Abstract
A series of new coumarin-based 1,2,3-triazole derivatives were designed, synthesized and evaluated for their antitubercular activity in vitro against Mycobacterium tuberculosis H37Ra, antioxidant activity by DPPH radical scavenging assay, antimicrobial activity in vitro against three gram-positive bacteria (Staphylococcus aureus, Micrococcus luteus and Bacillus cereus) and three gram-negative bacteria (Escherichia coli, Pseudomonas fluorescens and Flavobacterium devorans as well as three fungi (Aspergillus niger, Penicillium chrysogenum and Curvularia lunata). The bioactive assay showed that some synthesized coumarin triazoles displayed comparable or even better antitubercular, antioxidant, antibacterial and antifungal efficacy in comparison with reference drugs. Furthermore, docking study has been performed against DprE1 enzyme of M. tuberculosis that showed good binding interactions. Moreover, the synthesized compounds were also analyzed for ADME properties and showed potential to build up as good oral drug candidates.
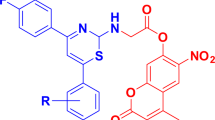
Graphical Abstract
New coumarin-based 1,2,3-triazole derivatives were designed, synthesized and evaluated for their antitubercular, antioxidant, antibacterial and antifungal activity. Some of the coumarin-based triazole derivatives displayed comparable or even better efficacy in comparison with reference drugs. Molecular docking study has been performed against DprE1 enzyme of Mycobacterium tuberculosis showed good binding interactions.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tuberculosis (TB) is the most important life-threatening infectious disease caused by the bacterial pathogen bacillus Mycobacterium tuberculosis (MTB) and characterized by tubercle lesions in the lungs (Russel et al., 2010; Dye and Williams 2010). According to the World Health Organization (WHO), in 2013, 9 million new TB cases and deaths of 1.5 million peoples were occurred due to the active TB (WHO Report, 2014). The mortality and spread of this disease have been further aggravated by its synergy with human immunodeficiency virus (HIV) (Guardiola-Diaz et al., 2001). Moreover, the appearance of multidrug-resistant strains (MDR) of MTB has led to the expansion of this disease and that is why the WHO gives priority immediately to control tuberculosis infection to prevent the spread of drug-resistant strains (WHO Report, 2000). However, there is a serious problem emergence as MTB developed resistance not only against the first-line drug, but also against the second-line drugs. Because of this, there is an emergence of MDR and extensively drug-resistant (XDR) strains of MTB all over the world (Singh, 2007).
In recent years, click chemistry has emerged as a fast and powerful approach to the synthesis of novel compounds with biological importance. The copper-catalyzed 1,3-dipolar azide and alkyne cycloaddition (CuAAC) reaction (Bock et al., 2006; Moses and Moorhouse, 2007; Meldal and Tornoe, 2008; Hein and Fokin, 2010) have emerged as the premier examples of “click chemistry” as it is virtually quantitative and easy to perform. The triazole formed is essentially inert to reactive conditions such as oxidation, reduction and hydrolysis. CuAAC is particularly useful for the synthesis of a variety of molecules ranging from enzyme inhibitors to molecular materials (Binder and Kluger, 2006). 1,2,3-Triazoles are important class of target molecules due to their interesting biological properties such as antitubercular (Boechat et al., 2011), antiallergic, antibacterial, anti-HIV activity (Agalave et al., 2011), antifungal (Lima-Neto et al., 2012) and α-glycosidase inhibitors (Senger et al., 2012). Currently, a few 1,2,3-triazole-based compounds are already in the market or in the final stages of clinical trials (Therrien and Levesque, 2000).
Literature survey reveals that coumarin backbone in assimilation with some nitrogen-containing heterocyclic moieties such as azetidine, thiazolidine, thiazole and triazole significantly increases the antimicrobial activity and broadens their antimicrobial spectrum (Ronad et al., 2010; Raghu et al., 2009; Shi and Zhou, 2011). Coumarin derivatives exhibit enormous amount of biological activities such as antioxidant, antimicrobial, anti-HIV, antibiotic, anticancer, muscle relaxant, anti-inflammatory, anticoagulant activity (Murakami et al., 2000) and potential antitumor agent (Zhang et al., 2014). In recent years, a library of coumarin derivatives conjugated with 1,2,3-triazole were synthesized and proved to possess different bioactivity. Zhang and coworkers discovered (Zhang et al., 2014) 4-(1,2,3-triazol-1-yl)coumarin derivatives exhibit cytotoxic activity against three human cancer cell lines, including human breast carcinoma MCF-7 cell, colon carcinoma SW 480 cell and lung carcinoma A549 cell. Similarly, there are various coumarin–triazole conjugated system which shows antimicrobial (Kushwaha et al., 2014), antimalarial (Pingaew et al., 2014), 5-lipoxygenase inhibitor (Ouellet et al., 2012) and anti-inflammatory activity (Stefani et al., 2012).
In continuation of our earlier work (Shingate et al., 2011; Kategaonkar et al., 2010a, b; Nikalje et al., 2015; Sangshetti et al., 2014) on synthesis and biological properties of heterocyclic moieties and considering the importance of coumarin and triazole moieties as a single molecular scaffold, we report herein the design and syntheses of new coumarin-linked triazole hybrids and evaluate their antitubercular, antioxidant, antibacterial and antifungal activities. The computational parameters like docking study for antitubercular activity and ADME prediction of titled compounds 4a–k were also performed.
Experimental
Materials and methods
All the solvents and reagents were purchased from commercial suppliers Spectrochem Pvt. Ltd., Sigma-Aldrich and Rankem India Ltd. and used without further purification. The key starting material 1a–f, 2a, 3a and 3b was synthesized according to the literature (Alvarez and Alvarez, 1997; Srinivasan et al., 2007; Naik et al., 2012; Anand et al., 2011). The progress of each reaction was monitored by ascending thin-layer chromatography (TLC) using TLC aluminum sheets, silica gel F254 precoated, Merck, Germany, and the spots are located using UV light as the visualizing agent or iodine vapors. Melting points were taken in open capillary method and are uncorrected. IR spectra were taken on Bruker FT-IR 4000, and the wave numbers were given in cm−1. 1H NMR spectra were recorded (CDCl3/DMSO-d6) on Bruker Avance 200 NMR Spectrometer. 13C NMR and DEPT 135 spectra were recorded (CDCl3/DMSO-d6) on Bruker Avance 200 NMR Spectrometer and JEOL ECX 400 NMR Spectrometer. Chemical shifts (δ) are reported in parts per million (ppm) using tetramethylsilane (TMS) as an internal standard. The splitting pattern abbreviations are designed as singlet (s), doublet (d), double doublet (dd), triplet (t), quartet (q) and multiplet (m).The mass spectra were recorded on Q-TOF micromass (YA-105) spectrometer in the ESI (electrospray ionization) modes.
General procedure for the synthesis of 4a–f and 4g–k
To the stirred solution of 4-Methyl-7-(prop-2-yn-1-yloxy)-2H-chromen-2-one (3a) or 4-(Prop-2-yn-1-yloxy)-2H-chromen-2-one (3b) (0.5 mmol), substituted benzyl azide 1a–f for 3a and 1a–e for 3b (0.5 mmol) and copper diacetate (Cu(OAc)2) (20 mol%) in t-BuOH-H2O (3:1, 8 mL) were added and the resulting mixture was stirred at room temperature for 16–22 h. The progress of the reaction was monitored by TLC using ethyl acetate/hexane as a solvent system. The reaction mixture was quenched with crushed ice and extracted with ethyl acetate (2 × 15 mL). The organic extracts were washed with brine solution (2 × 15 mL) and dried over anhydrous sodium sulfate. The solvent was evaporated under reduced pressure to afford the corresponding crude compounds. The obtained crude compounds were recrystallized using ethanol.
4-Methyl-7-((1-(4-nitrobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4a )
The compound 4a was obtained as a off-white solid via 1,3-dipolar cycloaddition reaction between azide 1a (89 mg) and alkyne 3a (107 mg) in 18 h with 92 % yield. Mp 146–147 °C. IR (KBr, cm−1): υ max 2916 (C–H), 1714 (C=O), 1612 (C=C), 1518 (N–O), 1388 (C–CH3), 1340 (C–N) and 1143 (C–O). 1H NMR (200 MHz, CDCl3, ppm): δ H 2.40 (s, 3H), 5.27 (s, N–CH2), 5.69 (s, O–CH2), 6.15 (s, 1H), 6.90–6.96 (m, 2H), 7.42–7.55 (m, 3H), 7.71 (s, 1H) and 8.21–8.25 (d, 2H). 13C NMR (50 MHz, CDCl3, ppm): δ 18.7 (–CH3), 53.3 (N–CH2), 62.2 (O–CH2), 102.1, 112.4, 114.2, 124.4, 125.8, 128.7, 141.4, 152.5 and 161.1 (C=O). HRMS calculated [M + H]+ for C20H17N4O5: 393.1193, found: 393.1190 and [M + Na]+ for C20H16N4O5Na: 415.1013, found: 415.1013.
4-Methyl-7-((1-(3-nitrobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4b )
The compound 4b was obtained as a pale pink solid via 1,3-dipolar cycloaddition reaction between azide 1b (89 mg) and alkyne 3a (107 mg) in 18 h with 92 % yield. Mp 173–174 °C. IR (KBr, cm−1): υ max 2916 (C–H), 1713 (C=O), 1621 (C=C), 1514 (N–O), 1386 (C–CH3), 1357 (C–N) and 1147 (C–O). 1H NMR (200 MHz, CDCl3, ppm): δ H 2.43 (s, 3H), 5.27 (s, N–CH2), 5.71 (s, O–CH2), 6.16 (s, 1H), 6.94–6.99 (d, 2H), 7.36–7.65 (m, 3H), 7.85 (s, 1H) and 8.19-8.26 (m, 2H). 13C NMR (50 MHz, CDCl3, ppm): δ 19.5 (–CH3), 54.1 (N–CH2), 62.9 (O–CH2), 103, 112.9, 113.7, 115.1, 123.9, 124.3, 124.7, 126.9, 131.3, 135, 137.5, 143.7, 154.3, 159.5 and 162.1 (C=O). HRMS calculated [M + H]+ for C20H17N4O5: 393.1193, found: 393.1193 and [M + Na]+ for C20H16N4O5Na: 415.1013, found: 415.1016.
4-Methyl-7-((1-(4-chlorobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4c )
The compound 4c was obtained as a white solid via 1,3-dipolar cycloaddition reaction between azide 1c (84 mg) and alkyne 3a (107 mg) in 17 h with 90 % yield. Mp 162–163 °C. IR (KBr, cm−1): υ max 2916 (C–H), 1692 (C=O), 1612 (C=C), 1390 (C–CH3), 1157 (C–O) and 754 (C–Cl). 1H NMR (200 MHz, CDCl3, ppm): δ H 2.40 (s, 3H), 5.24 (s, N–CH2), 5.53 (s, O–CH2), 6.15 (s, 1H), 6.92–6.96 (d, 2H), 7.21–7.25 (m, 2H), 7.35–7.39 (m, 2H) and 7.49–7.53 (d, 2H). 13C NMR (50 MHz, CDCl3, ppm): δ 18.7 (–CH3), 51.7 (N–CH2), 62.3 (O–CH2), 102.1, 112.3, 112.4, 114.1, 125.7, 129.4, 129.5, 132.8, 152.4, 155.1 and 161.1 (C=O). HRMS calculated [M + H]+ for C20H17ClN3O3: 382.0958, found: 382.0953 and [M + Na]+ for C20H16ClN3O3Na: 404.0778, found: 404.0768.
4-Methyl-7-((1-(3-chlorobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (4d )
The compound 4d was obtained as a off-white solid via 1,3-dipolar cycloaddition reaction between azide 1d (84 mg) and alkyne 3a (107 mg) in 18 h with 92 % yield. Mp 120–121 °C. IR (KBr, cm−1): υ max 2916 (C–H), 1720 (C=O), 1611 (C=C), 1385 (C–CH3), 1156 (C–O) and 679 (C–Cl). 1H NMR (200 MHz, CDCl3, ppm): δ H 2.38 (s, 3H), 5.23 (s, N–CH2), 5.68 (s, O–CH2), 6.11 (s, 1H), 6.87–6.96 (m, 2H), 7.19–7.51 (m, 5H) and 7.74 (s, 1H). 13C NMR (50 MHz, CDCl3, ppm): δ 18.7 (–CH3), 51.6 (N–CH2), 62.3 (O–CH2), 102.1, 112.2, 112.5, 114, 123.5, 125.7, 127.7, 130, 130.4, 130.5, 132.2, 133.5, 143.2, 152.6, 156.1 and 161 (C=O). HRMS calculated [M + H]+ for C20H17ClN3O3: 382.0958, found: 382.0953 and [M + Na]+ for C20H16ClN3O3Na: 404.0778, found: 404.0768.
4-Methyl-7-((1-(4-bromobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4e )
The compound 4e was obtained as a off-white solid via 1,3-dipolar cycloaddition reaction between azide 1e (106 mg) and alkyne 3a (107 mg) in 16 h with 95 % yield. Mp 159–160 °C. IR (KBr, cm−1): υ max 1697 (C=O), 1606 (C=C), 1386 (C–CH3), 1156 (C–O) and 752 (C–Br). 1H NMR (400 MHz, CDCl3, ppm):, δ H 2.40 (s, 3H), 5.25 (s, N–CH2), 5.52 (s, O–CH2), 6.15 (s, 1H), 6.92–6.96 (m, 2H), 7.15–7.19 (m, 2H), 7.28–7.37 (m, 2H) and 7.50–7.54 (m, 3H). 13C NMR (50 MHz, CDCl3, ppm): δ 18.7 (–CH3), 51.7 (N–CH2), 62.3 (O–CH2), 102.1, 112.3, 112.4, 114.1, 125.8, 129.4, 129.5, 132.9, 152.4, 155.1 and 161.1 (C=O). HRMS calculated [M + H]+ for C20H17BrN3O3: 426.0448, found: 426.0428 and [M + Na]+ for C20H16BrN3O3Na: 448.0268, found: 448.0266.
4-Methyl-7-((1-benzyl-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4f )
The compound 4f was obtained as a white solid via 1,3-dipolar cycloaddition reaction between azide 1f (67 mg) and alkyne 3a (107 mg) in 16 h with 92 % yield. Mp 134–135 °C. IR (KBr, cm−1): υ max 2915 (C–H), 1703 (C=O), 1649 (C=C) and 1155 (C–O). 1H NMR (400 MHz, CDCl3): δ H 2.39 (s, 3H), 5.23 (s, N–CH2), 5.55 (s, O–CH2), 6.14 (s, 1H), 6.89–6.94 (m, 2H), 7.37–7.58 (m, 7H). 13C NMR (100 MHz, CDCl3, ppm): δ 18.8 (–CH3), 54.4 (N–CH2), 62.4 (O–CH2), 102.1, 102.2, 112.3, 112.5, 112.8, 114.1, 123, 125.8, 128.3, 129, 129.3, 134.4, 143.5, 152.6, 155.2, 161.2 and 161.3 (C=O). HRMS calculated [M + H]+ for C20H18N3O3: 348.1343, found: 348.1346 and [M + Na]+ for C20H17N3O3Na: 370.1168, found: 370.1164.
4-((1-(4-Nitrobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (4g )
The compound 4g was obtained as a yellow solid via 1,3-dipolar cycloaddition reaction between azide 1a (89 mg) and alkyne 3b (100 mg) in 20 h with 89 % yield. Mp 195–196 °C. IR (KBr, cm−1): υ max 2916 (C–H), 1711 (C=O), 1620 (C=C), 1553 (N–O), 1102 (C–O) and 1339 (C–N). 1H NMR (200 MHz, DMSO-d 6 , ppm): δ H 5.41 (s, N–CH2), 5.65 (s, O–CH2), 6.15 (s, 1H), 7.32–7.47 (m, 6H), 7.60–7.74 (m, 2H) and 8.45 (s, 1H). 13C NMR (50 MHz, DMSO-d 6 , ppm): δ 52.1 (N–CH2), 62.7 (O–CH2), 91.3, 115, 116.4, 122.9, 124.2, 126.4, 128.8, 129.9, 132.8, 134.9, 141.4, 152.8, 161.5 (C=O) and 164.3. HRMS calculated [M + H]+ for C19H15N4O5: 379.1036, found: 379.1032 and [M + Na]+ for C19H14N4O5Na: 401.0856, found: 401.0853.
4-((1-(3-Nitrobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4h )
The compound 4h was obtained as a white solid via 1,3-dipolar cycloaddition reaction between azide 1b (89 mg) and alkyne 3b (100 mg) in 21 h with 87 % yield, Mp 153–154 °C. IR (KBr, cm−1): υ max 1703 (C=O), 1619 (C=C), 1544 (N–O), 1104 (C–O) and 1348 (C–N). 1H NMR (200 MHz, CDCl3, ppm): δ H 5.35 (s, N–CH2), 5.72 (s, O–CH2), 5.85 (s, 1H), 7.20–7.36 (m, 2H), 7.50–7.80 (m, 5H), and 8.20–8.27 (m, 2H). 13C NMR (50 MHz, CDCl3, ppm): δ 53.4 (N–CH2), 62.6 (O–CH2), 91.3, 116.8, 123, 124, 130.5, 132.6, 134, 136.3, 142.4, 150, 154.4, 162.5 (C=O) and 164.9. HRMS calculated [M + H]+ for C19H15N4O5: 379.1036, found: 379.1032 and [M + Na]+ for C19H14N4O5Na: 401.0856, found: 401.0853.
4-((1-(4-Chlorobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4i )
The compound 4i was obtained as a white solid via 1,3-dipolar cycloaddition reaction between azide 1c (84 mg) and alkyne 3b (100 mg) in 20 h with 88 % yield. Mp 194–195 °C. IR (KBr, cm−1): υ max 3061 (C–H), 1720 (C=O), 1619 (C=C), 1137 (C–O) and 751 (C–Cl). 1H NMR (200 MHz, CDCl3, ppm): δ H 5.35 (s, N–CH2), 5.58 (s, O–CH2), 5.88 (s, 1H), 7.21–7.36 (m, 4H), 7.55–7.62 (m, 3H) and 7.78–7.82 (m, 2H). 13C NMR (50 MHz, CDCl3, ppm): δ 53.8 (N–CH2), 62.7 (O–CH2), 91.3, 115.5, 116.9, 123.2, 123.4, 124, 129.6, 129.7, 132.7, 132.8, 135.2, 142.1, 153.4, 162.7 (C=O) and 165. HRMS calculated [M + H]+ for C19H15N3O3Cl: 368.0796, found: 368.0798 and [M + Na]+ for C19H14N3O3Cl Na: 390.0616, found: 390.0619.
4-((1-(3-Chlorobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4j )
The compound 4j was obtained as a white solid via 1,3-dipolar cycloaddition reaction between azide 1d (84 mg) and alkyne 3b (100 mg) in 22 h with 86 % yield. Mp 166–167 °C. IR (KBr, cm−1): υ max 1720 (C=O), 1620 (C=C), 1138 (C–O) and 748 (C–Cl). 1H NMR (200 MHz, CDCl3, ppm): δ H 5.36 (s, N–CH2), 5.76 (s, O–CH2), 5.89 (s, 1H), 7.21–7.59 (m, 7H) and 7.76–7.83 (d, 2H). 13C NMR (50 MHz, CDCl3, ppm): δ 53.9 (N–CH2), 62.7 (O–CH2), 91.3, 115.5, 116.9, 124, 129.9, 132.5, 132.7, 133.2, 153.4, 162.7 (C=O) and 165. HRMS calculated [M + H]+ for C19H15N3O3Cl: 368.0796, found: 368.0798 and [M + Na]+ for C19H14N3O3Cl Na: 390.0616, found: 390.0614.
4-((1-(4-Bromobenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one ( 4k )
The compound 4k was obtained as a off-white solid via 1,3-dipolar cycloaddition reaction between azide 1e (106 mg) and alkyne 3b (100 mg) in 20 h with 90 % yield. Mp 204–205 °C. IR (KBr, cm−1): υ max 2915 (C–H), 1721 (C=O), 1620 (C=C), 1137 (C–O) and 751 (C–Br). 1H NMR (200 MHz, CDCl3, ppm): δ H 5.35 (s, N–CH2), 5.58 (s, O–CH2), 5.88 (s, 1H), 7.21–7.40 (m, 4H), 7.55–7.65 (m, 3H) and 7.78–7.81 (d, 2H). 13C NMR (50 MHz, CDCl3, ppm): δ 51.8 (N–CH2), 62.7 (O–CH2), 91.2, 116.8, 123.2, 123.9, 127.8, 130.1, 130.6, 130.7, 132, 132.5, 133.7, 153.4, 162.6 (C=O), 165 and 169.4. HRMS calculated [M + H]+ for C19H15N3O3Br: 412.0291, found: 412.0292 and [M + Na]+ for C19H14N3O3BrNa: 436.0091, found: 436.0094.
Biological studies
Antitubercular activity
Mycobacterium tuberculosis H37Ra (ATCC 25177) was obtained from Astra Zeneca, India. It was grown in Difco Dubos medium and was used for further study. The stock culture was maintained at −70 °C and subcultured once in M. pheli medium before inoculation into experimental culture. MTB was grown to a logarithmic phase (up to OD620 −1.0) in a defined M. pheli medium. Isoniazid and pyrazinamide were purchased from Sigma-Aldrich. They were solubilized in dimethyl sulfoxide (DMSO) and stored in aliquots at −20 °C. XTT sodium salt powder (Sigma) was prepared as a 1.25 mM stock solution in sterile 1X PBS and used immediately. Even menadione (Sigma) was prepared as a 6 mM solution in DMSO and used immediately. The compounds 4a–k were screened for their inhibitory activity on MTB by following the XTT Reduction Menadione Assay (XRMA) protocol at 30, 10 and 3 ug/mL for primary screening. Active compounds were further evaluated for their dose–response activity using half dilution. Briefly, 2.5 µl of these inhibitor solutions was added into the 96-well plate. After that, total volume was made up to 250 µl by using M. pheli medium consisting of 1 % of 1 OD tubercular bacilli. The assay plates were incubated at 37 °C incubator. The incubation was terminated on the 8th day for MTB cultures. The XRMA was then performed to estimate the viable cells present in different wells of the assay plate. For that, in all wells of assay plate 200 µM XTT was added as a final concentration and incubated at 37 °C for 20 min. Then, 60 µM menadione was added as a final concentration and incubated at 37 °C for 40 min. The optical density (OD) was read on a microplate reader (SpectraMax plus 384 plate reader, Molecular Devices Inc.) at 470-nm filter against a blank prepared from cell-free wells. Absorbance given by cells treated with the vehicle DMSO alone was taken as 100 % cell growth. All experiments were carried out in triplicates, and the quantitative value was expressed as the average ± standard deviation, and IC50 values were calculated from their dose–response curves.
Antioxidant activity
1,1-Diphenyl-2-picrylhydrazyl (DPPH) radial scavenging activity
The hydrogen atom or electron donating ability of the compounds was measured from the bleaching of the purple-colored methanol solution of 1,1-diphenyl-1-picrylhydrazyl (DPPH). The spectrophotometric assay uses the stable radical DPPH as a reagent. One milliliter of various concentrations of the test compounds (5, 10, 25, 50 and 100 μg/mL) in methanol was added to 4 mL of 0.004 % (w/v) methanol solution of DPPH. After a 30-min incubation period at room temperature, the absorbance was measured against blank at 517 nm. The percent inhibition (I %) of free radical production from DPPH was calculated by the following equation.
where ‘A control’ is the absorbance of the control reaction (containing all reagents except the test compound) and ‘A sample’ is the absorbance of the test compound. Tests were carried out in triplicate.
Antibacterial activity
The antimicrobial susceptibility testing of newly synthesized compounds was performed in vitro against bacterial strains viz. gram-positive Staphylococcus Aureus (ATCC No. 29737), Micrococcus Luteus (ATCC No. 398), Bacillus Cereus (ATCC No. 6630) and gram-negative Escherichia Coli (NCIM No. 2256), Pseudomonas Fluorescens (NCIM No. 2173) and Flavobacterium Devorans (ATCC No. 10829), respectively, to find out minimum inhibitory concentration (MIC). The minimum inhibitory concentration (MIC, μg/mL) was defined as the lowest concentrations of compound that completely inhibits the growth of each strain. Serial twofold dilutions of all samples were prepared in triplicate in microtiter plates and inoculated with suitably prepared cell suspension to achieve the required initial concentration. Serial dilutions were prepared for screening. Dimethylsulfoxide (DMSO) was used as solvent control. Ampicillin, kanamycin and chloramphenicol were used as a standard antibacterial drug. The concentration range of tested compounds and standard was 128–0.5 µg/mL. The plates were incubated at 37 °C for all microorganisms; absorbance at 595 nm was recorded to assess the inhibition of cell growth after 24 h. The compounds which are showing promising antibacterial activity were selected for minimum inhibitory concentration studies. The MIC was determined by assaying at 128, 64, 32, 16, 8, 4, 2, 1 and 0.5 µg/mL concentrations along with standards at the same concentrations.
Antifungal activity
The antifungal activity was evaluated against different fungal strains such as Aspergillus Niger (NCIM No. 1196), Penicillium Chrysogenum (NCIM No. 723) and Curvularia Lunata (NCIM No. 1131). Fluconazole, miconazole and amphotericin B were used as standard drugs for the comparison of antifungal activity. The plates were incubated at 37 °C for all microorganisms; absorbance at 410 nm was recorded to assess the inhibition of cell growth after 48 h. The lowest concentration inhibiting growth of the organisms was recorded as the MIC. DMSO was used as a solvent or negative control. In order to clarify any effect of DMSO on the biological screening, separate studies were carried out with solutions alone of DMSO and showed no activity against any microbial strains. The compounds which are showing promising antifungal activity were selected for minimum inhibitory concentration studies. The MIC was determined by assaying at 128, 64, 32, 16, 8, 4, 2, 1 and 0.5 µg/mL concentrations along with standards at the same concentrations.
Computational study
Molecular docking
The protein preparation wizard integrated in the package was used to clean and optimize the protein–ligand crystal structure. The protein structure was preprocessed by removing all the crystallographic water molecules (water without H-bonds) since no water molecule was found to be conserved, rectifying the mistakes in PDB file and optimizing the hydrogen bonds. The hydrogen atoms were added to the protein structure corresponding to the physiological pH 7.0 considering the appropriate ionization states for the acidic as well as basic amino acids. The most likely positions of the OH and SH hydrogen atoms, protonation states and tautomers of His residues and Chi “flip” assignments for Asn, Gln and His amino acid residues were selected. After assigning charge and protonation state finally, energy minimization with root-mean-square deviation (RMSD) value of 0.30 Å was carried out using optimized potentials for liquid simulations (OPLS-2005) force field. Thereafter, the 3D geometries of the ligands were optimized using the Schrodinger LigPrep utility (Schrodinger, LLC, USA). This utility generates a number of low-energy 3D structures, with various ionization states, tautomers, stereochemistries and ring conformations, from each molecule input. Finally, partial charges were ascribed to these geometry-optimized ligands by using the OPLS-2005 force field. The active site of DprE1 was defined by a bounding box (grid) size of 10 × 10 × 10 Å that was centered on the native ligand in the crystal complex. Extra-precision glide docking (Glide XP) which docks ligands flexibly was used to rank the docking poses and to gauze the binding affinity of these ligands toward the protein.
ADME prediction
A computational study of synthesized compounds 4a–k was performed for the prediction of ADME properties. In this study, we have calculated molecular volume (MV), molecular weight (MW), logarithm of partition coefficient (miLog P), number of hydrogen bond acceptors (n-ON), number of hydrogen bond donors (n-OHNH), topological polar surface area (TPSA), number of rotatable bonds (n-ROTB) and Lipinski’s rule of five using Molinspiration online property calculation toolkit. Absorption (% ABS) was calculated by % ABS = 109 − (0.345 × TPSA).
Result and discussion
Chemistry
We have described a protocol for the syntheses of a series of new 4-Methyl-7-((1-(substitutedbenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-ones 4a–f and 4-((1-(substitutedbenzyl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-ones 4g–k as potential antitubercular, antioxidant and antimicrobial agents from commercially available starting materials. These compounds were formed by the fusion of benzyl azides and coumarin-based alkynes via click chemistry approach. The syntheses of starting material benzyl azides 1a-f were prepared from corresponding benzaldehydes via NaBH4 reduction, bromination and nucleophilic substitution reaction of sodium azide according to the reported procedure (Alvarez and Alvarez, 1997) (Scheme 1).
The synthesis of 7-hydroxy-4-methyl coumarin 2a has been achieved via Pechmann condensation between resorcinol and ethyl acetoacetate in the presence of acid (Srinivasan et al., 2007) in 80 % yield (Scheme 2). The reaction of compound 2a and 2b with propargyl bromide in the presence of K2CO3 as a base in N, N-dimethylformamide (DMF) at room temperature afforded 4-Methyl-7-(prop-2-yn-1-yloxy)-2H-chromen-2-one 3a (Naik et al., 2012) and 4-(Prop-2-yn-1-yloxy)-2H-chromen-2-one 3b (Anand et al., 2011), respectively, in 95 % yield (Scheme 2). Finally, benzyl azides 1a–f and coumarin-based alkynes 3a and 3b, and the 1,3-dipolar cycloaddition reaction have been performed in the presence of Cu(OAc)2 in t-BuOH-H2O (3:1) at room temperature for 16 to 22 h to give the corresponding 1,4-disubstituted-1,2,3-triazole-based coumarin derivatives 4a–f and 4g–k, respectively, in quantitative isolated yield (86–95 %) (Scheme 2).
The regioselective formation of 1,2-disubstituted 1,2,3-triazole-based coumarin derivatives 4a–4k has been confirmed by physical data and spectroscopic methods such as IR, 1H NMR, 13C NMR and HRMS. According to the 1H NMR data of compound 4a, the signal at δ 2.40 ppm for three protons was present on the coumarin ring in the form of methyl group, and the signals at δ 5.27 ppm for two proton and δ 5.69 ppm for two protons indicate that the two methylene groups attached with nitrogen and oxygen heteroatom, respectively. In addition to this, the signal appeared at δ 7.71 ppm for one proton clearly indicates the formation of 1,4-disubstituted 1,2,3-triazole ring. In the 13C NMR spectrum of compound 4a, the signal at δ 18.7 ppm for the methyl carbon and the signals at δ 53.3 and δ 62.2 ppm indicate the presence of two methylene groups attached to the nitrogen of triazole and oxygen attached to the coumarin ring, respectively. Furthermore, the peak observed at δ 161.1 ppm indicates the presence of carbonyl carbon present in coumarin ring. It has been further confirmed for the formation of compound 4a by high-resolution mass spectrometry (HRMS). The calculated [M + H]+ for compound 4a is 393.1193, and observed [M + H]+ in HRMS at 393.1190 also[M + Na]+ peak came at 415.1013 and in HRMS [M + Na]+ peak observed at 415.1013. Similarly, for compound 4h, in the 1H NMR, the two peaks observed at δ 5.35 and δ 5.72 ppm indicate that the methylene groups attached to nitrogen of 1,4-disubstituted 1,2,3-triazole ring and oxygen attached to coumarin ring, respectively. The peak observed at δ 5.85 ppm was due to the olefinic proton present in the coumarin ring of compound 4h. According to the 13C NMR spectrum of compound 4h, the signals at δ 53.4 and δ 62.6 ppm indicate the presence of two methylene groups attached to the nitrogen and oxygen heteroatom, respectively. Furthermore, the peak observed at δ 162.5 ppm indicates the presence of carbonyl carbon present in coumarin ring. The calculated mass for compound 4h in the form [M + H]+ is 379.1036 and observed in HRMS is 379.1036, and mass of [M + Na]+ is 401.0856 and in HRMS [M + Na]+ peak observed at 401.0853. The regioselective formation of remaining 1,4-disubstituted 1,2,3-triazole-based coumarin derivatives has been confirmed by physical data and spectroscopic methods such as IR, 1H NMR, 13C NMR and HRMS.
Biological activities
Antitubercular activity
The newly synthesized coumarin-1,4-disubstituted 1,2,3-triazole-based compounds 4a–k were screened for their in vitro antitubercular activity against MTB H37Ra (ATCC 25177) following an established XTT Reduction Menadione Assay (XRMA) (Sarkar and singh, 2011) protocol using first-line antitubercular drug pyrazinamide (IC50 = 10 μg/mL) and isoniazid (IC50 = 0.0023 μg/mL) as reference standards. The IC50 values of compounds 4a–k that are in the range of 1.80–4.00 μg/mL imply their potential as promising antitubercular agents (Table 1). All the synthesized compounds 4a–k were more potent than the pyrazinamide; however, these compounds 4g–k were less active than isoniazid. Among the 4-Methyl-7-substituted coumarin–triazole conjugates 4a–f, the compound 4a (IC50 = 2.20 μg/mL) with nitro-group at para position of phenyl ring shows better activity than compound 4b (IC50 = 2.80 μg/mL) with nitro-group at meta position of phenyl ring. Introduction of chloro-group at para position of phenyl ring gave the least active compound 4c with IC50 = 4.00 μg/mL. However, changing the position of chloro-group at meta position of phenyl ring in compound 4d shows increase in activity by almost twofold (IC50 2.20 µg/mL) as compared to compound 4c. The compound 4e (IC50 = 2.40 μg/mL) having bromo-group at para position of phenyl ring shows moderate activity. Compound 4f without any substituent on phenyl ring exhibits most potent activity (IC50 = 1.80 μg/mL) as compared to the standard drug pyrazinamide.
Among the 4-substituted coumarin analogues 4g–k, compound 4h (IC50 = 2.20 μg/mL) with nitro-group at meta position of phenyl ring is more active than compound 4g (IC50 = 2.60 μg/mL) with nitro-group at para position of phenyl ring. However, chloro-group at para position in compound 4i (IC50 = 2.30 μg/mL) has been 1.5 times more potent than compound 4j (IC50 = 3.20 μg/mL) with chloro-substituent at meta position of phenyl ring. Similarly, compound 4k with bromo-group at para position shows better activity than the reference drug pyrazinamide.
Antioxidant activity
1,1-Diphenyl-2-picrylhydrazyl (DPPH) radial scavenging activity
However, in the present study, antioxidant activity of the synthesized compounds has been assessed in vitro by the 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenging assay (Burits and Bucar, 2000), and the results were compared with standard antioxidants including natural ascorbic acid and synthetic antioxidant BHT (butylated hydroxytoluene). All the synthesized compounds 4a–k show good to moderate antioxidant activity as compared to the standard drugs ascorbic acid and BHT (Table 1). Coumarin–triazole conjugates derived from 4-Methyl-7-(prop-2-yn-1-yloxy)-2H-chromen-2-one (3a), compounds 4c and 4d having chloro-substituent on phenyl ring show potent activity (IC50 12.48 and 16.30 µg/mL, respectively) as compared to the standard drug BHT and ascorbic acid. The compounds 4a, 4b and 4e with nitro- and bromo-substituents on phenyl ring exhibit less activity as compared to standard drugs. The coumarin-based triazole derivatives 4g–k derived from 4-(Prop-2-yn-1-yloxy)-2H-chromen-2-one (3b) demonstrate good to excellent antioxidant activity. However, the compound 4g (12.55 µg/mL) with nitro-group at para position of phenyl ring, 4j (13.82 µg/mL) with chloro-group at meta position of phenyl ring and 4k (11.28 µg/mL) with bromo-group at para position of phenyl ring show excellent antioxidant activity as compared to the BHT and ascorbic acid. The compounds 4h and 4i having nitro-group at meta position of phenyl ring and chloro-group at para position of phenyl ring in the respective conjugates exhibit less potency as compared to standard drugs BHT and ascorbic acid.
Antibacterial activity
Minimum inhibitory concentration (MIC) values for bacteria were determined according to the twofold broth microdilution method using Muller-Hinton broth in 96-well microtest plates recommended by National Committee for Clinical Laboratory Standards (NCCLS) guidelines (NCCLS, 1997; NCCLS, 1998; NCCLS, 2000). All the tested coumarin-1,4-disubstituted 1,2,3-triazole-based derivatives 4a–k do not demonstrate significant activity against S. aureus (Table 2 ). It can be seen that the compounds 4a, 4f, 4g, 4h, 4i, 4j and 4k show excellent inhibitory activity with 4 µg/mL as MIC value against M. luteus, which is fourfold more potent than the clinical drug ampicillin (MIC 16 µg/mL). However, the compounds 4b, 4d and 4e also possess equivalent antibacterial effect against M. luteus with MIC value 16 µg/mL. All the synthesized compounds show considerable activity against B. cereus, except compounds 4d, 4f, 4i, 4j and 4k. In particular, compounds 4a, 4b, 4c, 4e, 4g and 4h show good activity at 4 µg/mL as a MIC value against B. cereus. It can be seen that all the synthesized coumarin-1,2,3-triazole-based derivatives 4a–k possess comparable activity against E. coli as compared to standard drug ampicillin. Among all the synthesized compounds 4a–k, compounds 4a and 4g with nitro-group at para position of phenyl ring show promising activity, i.e., twofold more activity as compared to ampicillin, also comparable with standard drugs kanamycin and chloramphenicol with 2 µg/mL as a MIC value against E. coli. Compounds 4a, 4b and 4g also possess comparable activity against gram-negative bacterial strain P. fluorescens with 2 µg/mL as a MIC value as compared to standard drugs ampicillin, kanamycin and chloramphenicol. It is due to the presence of nitro-group on phenyl ring of compounds 4a, 4b and 4g. It has been observed that E. coli and P. fluorescens bacteria are more sensitive to the compounds 4a and 4g. Most of the synthesized compounds are active against the bacterial strain F. devorans. In particular, the compounds 4c, 4d and 4g show twofold more activity with 2 µg/mL as MIC value against F. d2vorans with reference to standard drug ampicillin and comparable with kanamycin and chloramphenicol. The compounds 4e, 4f, 4i and 4k having 4 µg/mL as a MIC value show comparable activity against the bacterial strain F. devorans with standard drug ampicillin.
Antifungal activity
Fungi were sub-cultured in potato dextrose broth medium. MIC of the synthesized compounds was determined using potato dextrose broth in 96-well microtest plates recommended by NCCLS guidelines (NCCLS, 1997; NCCLS, 1998; NCCLS 2000). In case of antifungal activity, all the synthesized coumarin-1,4-disubstituted 1,2,3-triazole-based derivatives 4a–k show good to moderate activity against A. niger, P. chrysogenum and C. lunata strains (Table 2).
All the synthesized compounds 4a–k show equivalent or higher activity against the fungicidal strain A. niger as compared to standard drug miconazole and less potent than amphotericin B and fluconazole. In particular, compounds 4c, 4d and 4i with MIC value of 4 µg/mL show fourfold and compounds 4a, 4e, 4g and 4k with MIC value of 8 µg/mL show twofold antifungal activity against A. niger as compared to standard drug miconazole. The activity of compounds 4a–d, 4g and 4i was higher to that of miconazole against P. chrysogenum as compared to miconazole. In particular, compounds 4d and 4g with a MIC value of 4 µg/mL show comparable potent activity as compared to amphotericin B and twofold more active than miconazole and less active as compared to fluconazole. Most of the synthesized compounds are more active against the fungicidal strain C. lunata as compared to standard drugs miconazole and amphotericin B. In particular, compounds 4a and 4c with MIC value 4 µg/mL exhibit fourfold and compounds 4b, 4d, 4e, 4g and 4i with 8 µg/mL as a MIC value against C. lunata possess twofold more activity as compared to standard drug miconazole and amphotericin B. It has been revealed that the compound 4d having chloro-substituent at meta position of phenyl ring shows promising activity as compared to the standard drugs miconazole and amphotericin B.
Molecular docking
The triazole derivatives have been reported to inhibit DprE1 (decaprenylphosphoryl-β-D-ribose-2′-epimerase) enzyme of MTB (Mir et al., 2014; Stanley et al., 2012; Singh et al., 2008). DprE1 is involved in the biosynthesis of decaprenylphosphoryl-D-arabinose (DPA), which is an essential component of the mycobacterial cell wall (Giovanna et al., 2013). Molecular docking study was performed using the Glide (grid-based ligand docking with energetics) program incorporated in the Schrodinger molecular modeling package (Friesner et al., 2004; Halgren et al., 2004). In order to scrutinize, binding interactions of the synthesized compounds against DprE1 enzyme (PDB ID:4FDO (Batt et al., 2012) and to gain insights into their experimental inhibition pattern.
The most straightforward method for evaluating the accuracy of a molecular docking protocol is by extracting the native ligand from the binding site and re-docking it into the crystal complex to determine how closely the lowest energy pose (binding conformation) predicted by the scoring function resembles the experimentally determined binding mode by X-ray crystallography. In the present study, the docking protocol was validated by docking the native ligand back into the active site of DprE1 (PDB ID:4FDO). The root-mean-square deviation (RMSD) between the pose of native ligand obtained by docking and the observed X-ray crystallographic conformation was found to be <1 Å representing the reliability of the docking procedure in reproducing the experimentally observed binding mode for molecules investigated herein (Fig. 1).
The results obtained for the binding affinity of ligands 4a–k for DprE1 are discussed on the basis of four major parameters—Glide score, Glide energy, H-bonds and non-bonded interactions (van der Waals and Coulombic). The more negative value of Glide score signifies good binding affinity of the ligand with target enzyme. Also the minimum energy for the formation of complex between ligand and receptor (Glide energy) indicates a good binding affinity. Ten different binding conformations obtained from docking simulations have been retained for each of these ligands. The experimental values (IC50) of the biological activity of these molecules and the corresponding intermolecular interaction energy values between inhibitor and enzyme are obtained with the molecular docking shown in Tables 3 and 4.
A plot of the IC50 values versus the Glide docking score of these compounds 4a–k is given in Figure S1 (Supporting information). A general trend was observed between the docking scores of these compounds and there corresponding IC50 values where the active compounds possess high docking score, while compounds with relatively low inhibition were also predicted to show lower docking score. Analysis of the docking pose shows that all the inhibitors snugly fit into the active site of DprE1 in positions very close to that of native ligand in the crystal structure of its complex with DprE1 making various close contacts with the residues lining this site.
All the molecules are showing very good affinity toward DprE1 (average docking score −16.34) with a very similar topology of binding. However, the per-residue interaction analysis between the ligand and amino acids forming the active site could provide the quantitative explanation for the observed difference in binding affinity (Tables 3, 4).
The per-residue interaction analysis shows that the van der Waals contacts were more prevalent over the electrostatic contribution in the binding of molecules 4a–k to DprE1. Extensive van der Waals interactions have been observed with residues Lys418, Tyr415, Asp389, Cys387, Asn385, Phe369, Lys367, Val365, Leu363, Gln336, Gly321, Phe320, Leu317, Trp230, Lys134, Gly133, His132, Ile131, Thr118, Gly117, Pro116 and Tyr60 lining the active site of DprE1. It is also involved in favorable electrostatic contacts with Lys418, Asp389, Lys367, Asp318, Lys134 and His132. The enzyme–inhibitor complex was found to be further stabilized by strong H-bonding interaction observed with the amino acid residues—Lys418, Tyr415, Ser228, His132 and Tyr60 in the enzyme active site. The binding mode of the most active compound 4f in the active site of DprE1 enzyme (PDB ID:4FDO) obtained by molecular docking is shown in Fig. 2.
The per-residue interaction analysis as well as the glide score and the glide energy suggest that 4f interacts relatively more strongly with DprE1 enzyme than other ligands which is in agreement with the observed antitubercular activity. The most significant driving force for mechanical interlocking between these ligands 4a–k and the DprE1 enzyme was observed to be the steric complementarity between the ligands and the receptor site as evidenced from the relatively higher contribution of van der Waals interaction over other components in the overall binding of these compounds to DprE1 enzyme.
The binding pattern predicted by Glide complemented with a detailed per-residue interaction analysis clearly indicates that these triazole derivatives have a high affinity toward active site of DprE1 enzyme which provides a strong platform for new structure-based design efforts.
ADME prediction
A computational study of synthesized compounds 4a–k was performed for prediction of ADME properties using Molinspiration online property calculation toolkit (http://www.molinspiration.com/cgi-bin/properties 2014), and the values are presented in Table 5. The absorption (% ABS) was calculated by % ABS = 109 − (0.345 × TPSA) (Zhao et al., 2002). It is observed that all the synthesized coumarin-1,4-disubstituted 1,2,3-triazole-based derivatives exhibit a good % ABS ranging from 68.98 to 84.79 %. Furthermore, none of the synthesized compounds violated Lipinski’s rule of five (Lipinski et al., 2001), thus showing good drug-like properties. A molecule likely to be developed as an orally active drug candidate should not violate more than one of the following four criteria: miLog P (octanol–water partition coefficient) ≤5, molecular weight ≤500, number of hydrogen bond acceptors ≤10 and number of hydrogen bond donors ≤5 (Ertl et al., 2000). All the compounds 4a–k follow the criteria for orally active drug, and therefore, these compounds may have a good potential for eventual development as oral agents.
Conclusions
In conclusion, we have synthesized new triazole-based coumarin derivatives via click chemistry and evaluated biological activity. The synthesized compounds show promising antitubercular, antioxidant and antimicrobial activity as compared to the respective standard drugs. Among all the compounds, 4f exhibited interesting and most promising antitubercular activity (MIC = 1.80 µg/mL) for MTB H37Ra strain. Compound 4k shows potential antioxidant activity (IC50 = 11.28 µg/mL) when compared to standards BHT and ascorbic acid. Compounds 4g and 4d show significant antibacterial and antifungal activity as compared to the standard antibacterial drugs such as kanamycin, ampicillin, chloramphenicol and antifungal drugs such as miconazole, fluconazole and amphotericin B, respectively. In addition to this, molecular docking study of these synthesized triazole derivatives has a high affinity toward the active site of DprE1 enzyme which provides a strong platform for new structure-based design efforts. Furthermore, analysis of the ADME parameters for synthesized compounds showed good drug-like properties and can be developed as oral drug candidate, thus suggesting that compounds from present series 4f (antitubercular activity), 4k (antioxidant activity), 4g (antibacterial activity) and 4d (antifungal activity) can be further optimized and developed as a lead molecule.
References
Agalave SG, Maujan RS, Pore VS (2011) Click chemistry: 1,2,3-triazoles as pharmacophores. Chem Asian J 6:2696–2718
Alvarez SG, Alvarez MT (1997) A practical procedure for the synthesis of alkyl azides at ambient temperature in dimethyl sulfoxide in high purity and yield. Synthesis 4:413–414
Anand N, Jaiswal N, Pandey SK, Srivastava AK, Tripathi RP (2011) Application of click chemistry towards an efficient synthesis of 1,2,3-1H-triazolyl glycohybrids as enzyme inhibitors. Carbohydr Res 346:16–25
Batt SM, Jabeen T, Bhowruth V, Quill L, Lund PA, Eggeling L, Alderwick LJ, Futterer K, Besra GS (2012) Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors. Proc Natl Acad Sci 109:11354–11359
Binder WH, Kluger C (2006) Azide/Alkyne click reactions: applications in material science and organic synthesis. Curr Org Chem 10:1791–1815
Bock VD, Hiemstra H, van Maarseveen JH (2006) CuI-catalyzed alkyne-azide clicks cycloadditions from a mechanistic and synthetic perspective. Eur J Org Chem 2006:51–68
Boechat N, Ferreira VF, Ferreira SB, Ferreira MLG, da Silva FC, Bastos MM, Costa MS, Lourenço MCS, Pinto AC, Krettli AU, Aguiar AC, Teixeira BM, da Silva NV, Martins PRC, Bezerra FAFM, Camilo ALS, da Silva GP, Costa CCP (2011) Novel 1,2,3-triazole derivatives for use against Mycobacterium tuberculosis H37Rv (ATCC 27294) strain. J Med Chem 54:5988–5999
Burits M, Bucar F (2000) Antioxidant activity of nigella sativa essential oil. Phytother Res 14:323–328
Dye C, Williams BG (2010) The population dynamics and control of tuberculosis. Science 328:856–861
Ertl P, Rohde B, Selzer P (2000) Fast calculation of molecular polar surface area as a sum of fragment based contributions and its application to the prediction of drug transport properties. J Med Chem 43:3714–3717
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Giovanna R, Maria RP, Laurent RC, Giulia M, Andrea M, Claudia B (2013) The DprE1 enzyme, one of the most vulnerable targets of Mycobacterium tuberculosis. Appl Microbiol Biotechnol 87:8841–8848
Global tuberculosis control: WHO report (2014). http://apps.who.int/iris/bitstream/10665/137094/1/9789241564809_eng.pdf
Guardiola-Diaz H, Foster LA, Mushrush D, Vaz AND (2001) Azole-antifungal binding to a novel cytochrome P450 from Mycobacterium tuberculosis: implications for treatment of tuberculosis. Biochem Pharm 61:1463–1470
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759
Hein JE, Fokin VV (2010) Copper-catalyzed azide–alkyne cycloaddition (CuAAC) and beyond: new reactivity of copper (I) acetylides. Chem Soc Rev 39:1302–1315
Kategaonkar AH, Pokalwar RU, Sonar SS, Gawali VU, Shingate BB, Shingare MS (2010a) Synthesis, in vitro antibacterial and antifungal evaluations of new α-hydroxyphosphonate and new α-acetoxyphosphonate derivatives of tetrazolo [1,5-a] quinoline. Eur J Med Chem 45:1128–1132
Kategaonkar AH, Shinde PV, Kategaonkar AH, Pasale SK, Shingate BB, Shingare MS (2010b) Synthesis and biological evaluation of new 2-chloro-3-((4-phenyl-1H-1,2,3-triazol-1-yl)methyl)quinoline derivatives via click chemistry approach. Eur J Med Chem 45:3142–3146
Kushwaha K, Kaushik N, Lata Jain SC (2014) Design and synthesis of novel 2H-chromen-2-one derivatives bearing 1,2,3-triazole moiety as lead antimicrobials. Bioorg Med Chem Lett 24:1795–1801
Lima-Neto RG, Cavalcante NNM, Srivastava RM, Mendonça FJB, Wanderley AG, Neves RP, dos Anjos JV (2012) Synthesis of 1,2,3-triazole derivatives and in vitro antifungal evaluation on candida strains. Molecules 17:5882–5892
Lipinski CA, Lombardo L, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Meldal M, Tornoe CW (2008) Cu-catalyzed azide-alkyne cycloaddition. Chem Rev 108:2952–3015
Mir F, Shafi S, Zaman MS, Kalia NP, Rajput VS, Mulakayala C, Mulakayala N, Khan IA, Alam MS (2014) Sulfur rich 2-mercaptobenzothiazole and 1,2,3-triazole conjugates as novel antitubercular agents. Eur J Med Chem 76:274–283
Molinspiration Chemoinformatics (2014) Brastislava, Slovak Republic. http://www.molinspiration.com/cgi-bin/properties
Moses JE, Moorhouse AD (2007) The growing applications of click chemistry. Chem Soc Rev 36:1249–1262
Murakami A, Gao G, Omura M, Yano M, Ito C, Furukawa H, Takahashi D, Koshimizu K, Ohigashi H (2000) 1,1-Dimethylallylcoumarins potently suppress both lipopolysaccharide-and interferon-γ-induced nitric oxide generation in mouse macrophage RAW 264.7 cells. Bioorg Med Chem Lett 10:59–62
Naik RJ, Kulkarni MV, Pai KSR, Nayak PG (2012) Click chemistry approach for bis-chromenyl triazole hybrids and their antitubercular activity. Chem Biol Drug Des 80:516–523
National Committee for Clinical Laboratory Standard (1997) Reference method for Broth dilution antifungal susceptibility testing of yeast approved standard. Document M27-A; National Committee for Clinical Laboratory Standards: Wayne, PA, USA
National Committee for Clinical Laboratory Standard (1998) Reference method for Broth dilution antifungal susceptibility testing of conidium forming filamentous fungi proposed standard. Document M38-P; National Committee for Clinical Laboratory Standard: Wayne, PA, USA
National Committee for Clinical Laboratory Standards (2000) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically approved standard, 5th edn. NCCLS: Villanova, PA, M7-A5
Nikalje APG, Ghodke MS, Khan FAK, Sangshetti JN (2015) CAN catalyzed one-pot synthesis and docking study of some novel substituted imidazole coupled 1,2,4-triazole-5-carboxylic acids as antifungal agents. Chin Chem Lett 26:108–112
Ouellet SG, Gauvreau D, Cameron M, Dolman S, Campeau LCS, Hughes G, O’Shea PD, Davies IW (2012) Convergent, fit-for-purpose, kilogram-scale synthesis of a 5-lipoxygenase inhibitor. Org Process Res Dev 16:214–219
Pingaew R, Saekee A, Mandi P, Nantasenamat C, Prachayasittikul S, Ruchirawat S, Prachayasittikul V (2014) Synthesis, biological evaluation and molecular docking of novel chalcone-coumarin hybrids as anticancer and antimalarial agents. Eur J Med Chem 85:65–76
Raghu M, Nagaraj A, Reddy CS (2009) Synthesis and in vitro study of novel bis-[3-(2-arylmethylidenimino-1,3-thiazol-4-yl)-4-hydroxy-2H-chromen-2-one-6 yl]methane and bis-[3-(2-arylidenhydrazo-1,3-thiazol-4-yl)-4-hydroxy-2H-chromen-2-one-6-yl]methane as potential antimicrobial agents. J Heterocycl Chem 46:261–267
Ronad PM, Noolvi MN, Sapkal S, Dharbhamulla S, Maddi VS (2010) Synthesis and antimicrobial activity of 7-(2-substituted phenylthiazolidinyl)-benzopyran-2-one derivatives. Eur J Med Chem 45:85–89
Russel DG, Barry CE, Flynn JL (2010) Tuberculosis: what we don’t know can, and does, hurt us. Science 328:852–856
Sangshetti JN, Shaikh RI, Khan FAK, Patil RH, Marathe SD, Gade WN, Shinde DB (2014) Synthesis, antileishmanial activity and docking study of N-substitutedbenzylidene-2-(6,7-dihydrothieno[3,2-c]pyridine-5(4H)-yl)acetohydrazides. Bioorg Med Chem Lett 24:1605–1610
Sarkar D, Singh U (2011) A novel screening method based on menadione mediated rapid reduction of tetrazolium salt for testing of anti-mycobacterial agents. J Micro Methods 84:202–207
Senger MR, Gomes LCA, Ferreira SB, Kaiser CR, Ferreira VF, Silva FP Jr (2012) Kinetics studies on the inhibition mechanism of pancreatic α-amylase by glycoconjugated 1H-1,2,3-triazoles: a new class of inhibitors with hypoglycemiant activity. Chem Bio Chem 13:1584–1593
Shi Y, Zhou CH (2011) Synthesis and evaluation of a class of new coumarin triazole derivatives as potential antimicrobial agents. Bioorg Med Chem Lett 21:956–960
Shingate BB, Hazra BG, Salunke DB, Pore VS, Shirazi F, Deshpande MV (2011) Stereoselective synthesis and antimicrobial activity of steroidal C-20 tertiary alcohols with thiazole/pyridine side chain. Eur J Med Chem 46:3681–3689
Singh MM (2007) 37th Union world conference on lung health, Paris—a report. Indian J Tuberc 54:55–57
Singh BK, Yadav AK, Kumar B, Gaikwad A, Sinha SK, Chaturvedi V, Tripathi RP (2008) Preparation and reactions of sugar azides with alkynes: synthesis of sugar triazoles as antitubercular agents. Carbohydr Res 343:1153–1162
Srinivasan KK, Neelima Y, Alex J, Sreejith G, Ciraj AM, Rao J (2007) Synthesis of novel furobenzopyrone derivatives and evaluation of their antimicrobial and antiinflammatory activity. Indian J Pharm Sci 69:326–331
Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, Fitzgerald M, Dai H, Zhang L, Hung DT (2012) Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem Biol 7:1377–1384
Stefani HA, Gueogjan K, Manarin F, Farsky SHP, Schpector JZ, Caracelli I, Rodrigues SRP, Muscara MN, Teixeira SA, Santin JR, Machado ID, Bolonheis SM, Curi R, Vinolo MA (2012) Synthesis, biological evaluation and molecular docking studies of 3-(triazolyl)-coumarin derivatives: effect on inducible nitric oxide synthase. Eur J Med Chem 58:117–127
Therrien C, Levesque RC (2000) Molecular basis of antibiotic resistance and β-lactamase inhibition by mechanism-based in activators: perspectives and future directions. FEMS Microbiol Rev 24:251–262
World Health Organization (2000) Global tuberculosis control. WHO Report, Geneva, Switzerland: WHO/CDS/TB/2000.27
Zhang W, Li Z, Zhou M, Wu F, Hou X, Luo H, Liu H, Han X, Yan G, Ding Z, Li R (2014) Synthesis and biological evaluation of 4-(1,2,3-triazol-1-yl)coumarin derivatives as potential antitumor agents. Bioorg Med Chem Lett 24:799–807
Zhao Y, Abraham MH, Lee J, Hersey A, Luscombe NC, Beck G, Sherborne B, Cooper I (2002) Rate limited steps of human oral absorption and QSAR studies. Pharm Res 19:1446–1457
Acknowledgments
The authors M. H. S. and D. D. S. are very much grateful to the Council of Scientific and Industrial Research (CSIR), New Delhi, for the award research fellowship. Authors are also thankful to the Head, Department of Chemistry, Dr. Babasaheb Ambedkar Marathwada University, for providing laboratory facility.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shaikh, M.H., Subhedar, D.D., Shingate, B.B. et al. Synthesis, biological evaluation and molecular docking of novel coumarin incorporated triazoles as antitubercular, antioxidant and antimicrobial agents. Med Chem Res 25, 790–804 (2016). https://doi.org/10.1007/s00044-016-1519-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-016-1519-9