Abstract
Many patients with chronic myeloid leukemia in deep remission experience return of clinical disease after withdrawal of tyrosine kinase inhibitors (TKIs). This suggests signaling of inactive BCR–ABL, which allows the survival of cancer cells, and relapse. We show that TKI treatment inhibits catalytic activity of BCR–ABL, but does not dissolve BCR–ABL core signaling complex, consisting of CRKL, SHC1, GRB2, SOS1, cCBL, p85a-PI3K, STS1 and SHIP2. Peptide microarray and co-immunoprecipitation results demonstrate that CRKL binds to proline-rich regions located in C-terminal, intrinsically disordered region of BCR–ABL, that SHC1 requires pleckstrin homology, src homology and tyrosine kinase domains of BCR–ABL for binding, and that BCR–ABL sequence motif located in disordered region around phosphorylated tyrosine 177 mediates binding of three core complex members, i.e., GRB2, SOS1, and cCBL. Further, SHIP2 binds to the src homology and tyrosine kinase domains of BCR–ABL and its inositol phosphatase activity contributes to BCR–ABL-mediated phosphorylation of SHC1. Together, this study characterizes protein–protein interactions within the BCR–ABL core complex and determines the contribution of particular BCR–ABL domains to downstream signaling. Understanding the structure and dynamics of BCR–ABL interactome is critical for the development of drugs targeting integrity of the BCR–ABL core complex.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disorder defined by the t(9;22)(q34;q11) translocation, which creates fusion protein containing part of the breakpoint cluster region (BCR) gene joined with the ABL gene, which encodes a tyrosine kinase. The resulting oncogene has a constitutive tyrosine kinase activity caused by oligomerization of the BCR, leading to autophosphorylation and activation of ABL kinase [1]. Two major isoforms of BCR–ABL are known, p190 and p210, that differ in 501 aa BCR region containing double homology and pleckstrin homology domains present only in p210 [2].
The BCR–ABL is necessary and sufficient to maintain CML, as the acquisition of active BCR–ABL generates a lethal leukemia in mice [3]. At the cellular level, BCR–ABL transforms cells by increasing proliferation and survival, and decreasing dependency on extracellular signals delivered by cytokines and growth factors [4]. This is mediated by the activation of large amounts of signaling intermediates, including members of RAS–ERK–MAP kinase pathway involved in cell proliferation, AKT signaling conferring resistance to apoptosis, and STAT signaling contributing to cytokine independence [5,6,7].
Suppression of BCR–ABL tyrosine kinase activity with small molecule tyrosine kinase inhibitors (TKI) has greatly improved CML prognosis, effectively turning once lethal leukemia into the chronic condition. Imatinib was the first TKI approved for CML, which represented a major therapeutic breakthrough [8]. Over a decade of clinical experience with imatinib demonstrated an estimated 85% survival rate for the first-line treatment patients. Failed imatinib response often involves BCR–ABL gene amplification, increased expression or occurrence of mutations causing imatinib resistance [9, 10]. This was overcome by the development of second-generation TKIs, nilotinib and dasatinib, which inhibit BCR–ABL with greater efficiency than imatinib, and target imatinib-resistant BCR–ABL mutants [11, 12]. Nilotinib and dasatinib provide a significant improvement in CML treatment over imatinib, inducing 2-year complete cytogenetics response in ~ 40% of imatinib-resistant patients [13]. Unfortunately, in many CML patients who failed to respond to imatinib, a T315I substitution in the BCR–ABL’s kinase domain occurs. T315I targets the gate-keeper residue controlling access to the hydrophobic cavity adjacent to the ATP-binding site, which is important for the proper TKI binding [14]. The resistance BCR–ABL–T315I to TKIs was successfully addressed by the development of ponatinib, which induces complete cytogenetic response in 46% of patients resistant to both nilotinib and dasatinib [15].
The TKI therapy generates impressive results in CML treatment; however, it fails primarily in three areas. First, some CML patients remain resistant to TKIs, suggesting that BCR–ABL tyrosine kinase activity is not necessary for CML persistence in these patients [16]. Second, resistance to TKIs eventually develops in significant percentage of CML cases. Novel mutations in BCR–ABL, resistant even to ponatinib, have been described, including the dual mutations affecting one BCR–ABL allele [17]. Third, TKIs suppress but not eradicate CML. The slowly proliferating cancer stem cells are poorly targeted [18], and capable to recapitulate CML even after the period of deep molecular remission. In TKI discontinuation trials, a return of clinical CML after TKI withdrawal is observed in ~ 50% of patients in deep remission [19], suggesting that blockade of BCR–ABL kinase activity by TKIs alone is not sufficient to cure CML. This together with side effects and high economic costs of the life-long TKI therapy necessitates the development of conceptually novel treatments for CML. First, we need to completely understand the mechanics of BCR–ABL signaling, as the BCR–ABL may play other roles beyond the constitutively active tyrosine kinase. The protein–protein interactions within the BCR–ABL signaling complex may remain preserved when its kinase activity is inhibited, leading to residual signaling sufficient for long-term survival of CML cells.
The main downstream signaling pathways utilized by BCR–ABL to regulate cell functions are well established. In contrast, the composition of BCR–ABL interactome, i.e., the pool of proteins associating directly with BCR–ABL, is only beginning to emerge. Active BCR–ABL auto-phosphorylates on tyrosines 177, 1127 and 1294 (Y245 and Y412, cABL1b numbering) [20] and others, providing docking sites for proteins containing SH3 and PTB domains. Several such proteins have been identified, which form a core complex of direct BCR–ABL interactors. These include adapters GRB2, CRK-I and SHC1, adapter/phosphatase SHIP2, p85a subunit of the lipid kinase PI3K (p85a-PI3K), ubiquitin ligase cCBL and phosphatase STS1 [21]. Currently, the nature of binding interfaces and protein–protein interactions in the BCR–ABL signaling complex remains unclear. Yet these interactions hold a key to the understanding of the structure of the BCR–ABL core complex. This study was carried out to map the protein–protein interactions within the BCR–ABL signaling complex in detail, and elucidate the dynamics of the BCR–ABL signaling complex in the active and TKI-inhibited state of BCR–ABL.
Results and discussion
Inhibition of BCR–ABL kinase activity does not dissolve the BCR–ABL signaling complex
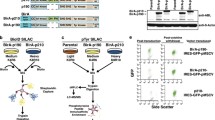
TKIs inhibit kinase activity of BCR–ABL but their effect on protein–protein interactions within the BCR–ABL core signaling complex (referred here as ‘core complex’) is not known. TKIs may not interfere with all protein–protein interactions within the BCR–ABL signaling complex, particularly those not mediated by phosphorylated tyrosine motifs, leaving the BCR–ABL complex partially intact. We asked whether the inhibition of BCR–ABL kinase activity results in the disintegration of the core complex. We expressed p190 BCR–ABL in 293T cells and separated the native BCR–ABL complexes by ultracentrifugation in 15–40% sucrose gradient. Treatment by 100 nM nilotinib leads to complete inhibition of BCR–ABL kinase activity, evidenced by the lack of autophosphorylation at Y412 (Fig. 1a). Inhibition of BCR–ABL kinase activity resulted in a partial shift in the BCR–ABL complexes towards lighter sucrose fractions, suggesting partial but not complete dissociation of the core complex (Fig. 1a; BCR–ABL blot). The members of core complex p85a-PI3K, GRB2, SHIP2, SHC1, SOS1 and cCBL [21] co-sedimented with BCR–ABL in sucrose gradient (Fig. 1a). No co-sedimentation was found with CRK, CRKL or GAB2, suggesting transient interaction not lasting through the experimental procedure.
Nilotinib causes partial dissolution of BCR–ABL signaling complex. a 293T cells were transfected with p190 BCR–ABL, native cell lysates were subjected to ultracentrifugation in the 15–40% sucrose gradient, and collected fractions were analyzed by western blot. The presence of BCR–ABL signal in more than one fraction suggests the existence of complexes of different compositions. Note the various degrees of co-sedimentation of BCR–ABL with p85a-PI3K, GRB2, SHIP2, SHC1, SOS1, SHP2 and cCBL; no co-sedimentation with CRK, CRKL or GAB2 was found. Inhibition of BCR–ABL kinase activity with 100 nM nilotinib resulted in a shift of a fraction of the BCR–ABL complexes towards lighter fractions, suggesting partial dissolution of the BCR–ABL signaling complex. b The western blot analysis of proteins co-sedimenting with BCR–ABL (p85a-PI3K, GRB2 and SHIP2) was quantified as described in “Materials and methods”. Note that portion of GRB2, but not SHIP2 or p85a-PI3K dissociated from the BCR–ABL complex after nilotinib treatment. Data represent a single experiment out of three independent experiments carried out. The fractions containing most of the p190 BCR–ABL are highlighted in red. Phosphorylation (p) at ABL Y412 was used to determine the degree of BCR–ABL inhibition using nilotinib; actin serves as a loading control in total cell lysates used for ultracentrifugation. c Cells were transfected with FLAG-tagged p190 BCR–ABL, V5-tagged GRB2 or SHIP2, treated with nilotinib, and subjected to PLA. The antibodies against protein tags were used in PLA (red); cABL antibody was used to counterstain the transfected cells (green). Cells transfected with BCR–ABL and an empty vector serve as the negative control. Number of PLA dots per cell was calculated and graphed (10–90 percentile). Statistically significant differences were highlighted (Student’s t test with Welch’s correction for unequal variances; *p < 0.05, **p < 0.01). Scale bars, 10 µm
Despite complete inhibition of the BCR–ABL activity, nilotinib did not cause exclusion of any of the endogenously expressed interactors from the co-sedimentation with BCR–ABL. Quantification of western blot analysis of proteins saturated on BCR–ABL, i.e., those which majority in 293T cells co-sedimented with BCR–ABL (p85a-PI3K, GRB2 and SHIP2) shows that portion of GRB2, but not SHIP2 nor p85a-PI3K dissociated from BCR–ABL after nilotinib treatment (Fig. 1b), suggesting only partial dissolution of BCR–ABL signaling complex in 293T cells treated with nilotinib. Similar data were obtained in 293T cells transfected with p210 BCR–ABL, and in K562 cells, a permanent cell line established from CML patient, which expresses endogenous BCR–ABL (Figs. S1, S2). Figure 1a shows a reduction in the BCR–ABL co-sedimentation with SHP2 and cCBL, induced by nilotinib. These changes may represent variability of the particular experiment because we were not able to replicate these results in cells expressing p210 BCR–ABL and in K562 cells (Figs. S1, S2). The association of BCR–ABL with GRB2 and SHIP2 in 293T cells expressing p190 BCR–ABL was probed by proximity ligation assay (PLA). In PLA analyses, nilotinib caused a statistically significant decrease in BCR–ABL interaction with co-transfected GRB2 or SHIP2, but more than 80% of the interaction was preserved for both partners, when compared to active BCR–ABL (Fig. 1c).
Next, the blue-native (BN)-PAGE was used to probe the composition of BCR–ABL complex in 293T cells (Fig. 2a). The protein lysates of 293T cells expressing p210 BCR–ABL were resolved by BN-PAGE to separate protein complexes, which were then analyzed by second-dimension SDS-PAGE to obtain their individual components (Fig. 2b, c). Immunoblotting revealed ~ 600-kDa protein complex containing BCR–ABL, SHIP2 and GRB2 (Fig. 2c). Quantification of the percentage of bound GRB2 and SHIP2 shows that inhibition of BCR–ABL catalytic activity with nilotinib did not inhibit SHIP2 association with BCR–ABL, while causing only weak reduction of GRB2 association (Fig. 2d). Approximately 30% of GRB2 still associated with BCR–ABL containing Y177F substitution, which is known to disable the GRB2-binding motif on BCR–ABL [22, 23]. Similar data were obtained with kinase-dead (KD) BCR–ABL mutant K271H [24].
SHIP2 and GRB2 associate with kinase-inactive BCR–ABL. a Scheme of used experimental procedure comprising native lysis, blue-native (BN)-PAGE, SDS-PAGE and western blot. The three members of protein complex are highlighted in color. b Cell lysates of 293T cells transfected with p210 BCR–ABL. KD, kinase-dead BCR–ABL; Y177, BCR–ABL Y177F mutant. The inhibition of BCR–ABL kinase activity by nilotinib is demonstrated by the lack of autophosphorylation (p) at Y412. Actin serves as loading control. No trans, non-transfected cells. c Merged second dimension BN-PAGE blots of cells transfected with BCR–ABL variants. The membranes have been probed sequentially for BCR–ABL, SHIP2 and GRB2, the BCR–ABL/SHIP2/GRB2 complexes are highlighted by yellow box. (D) Quantification of the percentage of bound GRB2 and SHIP2 to the BCR–ABL. Statistically significant differences are highlighted (Student’s t test, **p < 0.01; n.s., not significant). Data are representative of three independent experiments
Finally, immunoprecipitation was used to address the composition of BCR–ABL complex. 293T cells were transfected by p190 or p210 BCR–ABL and association with endogenously expressed SOS1, SHIP2, cCBL, SHC1 and p85a-PI3K was probed by co-immunoprecipitation. All studied interactors co-immunoprecipitated with both variants of BCR–ABL (Fig. 3a). Nilotinib reduced this association, but significant amounts of SOS1, SHIP2, cCBL and SHC1 still co-immunoprecipitated with BCR–ABL; no association of p85a-PI3K was found in cells treated with nilotinib (Fig. 3a, green arrows). Because endogenous STS1 was not expressed in 293T cells, and endogenous CRKL did not co-immunoprecipitate with BCR–ABL, a transient co-transfection of STS1 and CRKL constructs followed by immunoprecipitation was used to probe their interaction with BCR–ABL. Nilotinib suppressed, but did not abrogate STS1 and CRKL interaction with BCR–ABL (Fig. 3b, c; green arrows); similar results were obtained in experiments probing STS1 and CRKL association with KD BCR–ABL (Fig. 3b, c; right panels, blue arrows). The other known BCR–ABL interactors, such as adapters NCK1 and NCK2 [25, 26], also associated with nilotinib-inhibited BCR–ABL (Fig. S3A).
The BCR–ABL signaling complex is preserved after nilotinib treatment. 293T cells were transfected with p190 and p210 BCR–ABL alone (a) or together with STS1 (b), CRKL (c) and GRB2 (d). BCR–ABL was immunoprecipitated (IP) and binding of interaction partners was analyzed by western blot. The SHC1 isoforms are indicated (p46, p52, p66). Empty, transfection with empty plasmid. BCR–ABL kinase activity was determined by detecting autophosphorylation (p) at Y412. Note the co-immunoprecipitation of SOS1, SHIP2, cCBL, SHC1, STS1, CRKL and GRB2 with BCR–ABL in cells treated with nilotinib (green arrows). Also note the co-immunoprecipitation of STS1, CRKL, and GRB2 with kinase-dead (KD) BCR–ABL (blue arrows). Data are representative of three independents experiments (n = 3). Actin serves as a loading control in cell lysates used for IP
An endogenous GRB2 is difficult to detect in BCR–ABL immunocomplexes, due to co-migration with IgL in SDS-PAGE. Since transgenic GRB2 migrated above IgL, and could be detected by western blot, we transfected GRB2 into 293T cells and probed association with BCR–ABL by GRB2 immunoprecipitation. Figure 3d shows a significant association of GRB2 with BCR–ABL, kinase inactive due to nilotinib treatment (left panel, green arrows) or KD mutation (right panel, blue arrows).
Interaction of GRB2 with BCR–ABL
Secondary structure of p210 BCR–ABL features seven folded regions, namely the N-terminal coiled-coil (CC), double homology (DH), pleckstrin homology (PH), src homology domains SH3 and SH2, tyrosine kinase (TK) and F-actin binding (FAB) domains. Further, three intrinsically disordered regions (IDRs) are located between CC and DH (IDR1), PH and SH3 (IDR2), and TK and FAB domains (IDR3) (Figs. 4, 5a). To gain insight into the binding mode between BCR–ABL and its core complex partners, we used PepStar™ peptide microarray technology to mimic the interaction interface of BCR–ABL. To generate the peptide library, the BCR–ABL sequence was translated into 773 overlapping (with a sliding window of three amino acids) peptides (Fig. 5b). To account for the possibility of protein binding to phosphorylated sites in BCR–ABL, phosphorylated peptides’ analogs (corresponding to the known phosphorylation sites in BCR–ABL) were also included in the library. 13-mer peptides were spotted on microscopic glass plates in a form of microarrays, which were subsequently incubated with interacting protein of interest, and analyzed as described in “Materials and methods” (Fig. 5b). The peptide microarray approach is primarily suited for the identification of binding sites in intrinsically disordered regions (IDR) of a protein or biding sites that, despite being located in folded regions, have the characteristic of solvent-exposed linear motifs protruding from the structure. The peptide microarray approach was combined with conventional co-immunoprecipitation experiments, using a library of mutated and truncated (regional deletion) BCR–ABL constructs (Fig. 4).
The variants of p210 BCR–ABL used in the study. Schematic representation of p190 and p210 BCR–ABL. All truncated or point-mutated p210 BCR–ABL constructs contain N-terminal FLAG epitope. The amino acid substitutions are indicated in red; CC coiled-coil domain, DH double homology domain, PH pleckstrin homology domain, SH3, SH2 Src homology domain, TK tyrosine kinase domain, IDR intrinsically disordered region, FABD F-actin binding domain, NLS nuclear localization signal, PR proline-rich region. The numbering of BCR–ABL domains follows the total protein length (2030 aa in p210), numbering of individual residues is relative to the position of given amino acid in the individual sequence of BCR or cABL1b, with the exception of K271H, which is based on cABL1a
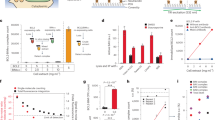
Interaction of GRB2, SOS1, cCBL, and SHC1 with BCR–ABL. a Secondary structure prediction of p210 BCR–ABL by IUPRED. Values above 0.5 indicate disordered regions IDR1 and IDR3 on the BCR–ABL N- and C-termini, involving Y177 and three NLS, respectively. Smaller disordered region IDR2 is located between domains PH and SH3. b Scheme of the microarray analysis. Thirteen amino acid long peptides corresponding to the primary sequence of p210 BCR–ABL were spotted on microarrays, incubated with protein of interest, primary and fluorescently labeled secondary antibodies, and scanned. Fluorescence intensity values for each spot were used to indicate the binding of protein to BCR–ABL peptides. c Microarrays indicate direct binding of GRB2 to phosphorylated Y177. Red lines on BCR–ABL scheme indicate potential binding sites. Graph shows averaged relative intensities for phosphorylated (red) and non-phosphorylated peptides involving peptides with Y177. Error bars indicate SD from three technical replicates shown in Fig. S2. d Co-immunoprecipitation (Co-IP) of BCR–ABL with GRB2 after expression in 293T cells; Y177F substitution abrogates GRB2 association with BCR–ABL as well as deleting the region (construct BT, bottom arrows). Side arrows indicate electrophoretic mobility shift GRB2 phosphorylated by BCR–ABL. e, f Co-immunoprecipitation of endogenous cCBL, SHC1 and SOS1 with transfected BCR–ABL in 293T cells. Please note the compromised SHC1 binding on BCR–ABL–BT, -ΔST, -ΔTK and KD variants (blue arrows). Y177F abrogates binding of SOS1 and largely limits the binding of cCBL (green arrows). Data are representative of three independent experiments (n = 3). g Quantification of SHC1 co-IP with BCR–ABL constructs from (f). SHC1 was normalized to BCR–ABL levels, error bars indicate SD from four independent experiments. Statistically significant differences are indicated (Student’s t test, *p < 0.05, ***p < 0.001; ns non-significant). h Scheme of the proposed interaction. GRB2 binds directly to phosphorylated Y177 and recruits SOS1. cCBL also requires GRB2 for recruitment. SHC1 requires TK domain and pleckstrin homology (PH) domain for binding
Microarray data indicated that GRB2 binds to Y177 in BCR–ABL, and that this interaction is highly specific for phosphorylated version of Y177; the same Y177 peptides lacking the phosphorylation showed no interaction with GRB2 (Fig. 5c). These results agree with previously published work [22, 23] and showed that GRB2 binds directly to Y177. Furthermore, microarrays revealed two additional potential binding sites (labeled G2 and G3) in IDR3 and at the interface between IDR3 and FAB domain, respectively (Figs. 5c; S4A). To verify these binding sites, we generated several truncated or mutated variants of p210 BCR–ABL (Fig. 4), and probed association of selected variants with V5-tagged GRB2 in 293T cells. Y177F substitution, mimicking non-phosphorylated Y177, completely abolished BCR–ABL binding to GRB2, as did deletion of N-terminal part of BCR–ABL containing Y177 (BCR–ABL–BT) (Fig. 5d; arrows). Interaction of KD BCR–ABL with GRB2 was largely limited, but still detectable.
The Y177 is known to be autophosphorylated by active BCR–ABL. However, Y177 can also be phosphorylated in trans by Src tyrosine kinases in KD BCR–ABL [27]. The phosphorylation of Y177 by other tyrosine kinases may thus allow GRB2 binding to TKI-inhibited BCR–ABL as shown by Fig. 3d. Deletion of IDR3 or FABD did not affect GRB2 binding (BCR–ABL–∆IDR3; Fig. 5d). These data demonstrate that IDR3 or FAB domain is not essential for GRB2 association with BCR–ABL (Fig. 5h). The observation that GRB2 did not bind to construct expressing only IDR3 sequence (Fig. S3B, blue arrows) suggested that the binding sites identified by microarrays were either false positives or genuine binding sites, but too weak to sustain the stable interaction. Previous in vitro study [28] described three proline-rich GRB2-binding sites in IDR3 region: PR1 (aa 1425–1433), PR2 (aa 1466–1476) and PR3 (aa 1509–1517). Notably, we observed positive microarray signals for all these three binding sites. However, these binding sites were formally excluded from our analysis as they were defined by less than three consecutive peptides providing above-threshold signal, a stringent criterium used for interpretation of our microarray data. For the same reason, we did not consider at first binding sites in IDR1 (353–365) and IDR3 (1661–1676) that contained PxxxR motif similar to consensus binding motif of GRB2 C-terminal SH3 domain PxxxRxxKP [29] (Fig. S4B, Table S3; g4–g9). Despite their formal exclusion in the process of microarray data interpretation, all these sites still need to be considered as candidate interaction sites.
Altogether, in agreement with previous studies, we show that GRB2 is direct interactor of BCR–ABL, and highlight the critical role of Y177 in mediating this interaction (Fig. 5h). We also confirmed the GRB2 association with PR regions in the IDR3 domain of BCR–ABL suggested before [29]. We further demonstrate that the latter interactions are not sufficient to mediate GRB2 association to BCR–ABL in the absence of phosphorylation at Y177.
Interaction of cCBL with BCR–ABL
cCBL is an E3 ubiquitin ligase known to negatively regulate receptor tyrosine kinase (RTK) signaling [30]. It can also act as an adaptor protein recruiting PI3K towards active RTKs, which mediates the activation of PI3K–AKT pathway [31]. For cCBL, the peptide microarray data yielded no potential binding sites despite extensive optimization and use of two different recombinant cCBL proteins (Fig. S5). Co-immunoprecipitation of endogenous cCBL with BCR–ABL revealed that Y177F substitution or elimination of N-terminal region containing Y177 (BCR–ABL–BT) almost entirely abolished cCBL interaction with BCR–ABL (Fig. 5e). These data mirrored the GRB2 interaction data (Fig. 5d), suggesting that cCBL might bind BCR–ABL indirectly, i.e., via GRB2 (Fig. 5h). This explanation is supported by the observation of interaction between cCBL and GRB2 [32,33,34,35]. As demonstrated by GST-SH2 pulldown assays, cCBL has capacity to directly bind to SH2 domain of BCR–ABL [36]. However, deletion of BCR–ABL SH2 or triple mutant containing Y177F, ΔSH2 and ΔPR1/PR2 was found to lower, but not to completely abolish cCBL binding to BCR–ABL in 32D cells [37]. In support of these data, we show that the deletion of SH2 domain does not abrogate cCBL binding (Fig. 5f).
Taken together, our data indicate that cCBL binding to BCR–ABL is indirect, and is likely mediated by GRB2 binding to (auto)phosphorylated Y177 (Fig. 5h). Similar data were obtained for the other GRB2 partner, the SOS1 [38,39,40,41], which required both Y177 motif and kinase activity of BCR–ABL for association (Fig. 5f).
Interaction of SHC1 with BCR–ABL
SHC1 is signaling adapter known to bind phosphotyrosines on various RTKs [42]. For SHC1, the peptide microarray data showed eleven putative binding sites. None of the identified binding sites contained phosphotyrosine(s) (Fig. S6; Table S4), suggesting an artifact because it is unlikely that protein with validated phosphotyrosine binding capacity would bind to such extent to non-phosphorylated sequences while omitting binding to their phosphorylated counterparts. In line with this discrepancy, deletion of IDR3, comprising of six putative SHC1-binding sites indicated by the microarray did not compromise SHC1 interaction with BCR–ABL (Fig. 5e), which marks the putative binding sites from microarray as false positives. The KD BCR–ABL, BCR–ABL–ΔST variants did not bind SHC1, indicating that SHC1 binding depends on BCR–ABL kinase activity (Fig. 5e, blue arrows). Observation that deletion of TK domain almost entirely abolished SHC1 binding only corroborates this interpretation (Fig. 5f, blue arrows). Further, SHC1 did not interact with BT construct that lacks CC domain. This can be explained by a decrease in BCR–ABL kinase activity resulting from loss of CC domain, which is vital for oligomerization and subsequent activation of catalytically active BCR–ABL [43].
Deletion of PH domain decreased SHC1 association by ~ 30% (Fig. 5f, g). To evaluate the binding within the SH-TK region more precisely, we used individual deletions of SH3, SH2 and TK domains, as well as W118A substitution, which disables SH3 ability to bind proline-rich motifs (BCR–ABL–XS3) and S173N substitution, which disables the SH2 ability to bind phosphotyrosine motifs (BCR–ABL–XS2) [44, 45] (Fig. 4). Removal of TK domain abolished the interaction almost completely. The individual deletions ∆SH3 and ∆SH2 also decreased the SHC1 binding by ~ 30% (Fig. 5f, g), but their inactivation by point mutations (W118A or S173N) produced no effect.
Overall, our data suggest that BCR–ABL kinase activity is necessary for SHC1 binding. SH3 and SH2 domains appear to partially contribute to SHC1 binding, as well as PH domain (Figs. 5h, 6f). However, SHC1 binds less only to PH mutant, but not to BCR–ABL that lacks the whole DH-PH domain. It remains unclear whether PH domain is a direct binding site, as PH domains are known to target proteins to plasma membrane, it seems more plausible that the decreased interaction is caused by altered localization of the ΔPH construct. Experiments carried out below indicate that SHC1 can bind also indirectly to BCR–ABL via SHIP2.
Interaction of CRKL and SHIP2 with BCR–ABL. a BCR–ABL scheme with red lines indicating potential CRKL binding sites identified by microarray. Graph shows binding intensities for phosphorylated (red) and non-phosphorylated peptides; strong binding is shown for phosphorylated Y89, Y134 and Y331. Sites c5 (PR1) and c6 (PR2) are not formally considered; however, blue bars indicate positive binding for respective peptides. b Co-immunoprecipitation (co-IP) of BCR–ABL with CRKL in 293T cells. Deletion of IDR3 and both PR1 and PR2 sites (constructs ΔPR1 and ΔPR1-2) limits the interaction of CRKL with BCR–ABL (arrows). Substituting Y89, Y134 and Y331 to phenylalanines (3YF) produced no effect on CRKL binding. c Quantification of CRKL co-IP with BCR–ABL constructs from (b, right panel). CRKL was normalized to BCR–ABL levels, error bars indicate SD from four independent experiments (n = 4). Statistically significant differences are indicated (Student’s t test, *p < 0.05, ***p < 0.001; n.s., non-significant). d Microarray analysis of SHIP2 binding to BCR–ABL shows multiple binding sites in the SH3–SH2–TK domains, association with the tyrosine-phosphorylated motifs is indicated in red. Binding site for Y488 was obtained in independent experiment and, therefore, different threshold applied to this site. e Immunoprecipitation of BCR–ABL constructs with SHIP2 in transfected 293T cells. Deletion of SH3–SH2–TK (ΔST), or TK domain (ΔTK) limits SHIP2 binding (arrows). Substituting Y89, Y134, Y158, Y276, Y331 and Y488 to phenylalanines (6YF) has no effect on SHIP2 binding. SHC1 associated with BCR–ABL in a manner similar to SHIP2, suggesting mutual interaction. f Scheme of proposed interaction. CRKL binds to region containing PR1 and PR2. SHIP2 binds to the TK region of ABL and also recruits SHC1 to BCR–ABL. n number of independent experiments
Interaction of CRKL with BCR–ABL
CRKL adapter is a major substrate of BCR–ABL that is heavily phosphorylated in CML cells [46,47,48]. For CRKL, the microarray data indicated four potential binding sites, all localized into structured regions of BCR–ABL. These sites included phosphopeptides with tyrosines Y89 (C1), Y134 (C2), and Y331 (C3) in SH3 domain, SH3–SH2 linker, and TK domain, respectively. The fourth binding site was found in FAB domain (C4) (Figs. 6a, S7; Table S5). Previous studies showed that CRK binds to proline-rich motifs APELPTKTR (PR1) and EPAVSPLLPRK (PR2) in IDR3 motif of cABL [28]. In the microarray, the peptides corresponding to the PR1 and PR2 sites were also positive (Fig. 6a, c5, c6, blue bars; Table S5). However, these sites were formally not considered as binding sites because they were defined by less than three consecutive peptides providing above-threshold signal.
Next, we co-expressed CRKL along with BCR–ABL variants, and used co-immunoprecipitation to probe the interaction. Deletion of IDR3 domain (involving PR1 and PR2) eliminated binding to CRKL (Fig. 6b). Deletion of SH3–SH2–TK domains (BCR–ABL–∆ST) also abolished interaction with CRKL. However, the fact that this deletion includes the PR1 site complicates interpretation. To narrow down the binding interface in IDR3, we used BCR–ABL constructs lacking both PR1 and PR2 sites (ΔPR1, ΔPR1-2); CRKL binding on these mutants was reduced by 40–50% (Fig. 6b, c; arrows).
To assess the importance of phosphorylated tyrosines Y89, Y134 (localized in SH3 domain) and Y331 for interaction between CRKL and BCR–ABL, the triple mutant with all three tyrosines changed for phenylalanines was generated (BCR–ABL–3YF). Binding of this mutant to CRKL was not affected (Fig. 6b), which is at variance with the microarray data (Figs. 6a, S7; Table S5). However, an individual deletion of SH3 and SH2 domains had small but significant effect on CRKL binding, which was reduced by 10–20% (Fig. 6b, c), suggesting contribution of SH domains to CRKL binding. Removal of BCR–ABL TK domain (involving also PR1) showed significant reduction in CRKL binding, comparable to removal of PR1 and PR1-2 (Fig. 6b). The interaction with FAB domain, identified by microarray, was not confirmed by co-immunoprecipitation (Fig. 6b, c). Altogether, we confirm CRKL as a direct BCR–ABL interactor (Fig. 6f). Furthermore, we provide support for involvement of proline-rich motifs PR1 and PR2 in BCR–ABL interaction with CRKL, as suggested before for CRK [28].
Interaction of SHIP2 with BCR–ABL
SHIP2 is an inositol phosphatase which converts phosphatidyl inositol triphosphates PI(3,4,5)P3 to PI(3,4)P2. SHIP2 associates with BCR–ABL, and is constitutively phosphorylated in primary CML cells or in BCR–ABL-expressing cells [21, 49, 50]. For SHIP2, the microarray data revealed thirteen potential binding sites located in structured domains, with an exception of the site SH1 corresponding to Y177 (Figs. 6d, S8; Table S6). Putative binding sites SH2–SH4 located in DH-PH domains. Binding sites SH5–SH12 in the SH3–SH2–TK region included tyrosines Y89, Y134, Y158, Y276, Y331 and Y488; SH13 was located in FAB domain. Preferential SHIP2 interaction with phosphorylated tyrosine was found for Y134, Y158, Y276, Y331 and Y488 (Fig. 6d).
Next, the selected BCR–ABL constructs were co-expressed with SHIP2 in 293T cells, and subjected to BCR–ABL co-immunoprecipitation. Deletion of SH3–SH2–TK domains essentially abolished interaction with SHIP2, similar to KD mutant (Fig. 6e; arrows). The Y177F substitution, as well as the deletion of DH-PH, IDR3 and FABD domains, had no effect on the interaction between BCR–ABL and SHIP2 (Fig. 6e). In contrast to the deletion of TK domain, which abolished the binding almost completely (Fig. 6e; right panel, arrow), deletions of SH3 and SH2 (ΔS3, ΔS2) had no effect on SHIP2 association with BCR–ABL (Fig. 6e, right panel). To address the importance of tyrosines within the SH3–SH2–TK domains, we created BCR–ABL mutant in which six tyrosines, implicated by the microarray, were mutated to phenylalanines (Y89F, Y134F, Y158F, Y276F, Y331F and Y488F) (BCR–ABL–6YF). SHIP2 and BCR–ABL–6YF co-immunoprecipitated normally (Fig. 6e, right panel) suggesting that phosphorylation at these sites is not critical for SHIP2 binding.
Interestingly, SHC1 interacted with BCR–ABL variants in a pattern similar to SHIP2 (Fig. 6e), suggesting cooperation between SHIP2 and SHC1 in BCR–ABL binding. The fact that BCR–ABL–ΔTK partially binds SHIP2 but not SHC1, however, demonstrates that SHIP2 binding to BCR–ABL is not dependent on SHC1 (Fig. 6f). Overall, we show that SHIP2 interacts with BCR–ABL directly, and that this interaction depends, to some extent, on catalytic activity of BCR–ABL. In contrast to the previous study indicating importance of SH3–SH2 for SHIP2 binding [50], our data highlight important role of BCR–ABL TK domain in mediating this interaction.
Interaction of p85a-PI3K and STS1 with BCR–ABL
p85a is a regulatory subunit of phosphatidyl inositol 3-kinase (PI3K). Inhibiting p85a-PI3K expression or PI3K activity leads to inhibition of growth in BCR–ABL positive cells, demonstrating that PI3K contributes to oncogenic signaling of BCR–ABL [5, 51]. The microarray analysis revealed two potential p85a-PI3K binding sites on BCR–ABL, i.e., phosphotyrosines Y134 (p85a-1) and Y158 (p85a-2), located in the SH3 and SH2 domains, respectively (Figs. 7a, S9; Table S7). Double substitution of Y134F and Y158F (BCR–ABL–2YF) had no effect on p85a-PI3K binding, suggesting that phosphorylation at these tyrosines is not critical for p85a-PI3K interaction with BCR–ABL (Fig. 7b).
Interaction of p85a-PI3K and STS1 with BCR–ABL. a BCR–ABL scheme with potential p85a-PI3K binding sites identified by peptide microarray. Graph shows intensities for phosphorylated (red) and non-phosphorylated peptides containing Y134 and Y158. b Immunoprecipitation (IP) of BCR–ABL with p85a-PI3K in 293T cells. Substitutions of Y134 and Y158 to phenylalanines (2YF) had no effect on p85a-PI3K interaction with BCR–ABL. Empty, cells transfected with empty plasmid. c Deletion of SH3, SH2 and TK domains (ΔST) abolishes p85a-PI3K interaction with BCR–ABL, similar to KD BCR–ABL. Graph, quantification of p85a-PI3K co-immunoprecipitation with BCR–ABL constructs. p85a-PI3K was normalized to BCR–ABL levels, error bars indicate SD from three independent experiments (n = 3). Statistically significant differences are indicated (Student’s t test, ***p < 0.001; n.s., non-significant). d, e Immunoprecipitation of BCR–ABL with STS1. p190 BCR–ABL and ΔDP mutant of p210 BCR–ABL show impaired interaction with STS1. Deletion of SH3, SH2, and TK domains (ΔST) also impairs BCR–ABL interaction with STS1. Statistically significant differences are indicated (Student’s t test, ***p < 0.001; n.s., non-significant). f Scheme of the proposed interaction. p85a-PI3K interacts with multiple domains of BCR–ABL, whereas STS1 requires for association the SH3–SH3–TK domains and DH-PH domains (absent in p190 BCR–ABL). Data are representative for 3-4 independent experiments (n)
In contrast to microarray data, co-immunoprecipitation experiments showed decreased p85a-PI3K binding to all tested BCR–ABL variants (Fig. 7c). In particular, deletion of SH3–SH2–TK domain containing Y134 and Y158 (∆ST) and KD BCR–ABL almost completely lost capacity to interact with p85a-PI3K. This complex mode of p85a-PI3K binding to BCR–ABL likely stems from the ability of p85a-PI3K to bind multiple components within the BCR–ABL complex, namely SHC1, cCBL, GAB2 and CRKL [52] (Fig. 7f). The co-immunoprecipitation experiments suggest that p85a-PI3K binding to BCR–ABL is indirect, mediated by other BCR–ABL interactors (Fig. 7f). Interestingly, we found about 50% reduction of p85a-PI3K binding to BCR–ABL with Y177F substitution (Fig. 7c). The Y177 was shown before to recruit p85a-PI3K via GAB2 [53].
STS1 is a tyrosine phosphatase which dephosphorylates BCR–ABL and thus is a negative regulator of BCR–ABL signaling [54]. Microarray data showed no specific association of STS1 with BCR–ABL, despite optimization and use of two different recombinant STS1 proteins (Fig. S10). Co-immunoprecipitation experiments demonstrate preferential STS1 association with p210 BCR–ABL, as p190 BCR–ABL showed ~ 50% less binding when compared to p210 (Fig. 7d, arrow). Decreased STS1 binding on p210 BCR–ABL with removed DH and PH domains (ΔDP) was also found. Deletion of PH domain only did not affect STS1 binding, suggesting that the entire region missing in p190 (aa 425–927) is necessary for STS1 interaction (Fig. 7d). These findings are in agreement with published proteomic data, which report preferential association of STS1 with p210 [55, 56]. However, it was shown that inactivating substitutions in PH domain of p210 lower interaction of STS1 in BaF3 cells [57]. In our hands, deleting PH domain did not significantly change binding to STS1 (Fig. 7d). We further show that kinase activity of BCR–ABL is essential for STS1 binding, as kinase-inactive KD BCR–ABL and BCR–ABL–ΔST showed significantly reduced STS1 binding (Fig. 7e). Altogether, our data point to the involvement of DH and PH domains in binding of STS1 to BCR–ABL and suggest that BCR–ABL catalytic activity is required for this interaction (Fig. 7f).
SHIP2 is required for BCR–ABL-mediated phosphorylation of SHC1
Because the precise role of SHIP2 in BCR–ABL signaling has not been established, we deleted endogenous SHIP2 by CRISPR/Cas9 in 293T cells (SHIP2Crispr cells), transfected these cells with BCR–ABL, and determined the effect of SHIP2 loss on BCR–ABL-mediated signaling. In contrast to wild-type 293T cells, SHIP2Crispr cells could not phosphorylate SHC1 on Y239 and Y240 (Fig. 8a; arrows). Add-back of wild-type SHIP2 into the SHIP2Crispr cells rescued SHC1 phosphorylation. However, co-transfecting catalytically inactive SHIP2 (phosphatase-dead, PD) only partially rescued SHC1 phosphorylation, suggesting that SHIP2 catalytic activity is important for BCR–ABL-mediated SHC1 phosphorylation (Fig. 8b). As SHIP2 and SHC1 are known interactors [50], we tested whether SHC1 interacts with BCR–ABL in SHIP2Crispr cells. SHC1 interacted with BCR–ABL normally in SHIP2Crispr cells (Fig. 8c). Lack of SHC1 phosphorylation in SHIP2Crispr cells is, therefore, not caused by defective recruitment of SHC1 to BCR–ABL. To validate the importance of SHIP2 catalytic activity in BCR–ABL signaling, we tested the effect of specific SHIP2 chemical inhibitor AS1949490 [58] on proliferation of CML cell lines K562, LAMA-84 and KYO-1. Treatment with AS1949490 inhibited proliferation in all three cell lines (Fig. 8d).
SHIP2 is required for BCR–ABL-mediated phosphorylation of SHC1. a, b Wild-type (wt) 293T cells and cells with SHIP2 deleted by CRISPR/Cas9 (SHIP2Crispr) were transfected by BCR–ABL, and analyzed for presence and phosphorylation (p) of given proteins by western blot. In contrast to wt cells, BCR–ABL could not phosphorylate SHC1 in SHIP2Crispr cells. Addition of wt SHIP2 into the SHIP2Crispr cells fully rescued BCR–ABL-mediated SHC1 phosphorylation, while the addition of catalytically inactive (phosphatase-dead, PD) had only minor effect. Same results were obtained with two independent SHIP2Crispr cell lines (not shown). b Quantification of SHC1 phosphorylation in (a), normalized to BCR–ABL levels. Bars represent averages from six experiments (n) with indicated SD, statistically significant differences are indicated (Student’s t test, ***p < 0.001). c SHC1 interacts with BCR–ABL in SHIP2Crispr 293T cells. BCR–ABL was transfected to wt and two clones of SHIP2Crispr 293T cells (1KF3, 3KF6), and immunoprecipitated (IP). SHC1 co-immunoprecipitates with BCR–ABL in SHIP2Crispr, suggesting that lack of SHC1 phosphorylation in SHIP2Crispr cells is not due to loss of interaction with BCR–ABL. Data are representative for 3 independent experiments (n). d K562, LAMA-84 and KYO-1 cells were seeded at 500 cells/μl, and treated with AS1949490 or DMSO. Cells were counted 48 and 96 h after treatment. Data represent averages ± SD from three independent experiments (n), with two technical replicates for each treatment. Statistically significant differences are indicated (Student’s t test, *p < 0.05, ***p < 0.001)
Conclusions
The BCR–ABL complex initiates signaling, governs the usage of downstream pathways and integrates BCR–ABL signal to processes ongoing in the cell. A precise mapping of the structure of BCR–ABL signaling complex is, therefore, essential for understanding the BCR–ABL function. Yet the in-cell characterization of protein–protein interactions within the complex does not always provide precise information about the hierarchy of these interactions. For instance, the co-immunoprecipitation of BCR–ABL with given interactor is the main approach to prove in-cell association, at the endogenous or forced levels of protein expression and without artifacts generated in cell-free systems (i.e., absence of competing interactors, excess of both partners, spatiotemporal aspects, solvents promoting interaction, and others). Successful co-immunoprecipitation, however, does not prove direct contact, because the whole signaling complex is pulled down, and thus the purified protein may be recruited to BCR–ABL via association with other interactors. This is a common theme in signaling, complicating data interpretation. For instance, recruitment of p85a-PI3K to the BCR–ABL complex may be direct, or maybe mediated by CRKL, GAB2, cCBL and SHC1 instead (Fig. 7f) [53, 59, 60].
Equally important to composition is dynamics of the complex, which allows for response to changing intracellular conditions during cell proliferation and differentiation, integration of BCR–ABL signaling to other signaling pathways, and adaptation and resistance of BCR–ABL signal to inhibitors, such as TKIs. In light of these facts, a detailed characterization of BCR–ABL interaction with GRB2, SHC1, cCBL, CRKL, STS1, p85a-PI3K and SHIP2 carried out here may only serve as basic reference, as it does not appreciate nor address the dynamics of the complex. The modes of interaction of individual signaling mediators with BCR–ABL are likely to be more complex and diverse in terms of the exact mechanism. This opens up a question of dispersity of BCR–ABL complexes, i.e., simultaneous co-existence of complexes of different composition, demonstrated by Figs. 1a, S1A, S2A (BCR–ABL blots). As shown here, several core complex members compete for the same domains when associating with BCR–ABL, suggesting that qualitatively different BCR–ABL complexes exist in a given cell at the same time. As this aspect of BCR–ABL function is poorly explored, the following research should address the quantitative differences in composition of BCR–ABL complexes.
We demonstrate that the core complex members associate with BCR–ABL in cells treated with TKI or with BCR–ABL inactive due to the KD mutation. This suggests that kinase activity of BCR–ABL is not necessary for the formation of signaling complex. The exact mechanism of recruitment on inactive BCR–ABL is likely to be different for different members of the core complex. Some proteins do not require phosphorylated motifs on BCR–ABL, and thus may associate regardless of BCR–ABL activity, while other interactors may rely on motifs phosphorylated, on inactive BCR–ABL, by other cellular kinases. For instance, although Y177 is autophosphorylated, it can also be phosphorylated by SRC-family kinases [27, 61], creating a GRB2-binding site on kinase-inactive BCR–ABL. The formation of kinase-inactive BCR–ABL complex underlies the residual signaling of inactive BCR–ABL in CML cells treated with TKI. Understanding of these mechanisms is essential for the development of novel therapeutic approaches for CML.
Materials and methods
Cell culture, vectors, transfection and CRISPR/Cas9
293T cells were obtained from ATCC, and propagated in DMEM media, supplemented with 10% FBS and antibiotics (ThermoFisher). CML cell lines K562 were obtained from ECACC, LAMA-84 and KYO-1 were obtained from DSMZ. CML cells were propagated in RPMI1640 supplemented with 10% FBS and antibiotics (ThermoFisher). All expression vectors are listed in Table S1. Cells were transiently transfected using FuGENE HD, according to the manufacturer’s protocol (Promega). p210 isoform b3a2 and p190 BCR–ABL sequences (isoform b3a2) were cloned into pCR3.1 vector with N-terminal FLAG tag. Polymerase chain reaction (PCR) mutagenesis was used to generate all BCR–ABL variants. GRB2 (RC200469), p85a-PI3K (RG210544) and SHIP2 (RC214716) vectors were obtained from Origene, CRKL (HG11261-CH) and STS1 (HG13868-NF) vectors were obtained from Sino Biological. Their coding sequences were subcloned into modified pCMV6 entry vector, where C-terminal Myc-DDK tag was replaced by HIS-V5 tag using a NEBuilder HiFi DNA assembly kit (New England Biolabs). SHIP2 deletion in 293T cells was carried out by CRISPR/Cas9 technology [62]. CHOPCHOP tool was used to design sgRNAs for a pair of SpCas9n (D10A) nickases, which targeted 5ʹ-CGATGGCAGCTTCCTGGTCC-3ʹ, 5ʹ-GCGCTCTGCGTCCTGTGAGT-3ʹ sites in the first exon of the SHIP2 gene [63]. Successful targeting disrupted the open reading frame of SHIP2, which was detected as loss of SHIP2 in individual clones by western blot. Targeted locus was PCR amplified using 5ʹ-CCGGGCGGCCGCGGAGGAG-3ʹ, 5ʹ-TCTGGCGTCCCACCGCCCCAGAAAC-3ʹ, inserted into pGEM T-Easy vector (Promega) and sequenced for determination of SHIP2 genotype.
Western blot and immunoprecipitation (IP)
Cells were lysed for 30 min at 4 °C in lysis buffer containing 50 mM tris–HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, 2 mM EDTA, 1 mM Na3VO4 and protease inhibitors (Roche). For IP, 25 ul of Dynabeads protein G (ThermoFisher) were bound to FLAG antibody (F1804, Sigma-Aldrich) or V5 antibody (R96025, Invitrogen) according to the manufacturer’s protocol. Immunocomplexes were collected overnight at 4 °C. Proteins attached to the beads were eluted to 2 × Laemmli buffer. For western blot, samples were resolved by SDS-PAGE, transferred onto a PVDF membrane, and visualized by chemiluminescence using Pierce ECL (Thermo Fisher Scientific), Immobilon Western (Millipore), Clarity (Biorad) or SuperSignal West Femto (Thermo Fisher Scientific) substrates. Table S2 lists all antibodies used in the study.
Gradient ultracentrifugation, BN-PAGE and proximity ligation assay (PLA)
The gradient ultracentrifugation was done as described before [64]. Briefly, the native cell lysates (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.5% Igepal CA-630, 1 mM EDTA, pH 8, 0.25% sodium deoxycholate, 1 mM Na3VO4; proteinase inhibitors) were loaded on 15–40% sucrose gradient (1 mM Tris–HCl, pH 7.4, 150 mM NaCl, 3 mM MgCl2; proteinase inhibitors), and centrifuged at 40,000 rpm/4 °C/16 h using SW 40 Ti rotor (Beckman Coulter). Approximately 18 fractions were collected from each gradient, the proteins were precipitated with 10% TCA, dissolved in 2× Laemmli buffer and boiled. The respective fractions, as well as the cell lysates collected before ultracentrifugation were resolved by western blot, and co-sedimentation of the endogenous interaction partners with expressed BCR–ABL was analyzed by densitometry (ImageJ; http://imagej.nih.gov/ij/). To quantify western blots shown in Fig. 1a, the relative abundance of each of the proteins in the dominant p190 BCR–ABL fractions (#6 in control and #5 in nilotinib treated cells) was calculated by densitometry of all fractions, and plotted. As only proteins saturated on BCR–ABL were analyzed, any peaks exceeding the relative optical density of 1 (red dashed lines) after treatment with nilotinib would suggest dissociation of the protein from the BCR–ABL complex, as observed with GRB2. Analogically, the peak of p85a-PI3K above 1 in control cells and the absence of the protein in heavier BCR–ABL fractions suggest that not all BCR–ABL complexes involve p85a-PI3K. The BN-PAGE was carried out as described before [64]. The native cell lysates have been loaded on 4–15% native gels, after the native electrophoresis the lane sample strips were excised from the gel, denatured and resolved in 12% SDS-PAGE gels. For Duolink® PLA (Sigma), cells were fixed in paraformaldehyde, post-fixed in ice-cold methanol and stained according to the manufacturer’s protocol. Mouse FLAG (F1804; Sigma) and Goat V5 (sc-83849; Santa Cruz) antibodies were used for PLA; rabbit anti-c-ABL (2862S; Cell Signaling) was used to counterstain the transfected cells. Secondary antibodies conjugated with AlexaFluor488/594 were from Invitrogen. PLA counting analysis was done in Fiji (http://fiji.sc/Fiji) using maximum projections of Z-stacks.
Peptide microarrays
Pepstar™ microarray technology (JPT Peptide Technologies) was used to identify sites in BCR–ABL involved in the binding of individual core complex interactors. Peptide library was generated by dividing the entire p210 BCR–ABL protein sequence into 13-aa-long peptides (674) with ten residues overlapping between neighboring peptides. Peptides were synthesized and immobilized on a glass slide. To account for phosphorylations at known BCR–ABL sites, namely Y177, Y328, S354, Y360 in BCR and Y89, Y134, Y147, Y158, Y191, Y204, Y234, Y245, Y251, Y272, Y276, Y331, Y412, Y488, S465, S637–638, T754 in cABL1b, phosphorylated versions of the 99 corresponding peptides were also included in the microarrays. For each experiment, experimental and control microarrays were processed in parallel. Arrays were incubated with recombinant interactors. GRB2 (TP300469), SHC1 (TP304362), STS1 (TP303523), CRKL (TP308129) and cCBL (TP314069) were obtained from Origene, SHIP2 (P09-20G-10) and p85a-PI3K (P31–30H) were obtained from SignalChem. Signal was developed by incubation of arrays with primary antibodies against GST (G1160), FLAG (F1804, Sigma-Aldrich) and HIS (sc-8036, Santa Cruz) and secondary, Cy-5-coupled antibody (715-175-151, Jackson ImmunoResearch). Arrays were scanned using InnoScan 1100 AL fluorescence scanner and data were analyzed by Mapix software (Innopsys). Each microarray slide contained three entire peptide libraries, which were analyzed as technical replicates. Fluorescence intensities were plotted as a function of the peptide number (BCR–ABL primary sequence). First, the intensities from all spots in the control microarray were used to calculate the arithmetic average (∅c) and the standard deviation (σc). If a signal intensity for a peptide spot in the control microarray exceeded the value of (∅c + 1.σc) in all three replicates, the respective peptide (microarray spot) was excluded from further analysis. Subsequently, fluorescence intensities in negative control microarrays were subtracted from values in experimental microarrays and these values were used for all further analyses. To allow direct comparison of fluorescence intensities from the three technical replicates, relative fluorescence intensities for each peptide were calculated by dividing each signal by the biggest value in the respective microarray. Finally, relative fluorescence intensities from the three replicates were averaged, plotted as a function of the peptide number (BCR–ABL primary sequence) and arithmetic average (∅ER) and standard deviation (σER) were calculated from the averaged fluorescence intensities. Potential binding site in the microarray was considered when at least three consecutive peptides exceeded (∅ER + 1.σER) in all replicates.
References
Zhao X, Ghaffari S, Lodish H et al (2002) Structure of the BCR–ABL oncoprotein oligomerization domain. Nat Struct Biol 9:117–120. https://doi.org/10.1038/nsb747
Hantschel O (2012) Structure, regulation, signaling, and targeting of Abl kinases in cancer. Genes Cancer 3:436–446. https://doi.org/10.1177/1947601912458584
Voncken JW, Kaartinen V, Pattengale PK et al (1995) BCR/ABL P210 and P190 cause distinct leukemia in transgenic mice. Blood 86:4603–4611
Hazlehurst LA, Bewry NN, Nair RR, Pinilla-Ibarz J (2009) Signaling networks associated with BCR–ABL-dependent transformation. Cancer Control J Moffitt Cancer Cent 16:100–107. https://doi.org/10.1177/107327480901600202
Skorski T, Bellacosa A, Nieborowska-Skorska M et al (1997) Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3 k/Akt-dependent pathway. EMBO J 16:6151–6161
Shuai K, Halpern J, ten Hoeve J et al (1996) Constitutive activation of STAT5 by the BCR–ABL oncogene in chronic myelogenous leukemia. Oncogene 13:247–254
Steelman LS, Pohnert SC, Shelton JG et al (2004) JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR–ABL in cell cycle progression and leukemogenesis. Leukemia 18:189–218. https://doi.org/10.1038/sj.leu.2403241
Druker BJ, Tamura S, Buchdunger E et al (1996) Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of BCR–ABL positive cells. Nat Med 2:561–566
Gorre ME, Mohammed M, Ellwood K et al (2001) Clinical resistance to STI-571 cancer therapy caused by BCR–ABL gene mutation or amplification. Science 293:876–880. https://doi.org/10.1126/science.1062538
Modugno M (2014) New resistance mechanisms for small molecule kinase inhibitors of Abl kinase. Drug Discov Today Technol 11:5–10. https://doi.org/10.1016/j.ddtec.2013.12.001
Weisberg E, Manley PW, Breitenstein W et al (2005) Characterization of AMN107, a selective inhibitor of native and mutant BCR–ABL. Cancer Cell 7:129–141. https://doi.org/10.1016/j.ccr.2005.01.007
Shah NP, Nicoll JM, Branford S et al (2005) Molecular analysis of dasatinib resistance mechanisms in CML patients identifies novel BCR–ABL mutations predicted to retain sensitivity to imatinib: rationale for combination tyrosine kinase inhibitor therapy. Blood 106:1093
Hochhaus A, Baccarani M, Deininger M et al (2008) Dasatinib induces durable cytogenetic responses in patients with chronic myelogenous leukemia in chronic phase with resistance or intolerance to imatinib. Leukemia 22:1200–1206. https://doi.org/10.1038/leu.2008.84
Zhou T, Parillon L, Li F et al (2007) Crystal structure of the T315I mutant of Abl kinase. Chem Biol Drug Des 70:171–181. https://doi.org/10.1111/j.1747-0285.2007.00556.x
Cortes JE, Kim D-W, Pinilla-Ibarz J et al (2013) A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med 369:1783–1796. https://doi.org/10.1056/NEJMoa1306494
Bewry NN, Nair RR, Emmons MF et al (2008) Stat3 contributes to resistance toward BCR–ABL inhibitors in a bone marrow microenvironment model of drug resistance. Mol Cancer Ther 7:3169–3175. https://doi.org/10.1158/1535-7163.MCT-08-0314
Eide CA, Adrian LT, Tyner JW et al (2011) The ABL switch control inhibitor DCC-2036 is active against the chronic myeloid leukemia mutant BCR–ABLT315I and exhibits a narrow resistance profile. Cancer Res 71:3189–3195. https://doi.org/10.1158/0008-5472.CAN-10-3224
Corbin AS, Agarwal A, Loriaux M et al (2011) Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR–ABL activity. J Clin Invest 121:396–409. https://doi.org/10.1172/JCI35721
Rousselot P, Charbonnier A, Cony-Makhoul P et al (2014) Loss of major molecular response as a trigger for restarting tyrosine kinase inhibitor therapy in patients with chronic-phase chronic myelogenous leukemia who have stopped imatinib after durable undetectable disease. J Clin Oncol Off J Am Soc Clin Oncol 32:424–430. https://doi.org/10.1200/JCO.2012.48.5797
Mitra A, Sasikumar K, Parthasaradhi BVV, Radha V (2013) The tyrosine phosphatase TC48 interacts with and inactivates the oncogenic fusion protein BCR–ABL but not cellular Abl. Biochim Biophys Acta BBA Mol Basis Dis 1832:275–284. https://doi.org/10.1016/j.bbadis.2012.10.014
Brehme M, Hantschel O, Colinge J et al (2009) Charting the molecular network of the drug target BCR–ABL. Proc Natl Acad Sci 106:7414–7419
Pendergast AM, Quilliam LA, Cripe LD et al (1993) BCR–ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell 75:175–185
Goga A, McLaughlin J, Afar DE et al (1995) Alternative signals to RAS for hematopoietic transformation by the BCR–ABL oncogene. Cell 82:981–988
Preyer M, Vigneri P, Wang JYJ (2011) Interplay between kinase domain autophosphorylation and F-actin binding domain in regulating imatinib sensitivity and nuclear import of BCR–ABL. PLoS One 6:e17020. https://doi.org/10.1371/journal.pone.0017020
Coutinho S, Jahn T, Lewitzky M et al (2000) Characterization of Grb4, an adapter protein interacting with BCR–ABL. Blood 96:618–624
Preisinger C, Kolch W (2010) The BCR–ABL kinase regulates the actin cytoskeleton via a GADS/Slp-76/Nck1 adaptor protein pathway. Cell Signal 22:848–856. https://doi.org/10.1016/j.cellsig.2009.12.012
Meyn MA, Wilson MB, Abdi FA et al (2006) Src family kinases phosphorylate the BCR–ABL SH3–SH2 region and modulate BCR–ABL transforming activity. J Biol Chem 281:30907–30916. https://doi.org/10.1074/jbc.M605902200
Ren R, Ye Z-S, Baltimore D (1994) Abl protein-tyrosine kinase selects the Crk adapter as a substrate using SH3-binding sites. Genes Dev 8:783–795
Lewitzky M, Kardinal C, Gehring NH et al (2001) The C-terminal SH3 domain of the adapter protein Grb2 binds with high affinity to sequences in Gab1 and SLP-76 which lack the SH3-typical P-X-X-P core motif. Oncogene 20:1052–1062. https://doi.org/10.1038/sj.onc.1204202
Thien CB, Langdon WY (2001) Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol 2:294
Dombrosky-Ferlan PM, Corey SJ (1997) Yeast two-hybrid in vivo association of the Src kinase Lyn with the proto-oncogene product Cbl but not with the p85 subunit of PI 3-kinase. Oncogene 14:2019
Buday L, Khwaja A, Sipeki S et al (1996) Interactions of Cbl with two adaptor proteins, Grb2 and Crk, upon T cell activation. J Biol Chem 271:6159–6163
Donovan JA, Wange RL, Langdon WY, Samelson LE (1994) The protein product of the c-cbl protooncogene is the 120-kDa tyrosine-phosphorylated protein in Jurkat cells activated via the T cell antigen receptor. J Biol Chem 269:22921–22924
Meisner H, Conway BR, Hartley D, Czech MP (1995) Interactions of Cbl with Grb2 and phosphatidylinositol 3′-kinase in activated Jurkat cells. Mol Cell Biol 15:3571–3578
Panchamoorthy G, Fukazawa T, Miyake S et al (1996) p120 is a major substrate of tyrosine phosphorylation upon B cell antigen receptor stimulation and interacts in vivo with Fyn and Syk tyrosine kinases, Grb2 and Shc adaptors, and the p85 subunit of phosphatidylinositol 3-kinase. J Biol Chem 271:3187–3194
Bhat A, Kolibaba K, Oda T et al (1997) Interactions of CBL with BCR–ABL and CRKL in BCR–ABL-transformed myeloid cells. J Biol Chem 272:16170–16175
Johnson KJ, Griswold IJ, O’Hare T et al (2009) A BCR–ABL mutant lacking direct binding sites for the GRB2, CBL and CRKL adapter proteins fails to induce leukemia in mice. PLoS One 4:e7439. https://doi.org/10.1371/journal.pone.0007439
Buday L, Downward J (1993) Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell 73:611–620. https://doi.org/10.1016/0092-8674(93)90146-h
Egan SE, Giddings BW, Brooks MW et al (1993) Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature 363:45. https://doi.org/10.1038/363045a0
Li N, Batzer A, Daly R et al (1993) Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature 363:85–88. https://doi.org/10.1038/363085a0
Rozakis-Adcock M, Fernley R, Wade J et al (1993) The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature 363:83–85. https://doi.org/10.1038/363083a0
Trb T, Choi WE, Wolf G et al (1995) Specificity of the PTB domain of Shc for β turn-forming pentapeptide motifs amino-terminal to phosphotyrosine. J Biol Chem 270:18205–18208. https://doi.org/10.1074/jbc.270.31.18205
McWHIRTER JR, Galasso DL, Wang JY (1993) A coiled-coil oligomerization domain of Bcr is essential for the transforming function of BCR–ABL oncoproteins. Mol Cell Biol 13:7587–7595
Brasher BB, Roumiantsev S, Van Etten RA (2001) Mutational analysis of the regulatory function of the c-Abl Src homology 3 domain. Oncogene 20:7744–7752. https://doi.org/10.1038/sj.onc.1204978
Grebien F, Hantschel O, Wojcik J et al (2011) Targeting the SH2-kinase interface in BCR–ABL inhibits leukemogenesis. Cell 147:306–319. https://doi.org/10.1016/j.cell.2011.08.046
Oda T, Heaney C, Hagopian JR et al (1994) Crkl is the major tyrosine-phosphorylated protein in neutrophils from patients with chronic myelogenous leukemia. J Biol Chem 269:22925–22928
ten Hoeve J, Arlinghaus RB, Guo JQ et al (1994) Tyrosine phosphorylation of CRKL in philadelphia + leukemia. Blood 84:1731–1736
Nichols GL, Raines MA, Vera JC et al (1994) Identification of CRKL as the constitutively phosphorylated 39-kD tyrosine phosphoprotein in chronic myelogenous leukemia cells. Blood 84:2912–2918
Odai H, Sasaki K, Iwamatsu A et al (1997) Purification and molecular cloning of SH2- and SH3-containing inositol polyphosphate-5-phosphatase, which is involved in the signaling pathway of granulocyte-macrophage colony-stimulating factor, erythropoietin, and BCR–ABL. Blood 89:2745–2756
Wisniewski D, Strife A, Swendeman S et al (1999) A novel SH2-containing phosphatidylinositol 3, 4, 5-trisphosphate 5-phosphatase (SHIP2) is constitutively tyrosine phosphorylated and associated with src homologous and collagen gene (SHC) in chronic myelogenous leukemia progenitor cells. Blood 93:2707–2720
Skorski T, Kanakaraj P, Nieborowska-Skorska M et al (1995) Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood 86:726–736
Ren S, Xue F, Feng J, Skorski T (2005) Intrinsic regulation of the interactions between the SH3 domain of p85 subunit of phosphatidylinositol-3 kinase and the protein network of BCR/ABL oncogenic tyrosine kinase. Exp Hematol 33:1222–1228. https://doi.org/10.1016/j.exphem.2005.06.030
Sattler M, Mohi MG, Pride YB et al (2002) Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell 1:479–492
Mian AA, Baumann I, Liebermann M et al (2019) The phosphatase UBASH3B/Sts-1 is a negative regulator of BCR–ABL kinase activity and leukemogenesis. Leukemia. https://doi.org/10.1038/s41375-019-0468-y
Reckel S, Hamelin R, Georgeon S et al (2017) Differential signaling networks of BCR–ABL p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia 31:1502–1512. https://doi.org/10.1038/leu.2017.36
Cutler JA, Tahir R, Sreenivasamurthy SK et al (2017) Differential signaling through p190 and p210 BCR–ABL fusion proteins revealed by interactome and phosphoproteome analysis. Leukemia 31:1513–1524. https://doi.org/10.1038/leu.2017.61
Reckel S, Gehin C, Tardivon D et al (2017) Structural and functional dissection of the DH and PH domains of oncogenic BCR–ABL tyrosine kinase. Nat Commun. https://doi.org/10.1038/s41467-017-02313-6
Suwa A, Yamamoto T, Sawada A et al (2009) Discovery and functional characterization of a novel small molecule inhibitor of the intracellular phosphatase, SHIP2. Br J Pharmacol 158:879–887. https://doi.org/10.1111/j.1476-5381.2009.00358.x
Harrison-Findik D, Susa M, Varticovski L (1995) Association of phosphatidylinositol 3-kinase with SHC in chronic myelogeneous leukemia cells. Oncogene 10:1385–1391
Salgia R, Pisick E, Sattler M et al (1996) p130CAS forms a signaling complex with the adapter protein CRKL in hematopoietic cells transformed by the BCR/ABL oncogene. J Biol Chem 271:25198–25203
Warmuth M, Bergmann M, Priess A et al (1997) The Src family kinase Hck interacts with BCR–ABL by a kinase-independent mechanism and phosphorylates the Grb2-binding site of Bcr. J Biol Chem 272:33260–33270
Ran FA, Hsu PD, Wright J et al (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. https://doi.org/10.1038/nprot.2013.143
Montague TG, Cruz JM, Gagnon JA et al (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42:W401–W407. https://doi.org/10.1093/nar/gku410
Kunova Bosakova M, Nita A, Gregor T et al (2019) Fibroblast growth factor receptor influences primary cilium length through an interaction with intestinal cell kinase. Proc Natl Acad Sci 116:4316–4325. https://doi.org/10.1073/pnas.1800338116
Acknowledgements
This study was supported by Agency for Healthcare Research of the Czech Republic, project NV15-34405A. AN was supported by European Regional Development Fund OP RDE Project no. CZ.02.1.01/0.0/0.0/16_019/0000729.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gregor, T., Bosakova, M.K., Nita, A. et al. Elucidation of protein interactions necessary for the maintenance of the BCR–ABL signaling complex. Cell. Mol. Life Sci. 77, 3885–3903 (2020). https://doi.org/10.1007/s00018-019-03397-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03397-7