Abstract
The pathogenesis of liver fibrosis in nonalcoholic fatty liver disease (NAFLD) remains unclear and the effective treatments have not been explored yet. The activation of hepatic stellate cells (HSCs) is considered as the most critical factor in the progression of liver fibrosis and cirrhosis. Autophagy has recently been identified as a new mechanism to regulate HSC activation. Here, we found that liver macrophages were polarized toward type 2 (M2) during the progression of nonalcoholic steatohepatitis (NASH) and liver fibrosis in both patients and NAFLD mice. Using the methionine–choline-deficient (MCD) diet NAFLD murine model and the in vitro cell culture system, we identified that the M2 macrophages promoted HSC autophagy by secreting prostaglandin E2 (PGE2) and binding its receptor EP4 on the surface of HSCs, which consequently enhanced HSC activation, extracellular matrix deposition, and liver fibrosis. Mechanistically, PGE2/EP4 signals enhanced HSC autophagy through the Erk pathway. A specific PGE2/EP4 antagonist E7046 significantly inhibited M2 macrophage-mediated HSC autophagy and improved liver fibrosis and histopathology in NAFLD mice. Our study provides novel mechanistic insights into the regulation of HSC activation and liver fibrosis. Our findings suggest that the PGE2/EP4 pathway is a promising therapeutic target to prevent NASH progression into cirrhosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nonalcoholic fatty liver disease (NAFLD), recently renamed metabolic associated fatty liver disease (MAFLD), is the most common form of chronic liver disease worldwide [1]. It encompasses a disease spectrum ranging from nonalcoholic simple fatty liver (steatosis), nonalcoholic steatohepatitis (NASH), fibrosis, and cirrhosis with its clinical consequences, such as hepatocellular carcinoma (HCC) [2]. Globally, approximately 25–30% of adults develop NAFLD [3]. Although most NAFLD patients are often clinically silent, about 20% of patients with NAFLD develop to NASH over time, and eventually, about 20% of patients with NASH progress to cirrhosis in their lifetime [4]. Once cirrhosis is advanced, the risk of developing hepatocellular carcinoma is significantly increased, even with standard anti-cirrhosis drugs. Therefore, the prevention of liver fibrosis in the NASH stage is the key to delaying the progression of cirrhosis and HCC. Unfortunately, there are currently no approved treatments for liver fibrosis [5]. These facts highlight the need for a deeper comprehension of the pathogenesis of liver fibrosis and the translation of this knowledge to novel treatments.
The core pathogenesis of liver fibrosis is the activation of hepatic stellate cells (HSCs). Under the stimulations of injury and inflammatory factors, quiescent HSCs are activated and transdifferentiated into myofibroblasts and generate a large amount of extracellular matrix (ECM), which is the characteristic change of liver fibrosis [6, 7]. Among the factors that activate HSCs, transforming growth factor-beta (TGF-β) is one of the most deeply studied cytokines, which is mainly produced by liver macrophages including Kupffer cells and monocyte-derived macrophages [8,9,10,11]. Macrophages can polarize into either a classical activation (pro-inflammatory M1) or alternative activation (anti-inflammatory M2) phenotype in response to different stimuli, the latter is the major source of TGF-β [12, 13]. However, long-term depletion of TGF-β is associated with severe adverse effects such as poor wound healing and increased carcinogenesis in mice [14]. In addition, there are TGF-β-independent pathways in HSC transformation, suggesting other mechanisms may involve in the development of liver fibrosis [15,16,17,18].
In recent years, autophagy has been identified as a new mechanism to regulate HSC activation. Autophagy degrades lipid droplets stored in HSCs and, consequently, provides vital cellular energy and promotes fibrogenic cell functions [19, 20]. Studies have confirmed that in carbon tetrachloride (CCl4)- or thioacetamide-induced fibrosis in mice, autophagy enhanced the activation of HSCs and promoted hepatic fibrosis [19, 21, 22]. Using siRNA to interfere with the expression of autophagy genes (Atg5 or Atg7) or using specific chemicals to block autophagy can suppress the activation of HSCs [8, 9]. Accumulating studies provide specific evidence to pinpoint the fundamental role of autophagy in HSC activation [21, 23, 24]. Therefore, further revealing signals triggering the autophagy of HSCs will provide new insights into the intervention of NAFLD-related liver fibrosis.
In the present study, to explore the underlying regulation of HSC autophagy in the progression of liver fibrosis, we adopted a widely used NAFLD model by methionine-choline deficient (MCD) diet for weeks. We identified that macrophages were polarized toward M2 in the development of NAFLD in human patients and MCD-diet mice, accompanied with autophagy and activation of HSCs. In vitro and in vivo studies demonstrated that M2 macrophages promoted HSC autophagy by secreting prostaglandin E2 (PGE2) and binding its receptor EP4 on the surface of HSCs. It is a newly discovered cytokine pathway that activates HSCs independent of TGF-β. More importantly, by using a small molecule inhibitor to block the binding of PGE2/EP4, we found a new approach to improve NASH and related liver fibrosis.
Results
Liver macrophages polarized toward M2 accompanied by HSC autophagy and activation during the progression of NAFLD
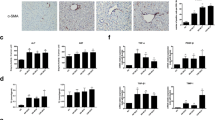
First, we observed an enhanced expression of MRC1 in the liver of human patients with NASH compared to those with nonalcoholic steatosis, indicating an elevated proportion of M2 macrophages in the liver during the progression from NAFLD to NASH (Fig. 1a, Supplementary Fig. 1a and 1b, and Supplementary Table 1). To confirm the observation in human liver, we construct a murine model of NAFLD by feeding an MCD diet to C57BL6/J mice for 8 weeks. Compared with the mice fed with methionine-choline sufficient (MCS) diet, MCD diet-fed mice developed significant characteristics of NASH and liver fibrosis, the serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were markedly increased and the body weight was decreased (Fig. 1b, c). MCD diet-fed mice had a markedly rise in NAFLD Activity Score (NAS) calculated by steatosis, lobular inflammation, and hepatocyte ballooning observed in hematoxylin–eosin (HE) staining, accompanied by lipid drop accumulation and collagen fiber deposition measured by Oil Red and Sirius Red staining (Fig. 1d). Then we stained the liver of mice with antibodies against CD80 and Mannose Receptor C-Type 1 (MRC1, also named CD206) to identify the polarization of macrophages. The immunofluorescence staining results revealed a growing infiltration of total macrophages and M2 macrophages (MRC1+) into the liver of MCD mice and the fibrosis progression (Fig. 1e, Supplementary Fig. 2A and 2B). We further isolated nonparenchymal cells from the liver of mice and determined macrophage polarization with flow cytometry (Fig. 1f). Compared to MCS mice, MCD mice had gradually elevated percentages of CD45+F4/80+MRC1+ M2 macrophages in the liver from 3 to 8 w post diet (Fig. 1g). From 5 w post diet, the percentage of M2 macrophages surpassed M1 macrophages and predominated in the liver of MCD mice (Fig. 1g). These results demonstrated an elevated proportion of M2 macrophages in the liver during the progression of NASH and fibrosis.
Liver macrophages polarized toward M2 phenotype during the progression of fibrosis in MCD-fed mice and human patients with NASH. a Immunofluorescence pictures of MRC1 (red) and DAPI (blue) staining in the liver of human patients with nonalcoholic steatosis or NASH. Integrated density of MRC1 signals in the staining pictures were quantified and compared between groups. Scale bar = 100 μm. b Serum concentrations of ALT and AST in the mice fed with MCS or MCD diet for 8 weeks. c The body weight of mice. d Representative histopathologic staining pictures of hematoxylin and eosin (HE), Oil red and Sirius red in the liver of mice. NAFLD Activity Score (NAS) was calculated according to the performance of HE staining. Integrated density of positive staining signals was quantified with ImageJ software and compared between groups. HE and Oil red staining picture, scale bar = 25 μm. Sirius red staining picture, scale bar = 100 μm. e Representative immunofluorescence pictures of CD80 (green) and DAPI (blue) staining in the upper panels, or MCR1 (green) and DAPI (blue) staining in the lower panels in the liver of mice. Mice fed with MCS or MCD diet for 8 weeks. Integrated density of positive signals in the staining pictures were quantified and compared between groups. Original photo, scale bar = 20 μm; Zoomed photo, scale bar = 100 μm. f Gating strategy for mouse liver macrophages by flow cytometry. Macrophages were gated as CD45+7aad−F4/80+. g Representative dot plots displaying the expression of CD80 (upper) and MRC1 (lower) gated on CD45+F4/80+ macrophages from liver of mice. Percentage of CD80+ (M1 phenotype) and MRC1+ (M2 phenotype) cells in macrophages. Data represent mean ± standard deviation (SD) (n = 4 in each group). P-values were obtained by unpaired t test (two groups) or one-way ANOVA (multiple groups). *P < 0.05, **P < 0.01 and ***P < 0.001 compared with MCS group
Next, we identified the activation and autophagy of HSCs in the liver of MCD diet-fed mice. Alpha Smooth Muscle Actin (α-SMA), Collagen 1 (COL1A1) and Collagen 3 (COL3A1), are considered reliable markers of HSC activation and the critical biomarkers for liver fibrosis. We found that the expression levels of α-SMA, COL1A1 and COL3A1 were significantly increased in the liver as the fibrosis progresses (Fig. 2a, b). Consistently, an enhanced expression of α-SMA protein was also observed in liver tissues of MCD mice detected by immunofluorescence staining (Fig. 2c). The expression of LC3B, a central protein in autophagy, was elevated and partially co-localized with α-SMA in MCD mice (Fig. 2c). To further confirm this result, we performed flow cytometry to precisely identify the autophagy in HSCs. According to a previous report, HSCs were defined as CD45−CD146−ultraviolet autofluorescence (UVAF)+ cells by flow cytometry (Fig. 2d) [25, 26]. In these cells, we found a coordinated increment in LC3B and α-SMA as the fibrosis progresses (Fig. 2d). These data collectively indicated that the autophagy and activation of HSCs was in sync with M2 macrophage polarization in the development of liver fibrosis in NAFLD mice.
The autophagy and activation of HSCs were enhanced in liver of mice during the progression of NASH. C57BL/6 mice were fed with MCS normal diet or MCD diet for 3, 5, and 8 weeks. a The mRNA levels of Acta2, Col1a1 and Col3a1 in the liver of mice were detected by real-time PCR assay. Data in bar graphs represent mean ± SD (n = 5 in each group). b The protein levels of α-SMA, COL1A1 and COL3A1 in the liver of mice were detected by Western blot. c Representative immunofluorescence staining pictures of α-SMA (green), LC3B (red) and DAPI (blue) in the liver of mice. Integrated density of positive signals and the percentage of positive cells in the staining pictures were quantified and compared between groups. Scale bar = 100 μm. d Representative dot plots displaying the expression of α-SMA (upper) and LC3B (lower) gated on CD45−CD146−UVAF+ HSCs in the liver of mice. Percentage of α-SMA+ and LC3B+ cells in HSCs from the liver of mice. Data in bar graphs represent mean ± SD (n = 5 in each group). P-values were obtained by one-way ANOVA. **P < 0.01 and ***P < 0.001 between indicated groups
M2 macrophages promoted autophagy and activation of HSCs in a cell contact-independent manner
Macrophages are vital regulatory cells in liver fibrosis and have plastic phenotypes according to signals derived from the hepatic microenvironment. To investigate the regulatory role of macrophages in HSC autophagy, we constructed co-culture systems of polarized macrophages and HSCs. First, we cultured bone marrow-derived macrophages (BMDMs) and induced them into M1 and M2 macrophages by interferon-gamma (IFN-γ)/lipopolysaccharide (LPS), and IL-4/IL-13, respectively. The phenotype and function were identified by real-time PCR, flow cytometry, and enzyme-linked immunosorbent assay (ELISA) (Supplementary Fig. 3a–d). Then we co-cultured human hepatic stellate cell line LX-2 with M0, M1, and M2 macrophages, respectively. We found that compared with M0 or M1 macrophage co-culture, co-culture with M2 macrophage significantly induced the expressions of fibrotic markers α-SMA, COL1A1 and COL3A1, as well as the conversion of LC3-I/II, in LX-2 cells (Fig. 3a, b).
M2 macrophages promoted HSC autophagy by the secretory pathway. a The mRNA levels of Acta2, Col1a1 and Col3a1 in LX-2 cells cultured with PBS, M0, M1 and M2 conditioned medium, respectively. Data represent mean ± SD (n = 3 in each group). b The protein levels of α-SMA, COL1A1, COL3A1 and LC3B in LX-2 cells were detected by Western blot. c Schematic diagram of transwell co-culture system. d, e The mRNA level of Acta2, Atg5, Atg7 and Becn1 in LX-2 cells were detected by real-time PCR. LX-2 cells were supplied with rapamycin, 3-MA or conditional medium as indicated for 24 h. f Autophagosome in LX-2 cells. Cells were stained with autophagy-specific dye and analyzed by microplate reader. Data represent mean ± SD (n = 4 in each group). P-values were obtained by one-way ANOVA. *P < 0.05, **P < 0.01 and ***P < 0.001 between indicated groups
Then we used a transwell culture system and cultured LX-2 in the lower plate and polarized macrophages in the upper insert to avoid direct interaction between LX-2 and macrophages (Fig. 3c). Similar to the treatment with rapamycin, an mTOR inhibitor known to induce autophagy, M2 macrophage co-culture significantly induced the expressions of Acta2, Atg5, Atg7, and Becn1 (encoding α-SMA, Atg5, Atg7, and Beclin-1, respectively) in LX-2 cells, which were attenuated by the autophagy inhibitor, 3-Methyladenine (3-MA; Fig. 3d, e). Subsequently, we induced polarization of M2 macrophages for 48 h and collected the cell culture supernatant to prepare M2 macrophage-conditioned medium. When supplemented with M2 macrophage-conditioned medium, the number of autophagosomes in LX-2 cells was markedly increased; however, the elevation was blocked in the presence of 3-MA (Fig. 3f). These results indicated that M2 macrophages played a regulatory role in HSC autophagy and activation independent of cell–cell contact.
M2 macrophages promoted HSC autophagy and activation via PGE2/EP4 pathway
M2 macrophages secrete multiple cytokines including TGF-β, platelet-derived growth factor (PDGF), insulin-like growth factor 1 (IGF1), epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), and PGE2, which have been reported to involve in chronic inflammation or fibrogenesis [27,28,29,30,31,32,33,34] (Fig. 4a). However, few studies showed which cytokine may specifically act on the autophagy of HSCs. We treated LX-2 cells with recombinant proteins of the above cytokines respectively and detected the expressions of autophagy-related genes with real-time PCR. The results showed that PGE2 stimulation significantly up-regulated the mRNA expression of Atg5, Atg7, and Becn1, while other cytokines had no significant effect on autophagy (Fig. 4b). Consistently, PGE2 stimulation also enhanced autophagic flux in LX-2 cells (Fig. 4c). We further detected that M2 macrophages synthesized and secreted much PGE2 compared to M0 or M1 macrophages (Fig. 4d). We adopted a recombinant adenovirus Ad-mCherry-GFP-LC3B to track autophagy in LX-2 cells under a laser confocal microscope. At resting state, LC3B existed in the cytoplasm in the form of diffuse yellow fluorescence; under the stimulation with recombinant PGE2, LC3B was concentrated on autophagosomes and appeared as yellow spots (Fig. 4e, left photo). The fluorescence intensity analysis results showed that the concentration of LC3B was significantly increased after PGE2 stimulation (Fig. 4e, right column chart). Western blot results further showed that PGE2 up-regulated the expression of α-SMA, COL1A1 and COL3A1 in LX-2 cells (Fig. 4f). These data indicated that PGE2 might be the critical cytokine to promote HSC autophagy and activation secreted by M2 macrophages.
M2 macrophages promoted activation and autophagy of HSCs by secreting PGE2. a, b The mRNA level of Acta2, Atg5, Atg7 and Becn1 in LX-2 cells were detected by real-time PCR. LX-2 cells were cultured with recombinant protein platelet derived growth factor (PDGF; 10 ng/mL), insulin-like growth factor-1 (IGF1; 100 ng/mL), transforming growth factor β (TGF-β; 10 ng/mL), epidermal growth factor (EGF; 10 ng/mL), vascular endothelial growth factor (VEGF; 10 ng/mL) and phenyl glycidyl ether 2 (PGE2; 100 ng/mL) for 48 h, respectively. c The autophagic activity in LX-2 cells. The autophagy flux was measured by the fluorescence-based autophagy assay kit. d The concentration of PGE2 in the media of macrophages was detected by ELIS(A) Macrophage polarization was induced by 40 ng/mL IFN-γ and 100 ng/mL LPS (for M1), or 40 ng/mL IL-4 and 20 ng/mL IL-13 (for M2) for 2 days. e Autophagosome in LX-2 cells was examined by laser scanning confocal microscope. Representative confocal images are shown. Integrated density of LC3B yellow fluorescence was quantified with ImageJ software and compared between groups. f The expression levels of α-SMA, COL1A1 and COL3A1 in LX-2 cells were detected by Western blot. Cells were cultured with or without 100 ng/mL PGE2 for 48 h. The levels of β-Actin were determined as internal reference. The intensity of α-SMA, COL1A1 and COL3A1 blot was quantified with ImageJ software and normalized to β-Actin. P-values were obtained by unpaired t test (two groups) or one-way ANOVA (multiple groups). *P < 0.05, **P < 0.01 and ***P < 0.001 compared with controls
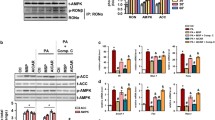
Gs protein-coupled prostaglandin receptor EP4 is a primary receptor for PGE2. We found that the expression level of EP4 mRNA was significantly increased in LX-2 cells after supplementing with M2 macrophage-conditioned medium (Fig. 5a). To further testify if M2 macrophages promoted HSC autophagy and activation via PGE2/EP4 pathway, we used a specific antagonist, E7046, to block the binding of PGE2 to its receptor EP4. Treating with E7046 significantly inhibited the up-regulation of α-SMA induced by both M2 macrophage-conditioned medium and recombinant PGE2 (Fig. 5b). We observed similar effects of E7046 on the mRNA expressions of ATG5, ATG7, and Beclin-1 (Fig. 5c). The PGE2-triggered converting from LC3B-I to LC3B-II was also restrained by E7046 (Fig. 5d). Consistently, E7046 prevented the increment of autophagosomes from both M2 macrophage-conditioned medium and recombinant PGE2 inductions (Fig. 5e). To strengthen the results, we silenced the EP4 gene in LX-2 cells with shRNA. As expected, knockdown of EP4 significantly reduced the activation and autophagy in LX-2 cells (Fig. 5f). We further used transmission electron microscopy (TEM) to inspect autophagy. As expected, we observed the visible presence of autophagosomes and lysosomes in PGE2-treated LX-2 cells. The phenomenon was similar to the stimulation with rapamycin (Fig. 5g). In contrast, it was challenging to observe autophagosomes or lysosomes after additional treatment with E7046 (Fig. 5g). These data consistently supported that M2 macrophages promoted HSC autophagy and activation through PGE2/EP4 pathway.
PGE2-induced autophagy in HSCs was restrained by EP4 inhibitor E7046. a The mRNA level of EP4 coding gene Ptger4 in LX-2 cells. LX-2 cells were cultured with M0- or M2-conditional medium (COND) for 48 h and total RNA was isolated for real-time PCR assay. b, c The mRNA levels of Acta2, Atg5, Atg7 and Becn1 in LX-2 cells were detected by real-time PCR. LX-2 cells were cultured with M2-conditional medium or 100 ng/mL PGE2, in the presence or absence of 1 μM E7046 for 48 h. d The expression levels of LC3B-I, LC3B-II and α-SMA in LX-2 cells were determined by Western blot. Cells were cultured with 100 ng/mL PGE2, in the presence or absence of 1 μM E7046 for 48 h. The intensity of α-SMA blot was quantified with ImageJ software. The conversion ratio of LC3B-I to LC3B-II was calculated and the expression level of α-SMA was normalized to β-Actin. e Autophagosome in LX-2 cells. Cells were stained with autophagy-specific dye and analyzed by microplate reader. f The expression levels of EP4, α-SMA, COL1A1, COL3A1, Beclin-1, and LC3B in LX-2 cells were detected by Western blot. LX-2 cells were transfected with EP4 shRNA and stimulated with recombinant PGE2. g The bilayer autophagosomes and lysosomes were observed with transmission electron microscope. Scale bar = 5 μm (top panel). The panels on the bottom are higher-magnification images of the cropped regions. Scale bar = 1 μm (bottom panel). P-values were obtained by unpaired t test (two groups) or one-way ANOVA (multiple groups). *P < 0.05, **P < 0.01 and ***P < 0.001 compared with controls
PGE2/EP4 signals enhanced HSC autophagy through an Akt/mTOR-independent Erk1/2 pathway
To explore the underlying mechanism of PGE2/EP4 regulating autophagy in HSCs, we analyzed the downstream signaling pathways to EP4. Phosphatidylinositol3-kinase (PI3K) is the central downstream molecule in response to PGE2/EP4 signal, activating both Akt/mTOR and Erk pathways and leading to opposite cellular effects. Akt/mTOR pathway is a classical pathway to inhibit autophagy, while the Erk pathway promotes autophagy by inducing the expressions of Beclin-1 and the conversion of LC3-I to LC3-II. Western blot results showed that recombinant PGE2 stimulation induced the phosphorylation of Akt. However, blocking the binding of PGE2 and EP4 with E7046 did not affect the phosphorylation of Akt and mTOR in LX-2 cells (Fig. 6). On the other side, the E7046 treatment significantly reduced the phosphorylation of Erk1/2, which was induced by PGE2 (Fig. 6). Consistently, PGE2-induced up-regulated expression of Beclin-1 was partly abolished by E7046 treatment (Fig. 6 and Supplementary Fig. 4). These data suggest that PGE2/EP4 signal promotes HSC autophagy by activating the Erk1/2 pathway independent of the Akt/mTOR pathway.
PGE2 activated Erk 1/2 /Beclin-1 pathway was inhibited by E7046. The expression levels of Akt, phosphorylated-Akt (p-Akt), mTOR, p-mTOR, Erk1/2, p-Erk1/2 and Beclin-1 in LX-2 cells were determined by Western blot. Cells were cultured with 100 ng/mL PGE2, in the presence or absence of 1 μM E7046 for 48 h. The intensity of blots was quantified with ImageJ software. The protein and phosphorylation levels were normalized by relative intensity. P-values were obtained by one-way ANOVA. **P < 0.01 and ***P < 0.001 compared with controls
EP4 blockade effectively alleviated HSC autophagy and liver fibrosis in mice
Consistent with the in vitro results, we found an elevated expression level of EP4 in the liver of MCD mice (Fig. 7a). Meanwhile, the immunofluorescence staining showed an increased number of HSCs with EP4 (Fig. 7b). To verify the regulatory roles of the PGE2/EP4 pathway in vivo, we orally administered MCD mice with 150 mg/kg of E7046 every other day from 5 to 7 w. Subsequently, we analyzed the autophagy and activation of HSCs in the liver of mice (Fig. 7c). As detected by flow cytometry, treating with E7046 for 2 w significantly reduced the percentages of LC3B+ HSCs and α-SMA+ HSCs in hepatic nonparenchymal cells in MCD mice (Fig. 7d). The immunofluorescence results also revealed that the expressions and co-localizations of LC3B and α-SMA were diminished in liver tissue by E7046 treatment (Fig. 7E, upper panel). Moreover, E7046 treatment reduced collagen fiber deposition as measured by Sirius red staining (Fig. 7E, lower panel). These results indicated that blocking the PGE2/EP4 pathway could effectively relieve liver damage and fibrosis in mice with NASH by hampering autophagy and activation of HSCs.
The autophagy of HSCs and liver fibrosis was improved by in vivo blockade of EP4 with E7046. a The expression of EP4 coding gene Ptger4 in the liver of MCD mice. The mice were fed with MCD diet for 8 weeks and the liver homogenate was subjected to real-time PCR assay. b Representative immunofluorescence staining pictures of α-SMA (green), EP4 (red) and DAPI (blue) in the liver of mice feeding with MCS or MCD diet for 8 w. Integrated density of positive signals and the percentage of α-SMA+EP4+ cells in the staining pictures were quantified and compared between groups. Scale bar = 100 μm. c Schematic diagram of E7046 treatment to MCD mice. d Representative dot plots displaying the expression of LC3B (upper) and α-SMA (lower) gated on CD45−CD146−UVAF+ HSCs in the liver of mice. Percentage of LC3B+ and α-SMA+ cells were calculated and compared between groups. Data in bar graphs represent mean ± SD (n = 5 in each group). e The upper pictures are representative immunofluorescence staining of α-SMA (green), LC3B (red) and DAPI (blue) in the liver of mice. Scale bar = 100 μm. The lower pictures are representative Sirius red staining in the liver of mice. Scale bar = 200 μm. The percentage of α-SMA+LC3B+ cells and the integrated density of Sirius red in the staining pictures were quantified and compared between groups. P-values were obtained by unpaired t test (two groups) or one-way ANOVA (multiple groups). ***P < 0.001 between indicated groups
The activated HSCs promoted M2 polarization in vitro
In addition, HSCs have been demonstrated to regulate the accumulation and proliferation of macrophages in the pathogenesis of NASH-driven fibrosis. We further speculated that HSCs might influence on the polarization of macrophages. To test this hypothesis, we co-cultured LX-2 cells with macrophages and induced macrophage polarization towards different phenotypes M0, M1, and M2 in vitro. Two days later, we analyzed the phenotypes and functions of macrophages by flow cytometry and ELISA. Compared to the absence of LX-2 cells, co-culture with LX-2 cells promoted up-regulation of M2 marker MRC1 and down-regulation of M1 marker CD80 (Fig. 8a, b). Accordingly, the secretions of M1-type cytokine IL-12p70 were decreased, and the M2-type cytokines IL-10 and PGE2 were increased in the presence of LX-2 cells (Fig. 8c). The results indicated the facilitation of HSCs on the polarization of macrophage towards M2 type.
LX-2 cells promoted M2 macrophage polarization and PGE2 secretion. BMDMs were cultured in the presence or absence of LX-2 cells for 48 h. BMDMs and the cell culture supernatant were collected to identify cell polarization phenotypes. a Representative flow cytometry plots displaying the expression of CD80 (M1 marker) and MRC1 (M2 marker) gated on F4/80+ BMDMs. b Percentage of CD80+MRC1− (M1) and CD80−MRC1+ (M2) subsets in BMDMs. c The cytokine concentrations in the culture media of BMDMs. Data represent mean ± SD (n = 4 in each group). P-values were obtained by Student t test or Mann–Whitney test. *P < 0.05, **P < 0.01 and ***P < 0.001 between indicated groups
Discussion
In this study, by performing the NAFLD mouse model and in vitro experiments, we revealed a novel regulatory mechanism of liver macrophages on HSC activation, which contributed significantly to the progression of liver fibrosis in NAFLD. First, liver macrophages polarize to type 2 phenotype (M2) during the progression from nonalcoholic steatosis to NASH. Next, M2 macrophages evoked HSC autophagy by secreting PGE2 to bind its receptor EP4 on the surface of HSCs; the autophagy further promotes the activation of HSCs and the trans-differentiation into myofibroblasts, which plays a cardinal role in liver fibrosis. Furthermore, the activated HSCs may feedback forward to boosting M2 macrophage polarization. Ultimately, by using a small molecule antagonist E7046 to block the binding of PGE2 and EP4, we observed a distinct improvement in liver fibrosis and histopathology in NAFLD mice, suggesting that the PGE2/EP4 pathway is a new promising therapeutic target to prevent the progression of NASH into cirrhosis (Fig. 9).
It has been reported that M2 macrophages are closely related to multiple fibrotic diseases. Romero et al. reported a significant infiltration of M2 macrophages into the pulmonary fibrosis area in patients with idiopathic pulmonary fibrosis [35]. M2 macrophages played a dominant role in the pathogenesis in renal interstitial fibrosis and were important therapeutic targets [36, 37]. Consistently, by using flow cytometry and immunofluorescence, we identified an increasing portion of M2 macrophages in the liver of NAFLD mice during the progression from steatosis to fibrosis. Moreover, we discovered that M2 macrophages contributed to HSC autophagy, activation, and transformation in an unrevealed mechanism (PGE2/EP4 pathway), which played an essential role in the pathology of NAFLD-related liver fibrosis. Our findings promoted the understanding of the pathologic mechanisms of liver fibrosis and provided a novel therapeutic target to prevent cirrhosis and ultimate end-organ failure.
Macrophage polarization is influenced by many factors, including metabolism, inflammation, and blood coagulation. Another important finding of this study is that activated HSCs could also promote the polarization of M2 macrophages. In a co-culture system, LX-2 cells induced the initial polarization of the M2 phenotype and enhanced the effect of cytokines (IL-4 and IL-13) to increase the proportion of M2 macrophages. Liver macrophages consist of ontogenically distinct populations termed Kupffer cells and monocyte-derived macrophages. Based on the results of our in vitro experiments, we speculated that both sedentary Kupffer cells and MDM could be induced to M2 polarization by HSC. Although only in vitro data are available in the present study, the positive feedback loop probably exists in the NASH microenvironment. Once this phenomenon is confirmed in vivo, it means the communication between macrophages and HSCs via a positive feedback loop makes a significant contribution to cirrhosis. Therefore, further studies on the activation and polarization of macrophages will make outstanding contributions to reveal the pathogenesis of NAFLD and the related diseases.
Here, we demonstrated the role of PGE2/EP4 axis in HSC activation and liver fibrosis. Since Cyclooxygenase 2 (COX-2) is the key enzyme that catalyzes the production of PGE2. Many studies have shown that COX-2 was closely related to the activation of HSCs, and promoted the onset and development of liver fibrosis [38]. For example, selective COX-2 inhibitors (celecoxib and NS-398) have been proved to ameliorate the severity of experimental steatohepatitis [39]. The inhibition of COX-2 could also repress the proliferation of HSCs and the progression of hepatic fibrosis in animals [40, 41]. Combining previous reports and our study, the COX-2/PGE2 signaling pathway is likely to play a crucial role in NAFLD and NASH.
Strong inflammation plays an important role in the development of NASH. Since the inflammatory mediators, such as TNF-α, IL-1β, and IL-6, are mainly secreted by M1 macrophages, we did not observe the levels of pro-inflammatory cytokines in NAFLD mice in the present study. Besides, in LX-2 cells after supplementation with M2 macrophage-conditioned medium, we only detected the expression of EP4 mRNA. In addition to EP4, there may be other molecules involve in HSC autophagy. A recent study reported that PGE2/EP4 axis promoted autophagy and repressed ER stress in hepatocytes of orthotopic liver transplantation [42]. In our study, we focused on the roles of the PGE2/EP4 axis in the autophagy of HSCs during the NASH progression. ER stress and autophagy are delicately balanced. So far, however, it is hard to answer whether PGE2 or pro-inflammatory cytokines activate oxidative or ER stress, and treatment with E7046 or other reagent could prevent induction of ER stress in a different stage of NAFLD.
We noticed that LC3B puncta appeared in other types of cells in the liver of MCD diet-fed mice by immunofluorescence staining (Figs. 2c, 7e). Based on the morphology and size of nuclei, we speculate that these cells are hepatocytes. Consistently, González-Rodríguez A et al. have proved that the LC3-II punctuates increased in hepatocytes from NASH model mice, as well as in biopsies from both non-alcoholic steatosis and NASH patients [43]. Nakamura et al. reported that PGE2/EP4 axis promoted autophagy of hepatocytes and played a liver-protective role in orthotopic liver transplantation [42], while in our present study, in vivo treatment with E7046 seems to not affect the autophagy in hepatocytes of NAFLD mice (Fig. 7e). In addition, PGE2 has been demonstrated to restrict autophagy in immune cells including neutrophils and macrophages [44, 45]. These findings collectively suggest that EP4 may play both promoting and inhibitory roles in autophagy depending on different pathophysiological conditions. Thus, the underlying mechanisms of how the PGE2/EP4 axis regulates autophagic flux in different cell types and different pathophysiological states need to be further studied.
EP4 is a Gs protein-coupled receptor preferentially activated by PGE2. When bound to PGE2 or other agonists, EP4 mobilizes Gs protein complex to dissociate into Gαs and Gβγ subunits and subsequently activate multiple signaling pathways including cAMP-ERK and PI3K [46]. This mechanism has been demonstrated to play a crucial role in promoting tumor growth and forming tumor microenvironment. The PI3K/Akt/mTOR signaling pathway has been broadly confirmed as a classical pathway regulating autophagy in mammalian cells. However, we found that E7046 failed to inhibit the phosphorylation of Akt induced by PGE2; meanwhile, neither PGE2 nor E7046 impacted on the phosphorylation of mTOR. On the other side, E7046 appropriately reduced the phosphorylation of ERK1/2 and the expression of Beclin-1 upon PGE2 stimulation. The ERK1/2 pathway has recently been identified to directly regulate the initiation of autophagy [47, 48]. Consistently, Ugland et al. revealed that cAMP-ERK could induce autophagosome formation by enhancing the perinuclear recruitment of Beclin-1 [49]. Beclin-1 is the first mammalian autophagy protein, which is essential for phagophore formation and triggering autophagic flux [50]. Therefore, we revealed for the first time that the binding of PGE2 and EP4 promoted autophagy through a non-classical pathway that is independent of mTOR signaling.
E7046 is a highly selective, small-molecule antagonist of PGE2 receptor EP4. According to the crucial roles of the PGE2/EP4 pathway in cell growth, inflammation, and immunoregulation, E7046 had displayed a robust anti-tumor activity by diminishing myeloid immunosuppression in vivo and in vitro [51]. Furthermore, a first-in-human phase 1 clinical trial of E7046 was launched in 2015 to treat patients with advanced cancers (NCT02540291). In patients with advanced malignancies, once-daily oral dosing of E7046 showed manageable tolerability, immunomodulatory effects, and disease stabilization [52]. Another phase 1b study of E7046 has also been launched in combination with radiotherapy/chemoradiotherapy in the preoperative treatment of subjects with rectum cancer (NCT03152370). These studies provide valuable insights into the safety of E7046 in clinical trials for the treatment of liver fibrosis.
Furthermore, our findings in the present study expanded the biological function of the PGE2/EP4 pathway and the application of E7046. Importantly, NAFLD is rapidly becoming a leading etiology underlying many cases of HCC. If E7046 shows therapeutic effects on the alleviation of HCC, it will become a multi-target drug to improve NAFLD and the associated diseases, including liver cirrhosis and HCC.
Improving or reversing liver fibrosis is the critical opportunity to prevent cirrhosis, liver cancer, and even death for most chronic liver diseases. Unfortunately, there are few effective therapies and fewer that target fibrogenesis precisely. TGF-β has been considered as a core molecular in virtually all types of fibrosis. Pirfenidone (PFD), targeting TGF-β, is the first drug used in a repeated, randomized, placebo-controlled phase 3 clinical trial to treat idiopathic pulmonary fibrosis. In phase 2 clinical trial in patients with cirrhosis due to hepatitis C virus (NCT02161952), treatment with PFD for 2 years reduced the grading of necroinflammation and decreased liver fibrosis [53]. Later, another clinical trial of PFD in advanced liver fibrosis had started up in 2019 and was recruiting. Ocaliva (Obeticholic acid, OCA) is another promising drug in treating NASH. OCA is a farnesoid X receptor agonist that indirectly inhibits the expression of the cytochrome 7A1 (CYP7A1) gene and blocks the synthesis of cholic acid. OCA is approved to treat primary biliary cirrhosis by FDA. In addition, it showed a curative effect on improving fibrosis and critical components of NASH disease activity in phase 3 clinical trial (NCT02548351) [54, 55]. Nevertheless, the new drug application of OCA for the treatment of liver fibrosis caused by NASH has not been approved yet.
Different from the above drugs, we identified a novel mechanism and potential therapeutic target for liver fibrosis associated with NASH in the present study. We found that PGE2/EP4 binding was a communication pathway for M2 macrophages to regulate HSCs, triggering autophagy through an Akt/mTOR-independent Erk1/2 pathway in HSCs. Although TGF-β also induced HSC activation, it showed no influence on HSC autophagy. These results indicate that multiple factors and mechanisms are contributing to HSC activation and liver fibrosis. Therefore, it is still urgently needed to elucidate the mechanisms of HSC activation for preventing cirrhosis systematically. Here we used a specific antagonist, E7046, to block the binding of PGE2 and its receptor EP4, and demonstrated that E7046 significantly repressed HSC activation and reduced collagen fiber deposition in the liver of mice with NASH by reducing autophagy of HSCs. E7046 is probably a very promising drug for the treatment of liver fibrosis. Considering that liver fibrosis is a pathological process mediated by multiple factors and mechanisms, E7046, combined with other drugs such as PFD or OCA, may have more excellent therapeutic effects on liver fibrosis.
Overall, our study provides novel mechanistic insights into the regulation of HSC activation and liver fibrosis. Our findings suggest that targeting PGE2/EP4 pathway is a promising therapeutic strategy to prevent NASH progression into cirrhosis.
Materials and methods
Patients
Liver tissue samples were collected from eight patients who underwent hepatic surgery or cholecystectomy. Clinical and pathological data were recorded from each patient’s file. Of these patients, three were diagnosed with NASH (2 males, 1 female; mean age, 43 years), the other five were diagnosed NAFLD (5 males; mean age, 35.2 years). All cases had abnormal liver function. The basic characteristics of all patients were provided in Supplementary Table 1. The immunocytochemistry staining pictures of all cases were provided in Supplementary Fig. 1b. Informed consent was obtained from patients or caregivers. This study was approved by the Ethics Committee of the Beijing Ditan Hospital, Capital Medical University (approval number: 2018-047-01).
Mice, NASH model and in vivo treatment
Male C57BL/6 mice (8 weeks old) were purchased from Beijing HuaFuKang Biotechnology Co., Ltd. and maintained under specific pathogen-free conditions in the Animal Care Facility of Capital Medical University. All the mice were used in compliance with the guidelines of Institutional Animal Care and Use Committee of Capital Medical University. To induce NASH, mice were fed with methionine-choline deficient (MCD) diet feeding for 7 weeks. E7046 (150 mg/kg body weight) or an equal amount of water was administered once daily by oral gavage during the last 2 weeks of the 7-week MCD diet. E7046 (#S6649) was purchased from Selleck Chemicals (Shanghai, China).
Histopathological staining and NAFLD activity scoring (NAS)
Livers were excised and immediately fixed with 10% buffered formalin. Samples were embedded in paraffin and cut into 5 μm sections. Liver sections were deparaffinized, rehydrated, and routinely stained with hematoxylin and eosin (H&E) or Sirius Red. Frozen liver sections were cut into 10 μm sections. Tissues were fixed in acetone for 10 min and then washed in PBS for 3 min. After fixation, tissue was washed with PBS and subjected to Oil red staining. The histology was scored by the NAFLD activity score (NAS) as previously reported [56].
Immunofluorescence staining
The paraffin sections were dewaxed and incubated with the antibodies overnight at 4 °C. After washing, the sections were incubated with the primary antibodies in the wet box for 40 min. Nuclei were counter-stained with 4',6-diamidino-2-phenylindole (DAPI) for 10 min. The primary antibodies against α-SMA (1:400, #GB111364), MRC1 (1:400, #GB113497), CD80 (1:200, #GB114055), and EP4 (1:500, #GB111814) were purchased from Servicebio Technology (Wuhan, China). The primary antibodies against LC3B (1:400, #43566) were purchased from Cell Signaling Technology (London, UK). Fluorophore-conjugated secondary antibodies were obtained from Abcam. Images were acquired using a laser scanning confocal microscope (Zeiss LSM 510 META; Carl Zeiss, Thornwood, NJ, USA).
Liver parenchymal cell isolation and flow cytometry
Liver was excised and minced in 1 mg/mL collagenase in PBS. Minced liver was incubated at 37 °C for 30 min. The cell suspension was filtered through a 100-μm filter and spun at 150g for 3 min. The supernatant was collected and centrifuged at 500g for 10 min. The pellet was resuspended in 40% Percoll and overlaid on top of 60% Percoll. The Percoll gradient was centrifuged at 2000g for 20 min. The two middle layers of the Percoll gradient were collected in 3% FBS in RPMI and centrifuged at 500g for 10 min. The supernatant was discarded, and the pellet was resuspended in FACS buffer and stained for flow cytometry. The mAbs used were LC3B-APC, α-SMA-AF488, CD45-APC-Cy7, CD146-PE (BioLegend), CD80-APC (BD Biosciences) and corresponding isotype controls. Data acquisition was performed on a LSR Fortessa flow cytometer (BD Biosciences) and data analysis was performed using FlowJo Software.
Measurement of serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST)
Mouse serum samples were collected and the activities of serum alanine transaminase (ALT) and aspartate transaminase (AST) were determined using an automatic analyzer (Model 7600 Series, Hitachi, Japan).
RNA isolation, cDNA synthesis, and real-time PCR
Total RNA from each sample was extracted with Total RNA Kit (Omega) and then reverse transcribed using the PrimeScript™ RT reagent Kit (TaKaRa). Amplification was performed using the Power SYBR® Green PCR Master Mix (Applied Biosystems) according to the manufacturer's instruction. The relative expression level of each transcript was normalized to murine GAPDH by using the 2(ΔΔCt) method. The primers were listed in Supplementary Table 2.
Western blot
Proteins were extracted with radioimmunoprecipitation (RIPA) lysis buffer with protease inhibitor cocktail (#04693132001, Roche Applied Science, Mannheim, Germany). Proteins were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), 12% or 15% gels were used for separating LC3B and 10% gels were used for other proteins. Then the proteins were electroblotted onto a polyvinylidene difluoride membrane (0.22 μm, Millipore, Billerica, MA, USA). The membranes were blocked with 5% BSA and incubated with specific antibodies overnight at 4 °C and then were incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. β-Actin was used as an internal control. The primary antibodies were as follows: α-SMA (1:2000, #GB111364) and COL3A1 (1:1000, #GB111323) were purchased from Servicebio Technology; COL1A1 (1:500, #sc-293182) was purchased from Santa Cruz Technology; Beclin-1 (1:2000, #ab207612) and EP4 (1:500, #ab217966) were purchased from Abcam company; LC3B (1:1000, #43566), Akt (1:1000, #4691), p-Akt (1:1000, #4060), mTOR (1:1000, #2983), p-mTOR (1:1000, 5536), Erk1/2 (1:1000, #4695), p-Erk1/2 (1:1000, #4370), β-Actin (1:1000, #3700) were purchased from Cell Signaling Technology. The anti-mouse (1:3000, #91196) and anti-rabbit (1:3000, #7074) secondary antibodies were purchased from Cell Signaling Technology. Immunoreactive bands were visualized by ECL and quantified by software Image J (NIH, USA).
Enzyme-linked immunosorbent assay (ELISA)
The levels of TNF-α, IL-12p70, and IL-10 were measured by ELISA kits (BD Biosciences), the level of PGE2 was measured by ELISA kits (Solarbio, Beijing, China) according to the manufacturer's protocols. The absorbance was measured at 450 nm with a microplate reader and the levels of TNF-α, IL-12p70, IL-10 and PGE2 were calculated by the optical density and standard curve.
Culture of bone marrow-derived macrophages (BMDMs)
Bone marrow-derived macrophages (BMDMs) were isolated from C57BL/6 male mice at the age of 6–8 weeks. BMDMs were fully resuspended with DMEM complete medium and evenly planted on 10 cm culture dishes. GM-CSF was added and adjusted to a final concentration of 10 ng/mL. On the third day of culture, fresh DMEM complete medium was replaced, and GM-CSF was added continuously. During the culture, the growth status of adherent cells was observed. Mature M0 macrophages were induced after the culture of 7 days. M1-type macrophages were induced by adding 100 ng/mL LPS and 40 ng/mL INF-γ to mature macrophages, or 40 ng/mL IL-4 and 20 ng/mL IL-13 were added to induce polarization to M2-type macrophages. The supernatant of macrophages with different polarization was collected to co-culture with LX-2 cells, and then the supernatant of co-culture system was retained for ELISA and cells were collected for flow cytometry.
Culture of LX-2 cell line
Human hepatic stellate cell line LX-2 cells were cultured with DMEM medium (GIBCO) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution (EuroClone). In the co-culture system, LX-2 cells were co-cultured with M0, M1 and M2 macrophages, respectively, in a conventional culture condition. In the transwell culture system (0.4 μm; Costar), LX-2 cells were cultured on the lower plate and macrophages were cultured on the upper insert.
RNA interference
LX-2 cells were transfected with EP4 shRNA plasmid (#sc-40173-SH, Santa Cruz) using Lipofectamine 3000 Transfection Reagent (L3000150, Thermo Fisher) according to the manufacturer’s instructions. After a 24-h transfection period, the cells were treated with recombinant PGE2 (#S3003, Selleck Chemicals, Shanghai, China) for another 48 h and analyzed by Western blot.
Fluorescence-based autophagosome detection
Fluorescence-based autophagosome was detected by the following two methods: autophagic vacuoles and autophagic flux in the live cells were selectively labeled with an Autophagy assay kit (#ab139484, Abcam) and measured with a fluorescent microplate reader according to the manufacturer's instruction. In another method, LX-2 cells were grown on 24-well plates and reached 50–70% confluence at the time of infection. After two washes, cells were infected with Ad-mCherry-GFP-LC3B adenovirus (Beyotime Institute of Biotechnology, Shanghai, China) at a multiplicity of infection of 100 for 24 h. The infected cells were stimulated with recombinant PGE2, autophagy flux was observed under laser scanning confocal microscope (Zeiss LSM 510 META). In the absence of autophagy, mCherry-GFP-LC3B was present in the cytoplasm as diffuse yellow fluorescence (the combined effect of mCherry and GFP). Under the condition of autophagy, mCherry-GFP-LC3B aggregated on the autophagosome membrane, presenting as a yellow dot or punctate. When the autophagosome and lysosome fuse, they appear as red spots due to partial quenching of GFP fluorescence.
Transmission electron microscopy (TEM)
LX-2 cells were fixed with 1% glutaraldehyde in PBS (pH 7.4). After rinsing, the suspension was loaded onto a formvar/carbon-coated grid, negatively stained with 3% (w/v) aqueous phosphotungstic acid for 1 min, and observed by transmission electron microscope.
Statistical analysis
All statistical analyses were performed using GraphPad (San Diego, CA) Prism 6.01 software. The Kolmogorov–Smirnov test was used to inspect the normality and homogeneity of variance of all data. For two-group comparison, P values were derived from the one-way Student t test to determine differences between groups with normally distributed data and Mann–Whitney nonparametric test with other data. For multi-group comparison, P values were derived from one-way ANOVA (continuous variables) or Chi-square test (categorical variables). For all comparisons, P < 0.05 was considered statistically significant.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Eslam M, Sanyal AJ, George J (2020) MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology 158:1999-2014e1991. https://doi.org/10.1053/j.gastro.2019.11.312
Matteoni CA, Younossi ZM, Gramlich T et al (1999) Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology 116:1413–1419. https://doi.org/10.1016/s0016-5085(99)70506-8
Huang DQ, El-Serag HB, Loomba R (2021) Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 18:223–238. https://doi.org/10.1038/s41575-020-00381-6
Sheka AC, Adeyi O, Thompson J et al (2020) Nonalcoholic steatohepatitis: a review. JAMA 323:1175–1183. https://doi.org/10.1001/jama.2020.2298
Rockey DC, Bell PD, Hill JA (2015) Fibrosis—a common pathway to organ injury and failure. N Engl J Med 372:1138–1149. https://doi.org/10.1056/NEJMra1300575
Higashi T, Friedman SL, Hoshida Y (2017) Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev 121:27–42. https://doi.org/10.1016/j.addr.2017.05.007
Widjaja AA, Singh BK, Adami E et al (2019) Inhibiting interleukin 11 signaling reduces hepatocyte death and liver fibrosis, inflammation, and steatosis in mouse models of nonalcoholic steatohepatitis. Gastroenterology 157:777-792e714. https://doi.org/10.1053/j.gastro.2019.05.002
Arab JP, Arrese M, Trauner M (2018) Recent insights into the pathogenesis of nonalcoholic fatty liver disease. Annu Rev Pathol 13:321–350. https://doi.org/10.1146/annurev-pathol-020117-043617
Cai X, Li Z, Zhang Q et al (2018) CXCL6-EGFR-induced Kupffer cells secrete TGF-beta1 promoting hepatic stellate cell activation via the SMAD2/BRD4/C-MYC/EZH2 pathway in liver fibrosis. J Cell Mol Med 22:5050–5061. https://doi.org/10.1111/jcmm.13787
Pradere JP, Kluwe J, De Minicis S et al (2013) Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 58:1461–1473. https://doi.org/10.1002/hep.26429
Li H, Zhou Y, Wang H et al (2020) Crosstalk between liver macrophages and surrounding cells in nonalcoholic steatohepatitis. Front Immunol 11:1169. https://doi.org/10.3389/fimmu.2020.01169
Kazankov K, Jorgensen SMD, Thomsen KL et al (2019) The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat Rev Gastroenterol Hepatol 16:145–159. https://doi.org/10.1038/s41575-018-0082-x
Parthasarathy G, Revelo X, Malhi H (2020) Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol Commun 4:478–492. https://doi.org/10.1002/hep4.1479
Kisseleva T, Brenner D (2021) Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol 18:151–166. https://doi.org/10.1038/s41575-020-00372-7
Svegliati-Baroni G, Inagaki Y, Rincon-Sanchez AR et al (2005) Early response of alpha2(I) collagen to acetaldehyde in human hepatic stellate cells is TGF-beta independent. Hepatology 42:343–352. https://doi.org/10.1002/hep.20798
Liu Y, Meyer C, Muller A et al (2011) IL-13 induces connective tissue growth factor in rat hepatic stellate cells via TGF-beta-independent Smad signaling. J Immunol 187:2814–2823. https://doi.org/10.4049/jimmunol.1003260
Czochra P, Klopcic B, Meyer E et al (2006) Liver fibrosis induced by hepatic overexpression of PDGF-B in transgenic mice. J Hepatol 45:419–428. https://doi.org/10.1016/j.jhep.2006.04.010
Sakai K, Jawaid S, Sasaki T et al (2014) Transforming growth factor-beta-independent role of connective tissue growth factor in the development of liver fibrosis. Am J Pathol 184:2611–2617. https://doi.org/10.1016/j.ajpath.2014.06.009
Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R et al (2012) Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 142:938–946. https://doi.org/10.1053/j.gastro.2011.12.044
Zhang Z, Zhao S, Yao Z et al (2017) Autophagy regulates turnover of lipid droplets via ROS-dependent Rab25 activation in hepatic stellate cell. Redox Biol 11:322–334. https://doi.org/10.1016/j.redox.2016.12.021
Tsuchida T, Friedman SL (2017) Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol 14:397–411. https://doi.org/10.1038/nrgastro.2017.38
Thoen LF, Guimaraes EL, Dolle L et al (2011) A role for autophagy during hepatic stellate cell activation. J Hepatol 55:1353–1360. https://doi.org/10.1016/j.jhep.2011.07.010
Lucantoni F, Martinez-Cerezuela A, Gruevska A et al (2021) Understanding the implication of autophagy in the activation of hepatic stellate cells in liver fibrosis: are we there yet? J Pathol. https://doi.org/10.1002/path.5678
Weiskirchen R, Tacke F (2014) Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology. Hepatobiliary Surg Nutr 3:344–363. https://doi.org/10.3978/j.issn.2304-3881.2014.11.03
Dunham RM, Thapa M, Velazquez VM et al (2013) Hepatic stellate cells preferentially induce Foxp3+ regulatory T cells by production of retinoic acid. J Immunol 190:2009–2016. https://doi.org/10.4049/jimmunol.1201937
Ogawa T, Tateno C, Asahina K et al (2007) Identification of vitamin A-free cells in a stellate cell-enriched fraction of normal rat liver as myofibroblasts. Histochem Cell Biol 127:161–174. https://doi.org/10.1007/s00418-006-0237-7
Ueshima E, Fujimori M, Kodama H et al (2019) Macrophage-secreted TGF-beta1 contributes to fibroblast activation and ureteral stricture after ablation injury. Am J Physiol Renal Physiol 317:F52–F64. https://doi.org/10.1152/ajprenal.00260.2018
Spiller KL, Anfang RR, Spiller KJ et al (2014) The role of macrophage phenotype in vascularization of tissue engineering scaffolds. Biomaterials 35:4477–4488. https://doi.org/10.1016/j.biomaterials.2014.02.012
Sprinzl MF, Puschnik A, Schlitter AM et al (2015) Sorafenib inhibits macrophage-induced growth of hepatoma cells by interference with insulin-like growth factor-1 secretion. J Hepatol 62:863–870. https://doi.org/10.1016/j.jhep.2014.11.011
Hsu LW, Huang KT, Nakano T et al (2020) MicroRNA-301a inhibition enhances the immunomodulatory functions of adipose-derived mesenchymal stem cells by induction of macrophage M2 polarization. Int J Immunopathol Pharmacol 34:2058738420966092. https://doi.org/10.1177/2058738420966092
Ramachandran P, Pellicoro A, Vernon MA et al (2012) Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci USA 109:E3186-3195. https://doi.org/10.1073/pnas.1119964109
Zeng XY, Xie H, Yuan J et al (2019) M2-like tumor-associated macrophages-secreted EGF promotes epithelial ovarian cancer metastasis via activating EGFR-ERK signaling and suppressing lncRNA LIMT expression. Cancer Biol Ther 20:956–966. https://doi.org/10.1080/15384047.2018.1564567
Yang C, Zeisberg M, Mosterman B et al (2003) Liver fibrosis: insights into migration of hepatic stellate cells in response to extracellular matrix and growth factors. Gastroenterology 124:147–159. https://doi.org/10.1053/gast.2003.50012
DeLeve LD (2015) Liver sinusoidal endothelial cells in hepatic fibrosis. Hepatology 61:1740–1746. https://doi.org/10.1002/hep.27376
Romero F, Shah D, Duong M et al (2015) A pneumocyte-macrophage paracrine lipid axis drives the lung toward fibrosis. Am J Respir Cell Mol Biol 53:74–86. https://doi.org/10.1165/rcmb.2014-0343OC
Deng X, Yang Q, Wang Y et al (2020) CSF-1R inhibition attenuates ischemia-induced renal injury and fibrosis by reducing Ly6C(+) M2-like macrophage infiltration. Int Immunopharmacol 88:106854. https://doi.org/10.1016/j.intimp.2020.106854
Wang YY, Jiang H, Pan J et al (2017) Macrophage-to-myofibroblast transition contributes to interstitial fibrosis in chronic renal allograft injury. J Am Soc Nephrol 28:2053–2067. https://doi.org/10.1681/ASN.2016050573
Yang H, Xuefeng Y, Shandong W, Jianhua X (2020) COX-2 in liver fibrosis. Clin Chim Acta 506:196–203. https://doi.org/10.1016/j.cca.2020.03.024
Yu J, Ip E, Dela Pena A et al (2006) COX-2 induction in mice with experimental nutritional steatohepatitis: role as pro-inflammatory mediator. Hepatology 43:826–836. https://doi.org/10.1002/hep.21108
Zhao Y, Wang Y, Wang Q et al (2012) Hepatic stellate cells produce vascular endothelial growth factor via phospho-p44/42 mitogen-activated protein kinase/cyclooxygenase-2 pathway. Mol Cell Biochem 359:217–223. https://doi.org/10.1007/s11010-011-1016-x
Planaguma A, Claria J, Miquel R et al (2005) The selective cyclooxygenase-2 inhibitor SC-236 reduces liver fibrosis by mechanisms involving non-parenchymal cell apoptosis and PPARgamma activation. FASEB J 19:1120–1122. https://doi.org/10.1096/fj.04-2753fje
Nakamura K, Kageyama S, Ito T et al (2019) Antibiotic pretreatment alleviates liver transplant damage in mice and humans. J Clin Invest 129:3420–3434. https://doi.org/10.1172/JCI127550
Gonzalez-Rodriguez A, Mayoral R, Agra N et al (2014) Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis 5:e1179. https://doi.org/10.1038/cddis.2014.162
Domingo-Gonzalez R, Martinez-Colon GJ, Smith AJ et al (2016) Inhibition of neutrophil extracellular trap formation after stem cell transplant by prostaglandin E2. Am J Respir Crit Care Med 193:186–197. https://doi.org/10.1164/rccm.201501-0161OC
Martinez-Colon GJ, Taylor QM, Wilke CA et al (2018) Elevated prostaglandin E2 post-bone marrow transplant mediates interleukin-1beta-related lung injury. Mucosal Immunol 11:319–332. https://doi.org/10.1038/mi.2017.51
Woodward DF, Jones RL, Narumiya S (2011) International Union of Basic and Clinical Pharmacology. LXXXIII: classification of prostanoid receptors, updating 15 years of progress. Pharmacol Rev 63:471–538. https://doi.org/10.1124/pr.110.003517
Li XF, Chen DP, Ouyang FZ et al (2015) Increased autophagy sustains the survival and pro-tumourigenic effects of neutrophils in human hepatocellular carcinoma. J Hepatol 62:131–139. https://doi.org/10.1016/j.jhep.2014.08.023
Jiang GM, Tan Y, Wang H et al (2019) The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol Cancer 18:17. https://doi.org/10.1186/s12943-019-0944-z
Ugland H, Naderi S, Brech A et al (2011) cAMP induces autophagy via a novel pathway involving ERK, cyclin E and Beclin 1. Autophagy 7:1199–1211. https://doi.org/10.4161/auto.7.10.16649
Gong C, Bauvy C, Tonelli G et al (2013) Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene 32:2261–2272. https://doi.org/10.1038/onc.2012.252
Albu DI, Wang Z, Huang KC et al (2017) EP4 Antagonism by E7046 diminishes myeloid immunosuppression and synergizes with Treg-reducing IL-2-Diphtheria toxin fusion protein in restoring anti-tumor immunity. Oncoimmunology 6:e1338239. https://doi.org/10.1080/2162402X.2017.1338239
Hong DS, Parikh A, Shapiro GI et al (2020) First-in-human phase I study of immunomodulatory E7046, an antagonist of PGE2-receptor E-type 4 (EP4), in patients with advanced cancers. J Immunother Cancer. https://doi.org/10.1136/jitc-2019-000222
Flores-Contreras L, Sandoval-Rodriguez AS, Mena-Enriquez MG et al (2014) Treatment with pirfenidone for two years decreases fibrosis, cytokine levels and enhances CB2 gene expression in patients with chronic hepatitis C. BMC Gastroenterol 14:131. https://doi.org/10.1186/1471-230X-14-131
Younossi ZM, Ratziu V, Loomba R et al (2019) Obeticholic acid for the treatment of non-alcoholic steatohepatitis: interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 394:2184–2196. https://doi.org/10.1016/S0140-6736(19)33041-7
Neuschwander-Tetri BA, Loomba R, Sanyal AJ et al (2015) Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet 385:956–965. https://doi.org/10.1016/S0140-6736(14)61933-4
Brunt EM, Kleiner DE, Wilson LA et al (2011) Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology 53:810–820. https://doi.org/10.1002/hep.24127
Acknowledgements
We sincerely thank Prof. Xiangying Kong from Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences for providing laboratory equipment, technical assistance and critically reading the manuscript and discussions.
Funding
This work was funded by grants from the National Natural Science Foundation of China (81600454, 81671940, 81871586 and 81770577), Beijing Municipal Excellent Talents Foundation (2018000021223ZK27) and Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (ZYLX201828).
Author information
Authors and Affiliations
Contributions
LZ, YC and JY designed the research; WM, RL, SD, LL, YZ, QQ, YZ, YY and YH conducted the research; MH and PL provided critical advice; LZ, WM, YC and RL analyzed the data; LZ, WM and YC wrote the paper; XZ provided clinical pathological resources. MH and WX provided essential materials.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing of interest.
Ethics approval and consent to participate
The study on human patients was approved by the Ethics Committee of the Beijing Ditan Hospital, Capital Medical University (approval number: 2018-047-01). All the individuals involved in the study have signed the written informed consent. The study on animals was conducted according to the 1998 XXVIII Hungarian law about animal protection and welfare.
Consent for publication
All authors read and approved the final paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cao, Y., Mai, W., Li, R. et al. Macrophages evoke autophagy of hepatic stellate cells to promote liver fibrosis in NAFLD mice via the PGE2/EP4 pathway. Cell. Mol. Life Sci. 79, 303 (2022). https://doi.org/10.1007/s00018-022-04319-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04319-w