8