Abstract
The urokinase receptor (uPAR) stimulates cell proliferation by forming a macromolecular complex with αvβ3 integrin and the epidermal growth factor receptor (EGFR, ErbB1 or HER1) that we name the uPAR proliferasome. uPAR transactivates EGFR, which in turn mediates uPAR-initiated mitogenic signal to the cell. EGFR activation and EGFR-dependent cell growth are blocked in the absence of uPAR expression or when uPAR activity is inhibited by antibodies against either uPAR or EGFR. The mitogenic sequence of uPAR corresponds to the D2A motif present in domain 2. NMR analysis revealed that D2A synthetic peptide has a particular three-dimensional structure, which is atypical for short peptides. D2A peptide is as effective as EGF in promoting EGFR phosphorylation and cell proliferation that were inhibited by AG1478, a specific inhibitor of the tyrosine kinase activity of EGFR. Both D2A and EGF failed to induce proliferation of NR6-EGFR-K721A cells expressing a kinase-defective mutant of EGFR. Moreover, D2A peptide and EGF phosphorylate ERK demonstrating the involvement of the MAP kinase signalling pathway. Altogether, this study reveals the importance of sequence D2A of uPAR, and the interdependence of uPAR and EGFR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The urokinase receptor (uPAR) is a multi-functional and multi-ligand molecule attached to the outer leaflet of the plasma membrane through a glycosyl-phosphatidyl-inositol (GPI) moiety. uPAR is primarily involved in binding of urokinase (uPA), a serine protease responsible of the activation of plasminogen, controlling the activity and localization of pericellular proteolysis. Since uPAR has no intracellular domain, signal transduction occurs through the intercession of other membrane receptors, which transmit uPAR-initiated signal to the cell [1,2,3,4,5,6]. Most important lateral partners of uPAR include G protein-coupled receptors (GPCRs), receptor tyrosine kinases (RTKs) or integrins [4]. However, uPAR is also capable of activating integrins distally through bidirectional signalling mediated by moesin [7]. This wide array of partners explains the physiological and physiopathological functions of uPAR in cell migration, adhesion and proliferation [8,9,10].
Interactions of uPAR with external soluble molecules and lateral partners are facilitated by its tridimensional structure [11,12,13,14]. uPA, the main ligand of uPAR, binds into the central pocket constituted by its three homologous domains, while other ligands bind on the back of the receptor where specific sites of interactions have been identified in particular in domains 2 and 3 [15,16,17,18,19].
uPA has been shown to stimulate cell proliferation [20,21,22,23]. These effects may be achieved via the activation and/or release of growth factors such as transforming growth factor-β and hepatocyte growth factor [24, 25]. Since integrins regulate cell proliferation and are partners of RTKs and uPAR [26, 27], the interaction of uPA with integrins may also be involved [8, 28, 29]. Better yet, uPA (or its catalytically inactive amino terminal fragment, ATF) can directly promote cell proliferation through binding to uPAR promoting the association of the latter with RTKs such as the EGF receptor (EGFR, ErbB1 or HER1) or the PDGF receptor-β [18,19,20,21, 23, 30,31,32]. Factor XII, another ligand of uPAR also promotes cell growth [19, 33, 34]. uPAR expression is upregulated by EGF and other mitogens [35, 36]. Moreover, uPAR−/− cells grow slower than wild-type cells underlining the importance of uPAR in the proliferative response [35, 37, 38]. However, to date, the role of uPAR in the regulation of cell proliferation is far from being completely known. Understanding the growth-promoting function of uPAR is also important because in the general view of cancer development, tumour growth precedes invasion and metastasis.
In this study, we report that uPAR stimulates cell proliferation through lateral interactions with integrins and EGFR (ErbB1 or HER1), forming a macromolecular complex that we called the uPAR proliferasome. EGFR is inactive in the absence of uPAR or when uPAR activity is inhibited. The first identified mitogenic sequence of uPAR corresponds to the D2A motif present in its domain 2 that we previously discovered for its functions in the regulation of cell migration [15]. NMR analysis showed that the derived D2A synthetic peptide has a specific three-dimensional (3D) structure. Lastly, D2A peptide transactivates EGFR and is as potent as EGF in stimulating cell growth.
Materials and methods
Materials
HT-1080 (a human fibrosarcoma cell line), HEK-293 and HEK-293-uPAR (transformed human kidney cell lines), NR6-EGFRwt and NR6-EGFR-K721A (derived from murine 3T3 cells) cells were cultured in DMEM plus 10% FCS. Medium of NR6-EGFRwt and NR6-EGFR-K721A cells was supplemented with 2 μg/ml puromycin. HT-29 (a human colorectal adenocarcinoma cell line) was cultured in McCoy 5A plus 10% FCS. All peptides have been previously described and provided by Eurogentec [15]: D2A peptide has been derived from sequence 130IQEGEEGRPKDDR142 harboured in the domain 2 of human uPAR; D2A-Ala peptide IQEGAAGRPKDDR is the inhibitory version of D2A peptide; scrambled D2A corresponds to DEIGQDKERPGRE; GEEG and GAAG are the minimal stimulatory and inhibitory peptides, respectively. The reagents used were: mouse monoclonal anti-αvβ3 (LM609) blocking antibody (Chemicon); monoclonal anti-phosphotyrosine clone 4G10 (Upstate); monoclonal anti-EGFR (Ab-1) clone 528 blocking antibody, inhibitors AG1478 and PD98059 (Calbiochem); polyclonal anti-EGFR and monoclonal anti-ERK 1/2 (C-9, sc-514302) antibodies (Santa Cruz Biotechnology); polyclonal anti-phosphorylated ERK 1/2 (9101, Cell Signaling Technology) and unrelated polyclonal and monoclonal antibodies (Sigma). Polyclonal SI517 and SI518 anti-D2A antibodies were produced in rabbit using standard procedure (Eurogentec). Anti-uPAR monoclonal R3 and polyclonal SI369 antibodies have been described [37, 39, 40]. Full-length human soluble uPAR (suPAR) was purified as previously described [41, 42]. EGF was from Millipore.
NMR analysis and three-dimensional modelling
D2A peptide was dissolved in 0.5 ml of 1H2O/2H2O (90/10 v/v) to yield concentrations in the range 2–3 mM. The pH was adjusted to 6.8 by adding HCl or NaOH. Deuterated water was obtained from CortecNet (France). Homonuclear 2D clean TOCSY and NOESY spectra were recorded at 280 K on an Avance 400 Bruker spectrometer operating at 400.13 MHz using an inverse multinuclear probehead fitted with gradients along the x-, y- and z-axes, by standard techniques and incorporating the excitation sculpting sequence for water suppression. We used a pulsed-field gradient double echo with a soft square pulse of 4 ms at the water resonance frequency, with the gradient pulses of 1 ms each in duration. 480 equally spaced evolution time-period t 1 values were acquired, averaging 32 transients of 2048 points, with 5208 Hz of spectral width. t 1 dimension was all zero-filled to 2048, yielding a digital resolution of 2.54 Hz/pt. Prior to Fourier transformation, resolution enhancement was applied with a Lorentz–Gauss window to both t 1 and t 2 dimensions for all the experiments. NOESY spectra were obtained at different mixing times (150 and 250 ms); TOCSY experiments were recorded with a spin-lock period of 68 ms, achieved with the MLEV17 sequence. Spectra were referenced to sodium 3-(trimethylsilyl)-[2,2,3,3-2H4]propionate. 3JNH-α values were estimated in a 1D spectrum acquired in 1H2O/2H2O (90/10 v/v). The time-domain was zero-filled to 131072 points yielding a resolution of 0.040 Hz/pt, and apodized with a strong Lorentz–Gauss window. The natural abundance 1H-13C HSQC spectra were recorded at 300 K on a DRX-600 Bruker spectrometer operating at 150.90 MHz for 13C, using an inverse TCI CryoProbe fitted with gradient along the z-axis. 128 equally spaced evolution time-period t 1 values were acquired, averaging 40 transients of 2048 points and using GARP for decoupling. t 1 dimension was zero-filled to 2048, and apodized before Fourier transformation by a shifted cosine window function in t 2 and in t 1. Spectra were referenced to methanol.
Migration assays
Chemotaxis assays were performed as previously described [15, 41] using chemotaxis microchamber (Neuro Probe). Filters (8 μm pore, Neuro Probe) were coated with collagen I (100 μg/ml) (Roche). 50,000 cells were added into the upper part of each well and allowed to migrate overnight at 37 °C. All experiments were performed at least three times in triplicate. Migration in the absence of chemoattractant is considered 100%.
Cell invasion was determined using 24-well plates and Transwell inserts (8 μm pore size, Corning) coated with thick layer of matrigel (100 μl per square centimetre of growth surface) (BD Biosciences). 200,000 cells were seeded on the top in serum-free medium. The molecule to be tested was diluted in serum-free medium and added in the well of the plate. After 24 h of invasion, cells remaining on the top of matrigel were scraped off, filters were fixed in 20% (v/v) methanol, stained using Diff-Quick solution (Medion Diagnostics) and cells were counted under the microscope (lens 40) in five high power fields per filter. The experiments were performed at least three times in duplicate. Invasion in the absence of stimulator (control) was given the value of 100%.
Cell proliferation assays
20,000 cells were added in each 2-cm2 well of a 24-well plate and cultured overnight in medium plus 10% FCS. Then, cells were washed twice with PBS pH 7.4, and further cultured in serum-free medium. The mitogenic agent to be tested was added in solution everyday in each well. Cell numbers were estimated by detaching the cells with trypsin/EDTA (Sigma) and counting the cells under the microscope using a Bürker chamber. Negative (cells cultured in serum-free medium) and positive controls (cells cultured in medium plus 10% FCS) were included. Experiments were performed at least three times in triplicate.
Immunoblotting
Subconfluent cell monolayers (50%) were serum-starved for 24 h, and challenged with the appropriate reagent as indicated. When present, inhibitors were added 15 min prior stimulation. Then, cells were washed with PBS, lysed in JS buffer: 1% Triton X-100, 150 mM NaCl, 1% Glycerol, 50 mM HEPES, pH 7.5, 1.5 mM MgCl2, 5 mM EGTA, 0.5% Na-deoxycholate, 0.1 M phenylmethylsulfonyl fluoride (PMSF), 500 mM sodium orthovanadate (all reagents from Sigma) and lysates incubated for 4 h at 4 °C, in the presence of 1 μg of specific antibody and protein G-Sepharose. Samples were boiled in reducing SDS-sample buffer; protein content of each sample was determined using Bradford assay (Pierce, Thermo Fisher Scientific) and equal amount of proteins (50 μg) loaded into the acrylamide gels using specific gel-loading pipet tips and separated in SDS-PAGE using 10% acrylamide gel. After protein transfer to the membrane but before immunological detection, the qualitative visualization of proteins present on the membrane was performed using red ponceau (Sigma) staining. Then, blots were incubated in blocking solution (5% non-fat dry milk in TBS-T or 3% BSA in TBS), incubated in the presence of the appropriate primary and HRP-conjugated secondary antibody (Santa Cruz Biotechnology) and then revealed using the SuperSignal West Pico chemiluminescent substrate (Pierce) and exposure to X-ray film. Immunoprecipitation was omitted for the detection of phosphorylated ERK and total ERK.
Statistical analysis
Student’s t test was employed for pairwise comparison of treatments. An ANOVA model was used for the evaluation of treatments with increasing doses of a reagent. Calculations were performed using Prism software.
Results
uPAR expression is required for EGF-induced cell proliferation and EGFR phosphorylation
We tested the capacity of EGF to stimulate the proliferation of parental HEK-293 cells, which do not express uPAR, and of uPAR-transfected cell line HEK-293-uPAR. EGF at doses ranging from 1 ng/ml (0.16 nM) to 100 ng/ml (16 nM) failed to promote HEK-293 cell proliferation (left panel, Fig. 1a). In contrast, HEK-293-uPAR cells expressing human uPAR proliferated when stimulated with EGF (right panel, Fig. 1a). These latter cells appeared to be extremely sensitive to EGF with a maximal response observed at very low doses of 1–2 ng/ml (0.16–0.32 nM) (Fig. 1a). However, EGF still promoted cell proliferation at the highest dose of 100 ng/ml. Thus, the mitogenic effect of EGF is dependent on uPAR expression. In an attempt to rescue HEK-293 cell proliferation, full-length soluble uPAR (10 pM) was added in the presence or in the absence of the same increasing doses of EGF (1–100 ng/ml). However, the addition of suPAR failed to restore HEK-293 cell proliferation in response to EGF (Fig. 1a). Furthermore, suPAR failed to influence HEK-293-uPAR cell proliferation either in the absence or in the presence of EGF (Fig. 1a). Therefore, the stimulation of cell proliferation by EGF requires the presence of membrane-bound uPAR.
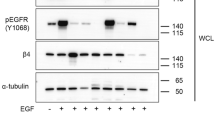
EGFR activity is dependent on uPAR expression. a The mitogenic effect of EGF depends on uPAR expression on the cell surface. Comparison of the effects of increasing doses of EGF in the presence or in the absence of suPAR on HEK-293 (devoid of uPAR) and HEK-293-uPAR cells (expressing uPAR). Cell numbers expressed as mean ± SD (n = 3) were determined at day 4. The number of cells cultured without EGF and suPAR is considered to be 100% cell proliferation (control). *p < 0.01 compared to control (white bar). b Serum-starved HEK-293 and HEK-293-uPAR cells were stimulated with either 1 ng/ml EGF or 100 pM D2A. When present, cells were pretreated for 15 min with the EGFR tyrosine kinase inhibitor AG1478 before being further stimulated for 10 min with EGF or D2A. Then, cells were lysed, and immunoprecipitated EGFR was blotted for phosphotyrosine (EGFR-P, upper pictures) and total EGFR (EGFR total, middle pictures) as described in the “Materials and methods” section. Each panel is representative of one independent experiment out of at least three. The bar graphs show the standardized densitometric analysis expressed as mean ± SD (n = 3, left panel; n = 4, right panel) of the ratio EGFR-P/EGFR total. Control (C, without AG1478) was given the arbitrary value of 100%. *p < 0.05 and **p < 0.01 compared to control
Since EGF is known to induce EGFR autophosphorylation, we examined the levels of EGFR phosphorylation in HEK-293 and HEK-293-uPAR cells. EGF induced EGFR phosphorylation in HEK-293-uPAR cells but not in parental HEK-293 cells (Fig. 1b). Moreover, AG1478, the specific inhibitor of the tyrosine kinase activity of EGFR, blocked EGF-induced phosphorylation of EGFR in HEK-293-uPAR cells but had no effect on parental HEK-293 cells (Fig. 1b). These data show that EGF-induced EGFR phosphorylation is due to the intrinsic tyrosine kinase activity of EGFR, which requires the expression of uPAR. The tyrosine kinase activity is defective and EGFR not functional when uPAR is not present on the cell surface.
Targeting uPAR blocks EGF-induced cell proliferation
To further establish the crucial role of uPAR in the mitogenic response to EGF, we used HT-29 cells derived from a colorectal adenocarcinoma expressing both EGFR and uPAR, in which EGF promoted proliferation in a dose-dependent manner (Fig. 2a). HT-29 cells are highly sensitive to EGF responding to doses as low as 1 ng/ml with a maximum at 5 ng/ml (0.80 nM) (Fig. 2a). The apparent inhibitory effect of higher doses (10–100 ng/ml = 1.6–16 nM) is due to the fact that at these doses EGF induced cell rounding and detachment from substrate (Online Resource 1).
EGF-induced cell proliferation is dependent on uPAR activity. a EGF promotes cell proliferation. HT-29 cells were treated with increasing doses of EGF ranging from 1 ng/ml (0.16 nM) to 100 ng/ml (16 nM). Cell number was estimated after 4 days of culture and is represented as mean ± SD from three experiments. Number of cells kept in serum-free media without EGF is referred to as 100% proliferation (control). *p < 0.05 and **p < 0.01 compared to control. b EGF-promoted cell proliferation is blocked by anti-uPAR antibodies. The effects of EGF (5 ng/ml = 0.80 nM) on HT-29 cell proliferation were determined in the presence or in the absence of either SI517 or SI518 (each at 40 μg/ml) anti-uPAR polyclonal antibody. An unrelated antibody (40 μg/ml) serves as negative control. Data are the mean ± SD (n = 3). Statistical significance is p < 0.05 in an ANOVA model, for 10% FCS, EGF and EGF + unrelated Ab conditions
In time-course experiments, EGF at the optimal dose of 5 ng/ml stimulated cell proliferation inducing a sixfold increase in cell number compared to day 0 or a 2.5-fold increase compared to negative control (0% FCS) at day 6 (Fig. 2b). To target uPAR, we used SI517 or SI518 anti-uPAR polyclonal antibody, which entirely inhibited EGF-induced cell growth, whereas an unrelated polyclonal antibody employed as control had no inhibitory effects (Fig. 2b). On their own, none of these three antibodies influenced cell proliferation. 0% FCS and 10% FCS served as negative and positive controls, respectively. Thus, EGF-induced cell proliferation mediated by EGFR is dependent on uPAR activity.
uPAR, through its D2A sequence, is capable of promoting cell proliferation
The above data prompted us to further investigate the functions of uPAR in the regulation of cell proliferation. Since SI517 and SI518 antibodies were raised against the D2A epitope of uPAR, we particularly explored the functions of this D2A sequence, which we previously identified [15].
We investigated the effects of D2A peptide on cell proliferation using HT-29 cells and HT-1080 cells isolated from a highly invasive fibrosarcoma, which also express both uPAR and EGFR. Firstly, we checked that HT-1080 cells proliferate in response to EGF challenge. Online Resource 2 shows that EGF induced proliferation of HT-1080 cells in a dose-dependent manner. However, HT-1080 cells are less sensitive than HT-29 cells displaying a maximal response at the dose of 50 ng/ml (8 nM), which was used thereafter (Online Resource 2). HT-29 cell growth was stimulated by D2A in a dose-dependent manner reaching a plateau at 100 pM (Fig. 3a). D2A also promoted proliferation of HT-1080 in a dose-dependent manner (Fig. 3b). Then, we tested the GEEG peptide, the shorter version the D2A peptide corresponding to 133GEEG136 sequence of human uPAR, which was previously proved to be as potent as D2A in stimulating cell migration [15]. HT-29 and HT-1080 cell proliferation were dose-dependently stimulated by GEEG peptide (Fig. 3c, d). In contrast, scrambled D2A (which has no GEEG sequence) failed to stimulate the proliferation of HT-29 and HT-1080 cells (Fig. 3e, f). In time-course experiments, D2A and GEEG equally stimulated proliferation of HT-29 and HT-1080 cells, while scrambled D2A and GAAG peptides (both harbouring no GEEG sequence) failed to promote cell growth (Fig. 4a, b). These data suggest that uPAR through its D2A sequence located in domain 2 is capable of promoting cell proliferation. Moreover, GEEG is the minimum mitogenic sequence of D2A.
Effects of peptide D2A and its shorter version GEEG on cell proliferation. Effects of increasing doses of peptide: D2A (a, b), GEEG (c, d) and scrambled D2A (e, f) on proliferation of HT-29 (a, c, e) and HT-1080 (b, d, f) cells. Cell numbers were determined at day 4. Values are the mean ± SD from three experiments. Control corresponds to the number of cells cultured in serum-free media without peptide (100% proliferation). *p < 0.01 compared to control
D2A and GEEG peptides are equipotent in stimulating cell proliferation. a, b Time-course experiments showing the effects of optimal dose (100 pM) of peptides D2A, GEEG, scrambled D2A (Scr. D2A) and GAAG on proliferation of HT-29 (a) and HT-1080 (b) cells. Cell proliferation was enhanced to similar levels by D2A and GEEG peptides, whereas scrambled D2A and GAAG peptides harbouring no GEEG sequence failed to promote cell proliferation. Number of cells cultured in serum-free media without peptide is referred to as 100% proliferation (control). Data are the mean ± SD from three experiments. The conditions 10% FCS, D2A, GEEG are significantly different (a p < 0.01; b p < 0.05) from control as determined by ANOVA test
The growth-promoting effect of peptide D2A matches the effect of EGF
Next, we compared the effects of D2A peptide on cell proliferation with those of EGF. D2A peptide stimulated cell proliferation to an extent similar to that of EGF demonstrating that peptide D2A is equally potent as EGF in inducing cell proliferation (Fig. 5a, b). However, the mitogenic effects of D2A and EGF were not additive, since the combination of optimal doses of peptide D2A (100 pM) and EGF (5 ng/ml or 0.80 nM) did not further increase HT-29 cell proliferation (Fig. 5c). These equivalent but not additive effects of EGF and peptide D2A led us considering whether EGFR was the actual mediator of the D2A-promoted mitogenic signal. A blocking monoclonal antibody against EGFR entirely inhibited D2A-induced cell growth. EGF-promoted cell proliferation was also blocked by this antibody while an unspecific control antibody had no effect (Fig. 5a). Either of these two antibodies alone had no effects. 0 and 10% FCS served as negative and positive controls, respectively (Fig. 5a). Moreover, AG1478 fully blocked D2A- and EGF-promoted cell proliferation suggesting that EGFR tyrosine kinase activity transduced both D2A- and EGF-induced mitogenic signals (Fig. 5b). To further support this latter idea, we used NR6 cells (derived from murine 3T3 cells) [43] lacking endogenous EGFR, which were transfected to express either wild-type human EGFR or K721A, a kinase-defective mutant of EGFR [41, 44, 45]. Peptide D2A and EGF were mitogenic for NR6-EGFRwt cells (Fig. 5d, f), but failed to induce the proliferation of NR6-EGFR-K721A cells (Fig. 5e, g). Altogether these data reveal that the EGF-matching growth-promoting effect of peptide D2A is transmitted to the cell through the tyrosine kinase activity of EGFR.
EGFR mediates both D2A- and EGF-induced cell proliferation. a EGFR is essential for the induction of cell proliferation by either D2A peptide or its main ligand EGF. HT-29 cell proliferation was stimulated with either 100 pM D2A or 5 ng/ml (0.80 nM) EGF with or without the addition of an anti-EGFR (1 μg/ml) blocking monoclonal antibody. An unrelated (1 μg/ml) antibody served as negative control. Data are the mean ± SD from three experiments. Number of cells cultured in serum-free media without D2A, EGF and antibody is referred to as 100% proliferation (control). The conditions 10% FCS, D2A, D2A + unrelated Ab, EGF, EGF + Unrelated Ab are significantly different (p < 0.01) from control. b Both peptide D2A- and EGF-promoted cell proliferations are inhibited by either AG1478 or PD98059 inhibitors. Proliferation of HT-29 cells was induced by the addition of 100 pM D2A or 5 ng/ml (0.80 nM) EGF in the absence or in the presence of either 1 μM of AG1478 (a specific inhibitor of EGFR tyrosine kinase activity) or 5 μM of PD98059 (an inhibitor of MEK). Data are the mean ± SD from three experiments. Number of cells cultured in serum-free media without D2A, EGF or inhibitor is referred to as 100% proliferation (control). The statistical significance of the results is p < 0.01 in an ANOVA model, for 10% FCS, D2A and EGF. c D2A and EGF mitogenic effects are not additive. Proliferation of HT-29 cells was stimulated with optimal dose of D2A (100 pM), EGF (5 ng/ml or 0.80 nM) or both mixed together as indicated. Cell numbers were determined at day 4. Data are the mean ± SD (n = 3). Control corresponds to the number of cells cultured in serum-free media without D2A and EGF (100% proliferation). **p < 0.01 compared to control. d–g Stimulation of cell proliferation by D2A or EGF is dependent on the intact tyrosine kinase activity of EGFR. NR6 cells expressing either wild-type EGFR (EGFRwt) or the tyrosine kinase-deficient mutant EGFR K721A were treated with increasing doses of either D2A peptide (d, e) or EGF (f, g). Cell number was estimated after 4 days of culture and is represented as mean ± SD (n = 3). Numbers of cells kept in serum-free media without D2A or EGF are referred to as 100% proliferation (control). *p < 0.05 and **p < 0.01 compared to control
Then, we compared the activation of EGFR by its ligand EGF with the effects triggered by D2A. Both EGF and D2A activated EGFR but the kinetics of EGFR phosphorylation were slightly different (Fig. 6a, b). EGF elicited a maximal response at 2 and 10 min (Fig. 6a), while D2A effect was visible after 1 min and lasted up to 60 min (Fig. 6b). The addition of the EGFR tyrosine kinase inhibitor AG1478 completely prevented both EGF- and D2A-induced EGFR activations (Fig. 6c). Therefore, D2A transactivates EGFR, which mediates the mitogenic signal to the cell.
Effects of peptide D2A and EGF on EGFR activation and ERK phosphorylation. a–c Serum-starved HT-29 cells were treated with either 5 ng/ml (0.80 nM) EGF (a), 100 pM D2A (b) for different times as indicated. In c, cells were pretreated or not for 15 min with the EGFR tyrosine kinase inhibitor AG1478 (1 μM) before being further stimulated for 10 min with 5 ng/ml (0.80 nM) EGF or 100 pM D2A. Then, cells were lysed, and immunoprecipitated EGFR was blotted for phosphotyrosine (EGFR-P, upper pictures) and total EGFR (EGFR, middle pictures). Each panel is representative of one independent experiment out of three. Bar graphs show the standardized densitometric analysis expressed as mean ± SD (n = 3) of the ratio EGFR-P/EGFR total. *p < 0.05 and **p < 0.01 compared to control. d, e Serum-starved HT-29 cells were stimulated with 5 ng/ml (0.80 nM) EGF (d) or 100 pM D2A (e) for increasing times as indicated. Then, phosphorylated ERK (ERK-P, upper pictures) and total ERK (ERK, middle pictures) were detected in cell lysates by immunoblotting. Each panel is representative of one independent experiment out of three. Bar graphs show the densitometric analysis of ERK activation (ERK-P) normalized to total ERK and expressed as mean ± SD (n = 3). *p < 0.05 and **p < 0.01 compared to control
D2A and EGF activate the MAP kinase pathway
We explored the effects of D2A on the mitogen-activated protein (MAP) kinase signalling pathway, which has been reported to be activated by uPA/uPAR [46] through EGFR [30]. Western blotting analysis showed that EGF activated ERK 1/2 with a peak of activity observed after 5–10 min (Fig. 6d), while D2A-induced ERK activation was maximal at 1–2 min (Fig. 6e). Furthermore, the MAP kinase kinase (MEK) inhibitor PD98059 entirely abolished the mitogenic effects of D2A and EGF (Fig. 5b). These data suggest that both peptide D2A and EGF activate EGFR that in its turn stimulates the MAP kinase signalling pathway, which is involved in the regulation of D2A- and EGF-promoted cell growth.
Mechanism of action of D2A: an involvement of uPAR, integrin αvβ3 and EGF receptor
We previously reported that peptide D2A binds to integrin αvβ3 [15]. Furthermore, integrins are capable of transactivating EGFR [30, 47, 48]. Therefore, we explored the involvement of αvβ3 in the mechanism of transduction of D2A-induced mitogenic signal. LM609, a blocking monoclonal anti-αvβ3 antibody totally inhibited D2A-promoted cell proliferation, while an unrelated control monoclonal antibody had no effect (Fig. 7a). Moreover, LM609 also blocked EGF-induced cell proliferation (Fig. 7a). The importance of uPAR in the transduction of the growth signal promoted by D2A or EGF was further demonstrated by the fact that two anti-uPAR antibodies R3 and SI369 completely prevented D2A- and EGF-induced cell proliferation (Fig. 7a). Moreover, the combination of D2A and EGF failed to restore the capacity of HEK-293 cells to proliferate in response to EGF or D2A (Fig. 7b). In contrast, HEK-293-uPAR proliferated when challenged with either D2A or EGF but in line with data shown above in Fig. 5c, the mitogenic effects of D2A and EGF were not additive (Fig. 7b). These results underline the prerequisite for the expression of uPAR at the cell surface. This idea is further supported by the fact that when added to increasing doses of D2A (0.1–106 pM), suPAR failed to rescue the proliferation of HEK-293 cells (Fig. 7c). Actually, suPAR does not appear to exert any effects on cell proliferation for either D2A alone or the mix D2A + suPAR led to similar levels of growth in HEK-293-uPAR cells (Fig. 7d). Thus, to be mitogenic D2A (like EGF) needs membrane-bound uPAR on the cell surface. This view is reinforced by the fact that D2A triggered the phosphorylation of EGFR in HEK-293-uPAR cells but not in HEK-293 cells devoid of uPAR (Fig. 1b). In HEK-293-uPAR cells, this effect of D2A was blocked by the EGFR tyrosine kinase inhibitor AG1478 (Fig. 1b). Therefore, D2A activates EGFR only when cells express uPAR (Fig. 1b). Taken together these results suggest that D2A promotes cell proliferation through interactions with the integrin αvβ3, which actually transactivates EGFR. Since uPAR expression is also required, it is reasonable to conceive that an uPAR-integrin αvβ3-EGFR macromolecular complex is responsible of the transduction of the mitogenic signal induced by D2A or EGF.
uPAR, integrin αvβ3 and EGFR form a functional signalling complex. a uPAR and integrin αvβ3 are involved in the transduction of the mitogenic signal promoted by D2A and EGF. Time-course experiment showing the effects of peptide D2A (100 pM) or EGF (5 ng/ml = 0.80 nM) in the absence or in the presence of various antibodies (each at 1 μg/ml): LM609, a blocking monoclonal anti-αvβ3; R3, a monoclonal anti-uPAR; SI369, a polyclonal anti-uPAR and an unrelated control antibody on proliferation of HT-29 cells. Data from three experiments are the mean ± SD. Number of cells cultured in serum-free media without D2A or EGF and in the absence of any antibody is referred to as 100% proliferation (control). The conditions 10% FCS, D2A, D2A + unrelated Ab, EGF and EGF + unrelated Ab are significantly different (p < 0.01) from control (0% FCS) as determined by ANOVA test. b The proliferation-promoting effects of D2A and EGF requires uPAR expression, however, D2A and EGF mitogenic effects are not additive. Proliferation of either HEK-293 or HEK-293-uPAR cells was stimulated with D2A (100 pM), EGF (1 ng/ml or 0.16 nM) or both mixed together as indicated. Data are the mean ± SD (n = 3) of cell numbers determined at day 4. Control corresponds to the number of cells cultured in serum-free media without D2A and EGF (100% proliferation). *p < 0.05 compared to control. c, d Cell proliferation in response to D2A challenge requires the expression of uPAR. Comparison of the effects of increasing doses of D2A in the presence or in the absence of suPAR on HEK-293 cells (devoid of uPAR) (c), and HEK-293-uPAR cells (expressing uPAR) (d). Cell numbers were determined at day 4. Number of cells cultured in serum-free media and in the absence of D2A and suPAR is considered to be 100% cell proliferation (control). Data shown as mean ± SD (n = 3), *p < 0.01 compared to control
D2A peptide has a specific three-dimensional structure
The 3D structure of D2A was investigated by NMR analysis. Assignment of proton spin systems for D2A peptide was obtained with the sequential methodology outlined by Wüthrich [49]. From the amide protons, TOCSY experiments allowed identification of alpha and beta protons of almost all of the amino acids. Residues with a long sidechain were identified by a combination of TOCSY and NOESY experiments. Individual spin systems were placed in the primary structure by identification of characteristic short-range NOE connectivities. From the analysis of D2A, NOESY spectra 32 sequential and 6 short- and medium-range unambiguous NOEs were obtained apart from the intraresidual ones. Sequential medium-strong NHi-NHi+1 NOEs were clearly identified for most of the amino acids from Gly4 to Arg13, while strong αCHi-NHi+1 effects were identified from Gln2 to Arg13, with only two interruptions due to the presence of Pro9 and to a cross-peak overlap. These effects were indicative of the absence of general helical structures. However, the presence of NHi–NHi+1 and medium-weak unambiguous βCHi-NHi+1 cross-peaks (Glu6-Gly7, Pro9-Lys10, Lys10-Asp11 and Asp12-Arg13) together with some unambiguous medium-range connectivities (Gln2α-Glu6NH, Glu6NH-Arg8NH, Arg8NH-Asp11NH, Gly7α-Lys10β, Arg8α-Asp11β, Lys10β-Asp12NH) were indicative of local structure and short-range order. No conformational heterogeneity was observed as far as the prolyl residue is concerned; in fact only the trans form was identified by the NHi-deltai+1 NOE cross-peaks.
Since the region spanning residues 4–11 appeared to be partially structured, the ten models compatible with the NMR constraints and the peptide in the extended conformation were superimposed pairwise on the average structure minimizing root mean square distances in the range 4–11. A stereo view is shown in Fig. 8 where the compatible models are represented as thin grey rod connecting the α backbone carbons (Cα), the average structure is represented in thick black rod and the extended peptide is represented as thick grey rod. These data show that D2A peptide presents a folded (average) 3D structure most likely relevant for its biological activity.
Model of the three-dimensional structure of the D2A peptide. The average 3D structure of D2A peptide, as obtained from NMR data, is shown in thick black rod while the extended conformation of the peptide is represented as thick grey rod. All the possible 3D structures of D2A compatible with NMR constraints are also displayed as thin grey rod. D2A peptide corresponds to sequence 130IQEGEEGRPKDDR142 of human uPAR
Discussion
In this report, we have studied the molecular mechanism that uPAR uses to regulate cell proliferation. We show that uPAR stimulates cell proliferation by forming a macromolecular complex with integrins and EGFR (ErbB1 or HER1) that we name the uPAR proliferasome. uPAR-initiated signal transactivates EGFR, which in its turn mediates uPAR-initiated mitogenic signal to the cell. Further investigations revealed that the mitogenic sequence of uPAR corresponds to the D2A motif present in its domain 2 [15]. NMR analysis showed that D2A synthetic peptide presents a well-defined 3D structure, which might explain its activity. In the X-ray structure of uPAR (PDB code 3BT1), the corresponding sequence forms a hairpin between the third and the forth strand in the second domain, GEEG (155–158 protein numbering) being in bend conformation. This peptide is as effective as EGF in promoting cell proliferation. Furthermore, uPAR is required for D2A- and EGF-induced EGFR phosphorylation and EGFR-dependent cell proliferation, which are blocked in the absence of uPAR expression or when uPAR activity is inhibited.
D2A sequence was previously reported to stimulate cell migration [15]. Herein, we show that this sequence also promotes cell proliferation. Differences between the effects on cell migration and proliferation suggest that the molecular mechanisms are distinct. Certainly, the most obvious difference resides in the two diverse optimal doses inducing cell migration (1 pM) and proliferation (100 pM) [15]. Furthermore, the kinetics of the two processes are counted in minutes for the former and in days for the latter.
Our present data evidence the co-involvement and the importance of uPAR, integrins and EGFR suggesting that a macromolecular complex is in fact responsible for the transduction of the cell proliferation signal induced by D2A and EGF. In agreement with the literature underlining the crucial role of macromolecular complexes involving uPAR and other receptors [18, 19, 30, 31, 41, 46, 48, 50,51,52], HKa, a ligand of uPAR, inhibits migration and invasion of cancer cell initiated by an uPAR-α5β1 integrin–EGFR complex [52]. Another ligand of uPAR, factor XII, reportedly induced endothelial cell proliferation and angiogenesis through an uPAR-β1 integrin–EGFR complex activating the MAP kinase pathway. The role of EGFR was not exactly determined [18]. However, it was previously reported that uPAR binds to α5β1 integrin that associates with and phosphorylates EGFR, which mediates uPAR-dependent signal [30]. An uncleavable mutant of uPAR was also reported to activate EGFR by forming an uPAR-α5β1 integrin–EGFR complex [48]. Moreover, pro-uPA regulates angiogenesis through an uPAR-β1 integrin–VEGFR2 complex [19].
The involvement of a macromolecular complex is strengthened by the fact that in the absence of uPAR, EGFR is not functional [38, 53]. HEK-293 cells do not express uPAR [54]; however, these cells express EGFR (ErbB1 or HER1) (~ 20,000 EGFRs/cell) [55]. Therefore, HEK-293 cells possess the required receptor to proliferate in response to EGF. Yet, in our hands, HEK-293 cells failed to proliferate when stimulated with D2A or EGF or with D2A + EGF, while their counterparts transfected to express uPAR grew normally. This no/yes proliferative response of HEK-293 and HEK-293-uPAR cells cannot be explained by an upregulation of EGFR when cells are transfected to express uPAR. Rather, the expression of uPAR and EGFR appears inversely correlated. Downregulation of uPAR enhances EGFR expression [56]. Conversely, EGFR overexpression suppresses uPAR expression [57]. This inversely correlated expression of uPAR and EGFR has also been proposed to be responsible of the development of EGFR resistance to tyrosine kinase inhibitors [56,57,58]. In addition, EGFR expression was similar in both uPAR−/− and uPAR+/+ mouse embryonic fibroblasts (MEFs); however, EGF was mitogenic only in uPAR+/+ MEFs demonstrating the necessary expression of uPAR for EGF-induced cell proliferation [38]. The specificity of the alteration of the proliferative response to EGF in the absence of uPAR is further supported by the fact that uPAR−/− MEFs grow faster in 10% FCS than their counterparts expressing uPAR [59]. Attempting to rescue the proliferation of HEK-293 cells, which do not express uPAR and do not produce uPA [54], with full-length suPAR failed because this form of suPAR binds very poorly to integrins due to an incorrect conformation [15]. Only uPA-bound suPAR associates with integrins [15]. In addition, suPAR did not further enhance either D2A- or EGF-promoted proliferation of HEK-293-uPAR. These data show that not only expression of uPAR but also its correct conformation are required for uPAR interaction with the integrins, and thus for initiating cell proliferation. Indeed, the conformation of uPAR is known to be crucial for the interactions with the integrins. Full-length uPAR has been reported to associate with the integrins but not D2D3-uPAR [30, 60]. Moreover, antibodies against uPAR blocked both D2A- and EGF-dependent cell proliferations. Interestingly, a blocking anti-EGFR antibody also inhibited D2A- and EGF-induced cell proliferation. These functional effects are closely related with the molecular activation state of EGFR. uPAR expression is required for D2A- and EGF-induced EGFR phosphorylation. Both D2A and EGF failed to phosphorylate EGFR when cells do not express uPAR, whereas D2A and EGF activated EGFR in cells expressing uPAR. Furthermore, D2A- and EGF-promoted cell growths as well as D2A- and EGF-induced EGFR phosphorylation were blocked by AG1478, a specific inhibitor of EGFR tyrosine kinase. Thus, our data imply that uPAR exerts a permissive effect on the tyrosine kinase activity and functions of EGFR. Our results are in line with previous reports suggesting a pivotal influence of uPAR on EGFR functioning, which depends on uPAR [31, 38, 53]. Furthermore, the phosphorylation of EGFR on Tyr845 and EGFR-dependent activation of Stat5b have also been shown to be dependent on uPAR [31, 38]. The fact that Iressa (ZD1839 or Gefitinib, a specific inhibitor of the EGFR tyrosine kinase activity) exerts its inhibitory effects on cell proliferation, migration and invasion in a uPAR-dependent manner further supports our conclusion [61].
In our model, the association of D2A with αvβ3 integrin triggers EGFR activation, which transmits the mitogenic signal to the cell (Fig. 9). However, the presence of uPAR on the cell membrane is prerequisite for D2A activity. When we discovered D2A, we hypothesized the existence of required interactions between domain 3 of uPAR and integrins [15], which were later reported [16, 17]. Therefore, to explain uPAR-integrin interactions and the signalling capacity of D2A, we suggest that the association of domain 3 of uPAR with the integrin stabilizes the uPAR–integrin complex while the connection between D2A and the integrins induces signalling (Fig. 9). The target of our D2A and GEEG peptides, the integrins, can transactivate EGFR [30, 62,63,64]. The fact that integrins actually transactivate EGFR is supported by two sets of data. Firstly, D2A peptide binds to integrins [15], and D2A-promoted cell proliferation is inhibited by LM609, a blocking anti-αvβ3 integrin antibody. Secondly, as shown by the requirement for uPAR expression, the induction of cell proliferation by D2A peptide necessitates other sequences of uPAR involved in creating contacts with the integrins [16, 17]. Furthermore, EGF-dependent stimulation of the MAP kinase pathway, i.e. ERK activation was observed only when integrins were occupied by ligand and aggregated [62], which can be induced by uPAR for this receptor is known to induce integrins clustering [41]. Yet, integrins activate many RTKs [65], thus we cannot exclude that other RTKs such as VEGFR2 might take the place of the EGFR in order to stimulate cell growth. For this reason, we suggest that uPAR–integrin–RTK represents the minimal configuration of the uPAR proliferasome. Targeting components of this macromolecular complex with blocking antibodies against either uPAR, αvβ3 integrin or EGFR, or with specific inhibitors AG1478 or PD98059 effectively inhibited uPAR- or EGFR-promoted cell growth. Therefore, these data indicate that blocking the signalling macromolecular complex represents an efficient strategy to inhibit biological processes such as cell proliferation and migration. This novel strategy may allow to control the activity of several valuable receptors and various signals. For example, targeting macromolecular complexes rather than receptors alone should let fewer alternatives to the cancer cells to counteract and escape by using other receptors/signalling pathways as substitute.
D2A promotes cell proliferation via the activation of EGFR. In cells constitutively expressing uPAR on the cell surface, the association of domain 3 of uPAR with the integrin stabilizes the uPAR–integrin complex. D2A binding to αvβ3 integrin of pre-formed uPAR–integrin complex initiates the formation of uPAR-αvβ3 integrin–EGFR complex leading to the dimerization and phosphorylation of EGFR, activation of ERKs and stimulation of cell proliferation
Both D2A and EGF stimulatory effects on cell proliferation are equivalent (see Figs. 5, 7), and both promoted a similar two-fold increase in the phosphorylation of EGFR (see Fig. 6). Furthermore, the mitogenic effects of D2A and EGF are not additive suggesting that both activates similar signalling pathways as shown in Fig. 6. Nevertheless, Western blotting analysis also revealed a subtle difference: D2A-induced phosphorylation of EGFR lasted longer, from 2 up to 30 min, than that promoted by EGF, which was only observed from 2 to 10 min. There are several possible explanations for this difference and all deserve further investigations and dedicated publications. Briefly, this difference may be due to (but not limited to): (1) D2A and EGF may induce different stoichiometry while forming the uPAR-αvβ3 integrin–EGFR complex. Indeed, it is known that dimerization and/or aggregation plays an important role in the functioning of uPAR, integrins and EGFR, thus upon D2A or EGF challenge the number of copy of each receptor present in the uPAR-αvβ3 integrin–EGFR complex may be different [1, 8,9,10, 26, 62, 66]. (2) There are numerous sites of phosphorylation within the EGFR cytoplasmic domain. Therefore, D2A and EGF may possibly induce EGFR phosphorylation on multiple but different sites. However, some sites may be also common as EGFR phosphorylation on Tyr845 was shown to be dependent on uPAR expression and/or activity, and required for uPA- and EGF-induced cell proliferation and Stat5b activation [31, 38]. (3) As shown here in Fig. 6, EGFR phosphorylation by either D2A or EGF exhibits different kinetics. Yet, the inactivation of EGFR also follows diverse kinetics. EGF-induced EGFR phosphorylation decreased abruptly after peaking at 10 min, whereas phosphorylation of EGFR promoted by D2A reached its maximum at 2 min and then declined slowly, only disappearing after 60 min. Since endocytosis is the major mechanism responsible of the inactivation of EGFR through dephosphorylation or receptor degradation, it is reasonable to conceive that EGFR internalization, thus EGFR inactivation, after D2A or EGF stimulation is not identical. (4) D2A and EGF challenges lead to distinct recruitment of intracellular adaptors and signalling components, which may influence EGFR phosphorylation. For example, the Src family of kinases (which is composed of nine members) is involved in EGFR- and uPAR-dependent signalling [41, 67]. Moreover, c-Src was reported to phosphorylate EGFR on a site, Tyr845, which is dependent on uPAR [31, 38, 68]. Thus, a diverse regulation of the Src family of kinases by D2A and EGF may produce the difference in the phosphorylation of EGFR reported above (see Fig. 6).
Nevertheless, phosphorylation of EGFR by D2A and EGF triggers the transient activation of ERKs (see Fig. 6). This result confirms previous studies showing that uPAR activates the MAP kinase signalling pathway [30, 41, 46, 69]. This result is also in line with the literature stating that cell proliferation is commonly produced by transient activation of ERK 1/2, whereas cell differentiation is usually the consequence of sustained activation of the same [70]. EGF binding to EGFR is known to induce a transient activation of ERKs, which is involved in the stimulation of cell proliferation. In this study, D2A also promoted a transient activation of ERKs that correlated with its mitogenic effects. Thus, these data agree with the fact that D2A is a mitogen (like EGF) and with the general rule: transient activation of ERKs leads to cell proliferation. Still, the slight difference in ERK activation by either D2A or EGF may be the consequence of the points 1–4 discussed above.
uPAR is highly expressed in numerous if not all malignant cells and a tumour marker of various types of cancer [71]. uPAR is involved in many processes regulating tumour development such as cell survival, tumour dormancy, angiogenesis, epithelial–mesenchymal transition, tumour invasion and metastasis [18, 19, 72,73,74,75]. However, as shown herein by permitting and maintaining EGFR activity, uPAR has now potentially acquired the capacity to regulate neo-plastic transformation as the EGFR gene is a proto-oncogene [58, 63]. Thus, uPAR may be involved in very early steps of tumourigenesis [59, 76]. Moreover, EGFR-dependent transformation occurs through the RAS/RAF/MEK/ERK pathway [77], which is also regulated by uPAR [46]. The connection between uPAR and EGFR is tight as uPAR-induced ERK activation is EGFR-dependent when cells express EGFR [30, 69].
Still, evidencing a clear relationship in cancer development between uPAR, integrin, EGFR and their ligands is far from being obvious for all players are connected and regulate each other [8]. EGFR−/− mice die shortly after birth showing defects in epithelial tissues, thereby primarily affecting cells in which EGFR is overexpressed in tumours, i.e. epithelial and glial cells [78, 79]. In contrast, EGF−/− mice are viable and do not exhibit any obvious phenotype [80]. Similarly, uPAR knockout mice are viable displaying no observable phenotype [81, 82]. However, metastasis was shown to be both dependent and independent of uPAR [83, 84]. In addition, uPAR expressed by the environment of the tumour also exerts an important role as evidenced using a syngeneic ovarian cancer model in which tumour growth and progression were reduced, whereas survival time was extended in uPAR−/− mice [85]. A similar conclusion was drawn for EGFR expression by stroma in colorectal cancer [86]. If there are numerous articles of the literature leaving no doubt on the important role of uPAR in cancer [1,2,3,4,5,6, 8,9,10, 83, 85], the generation of uPAR−/− mice suggested that uPAR may play a less important role than expected and/or a redundant role [87, 88]. Nonetheless, uPAR null mice display delayed wound healing due to impaired keratinocyte migration and proliferation [53]. Importantly, uPAR−/− keratinocytes failed to proliferate in response to EGF; a response that was rescued by expressing uPAR [53]. Thus, these results are in perfect agreement with our data. Inhibition of uPA, the main ligand of uPAR, blocks metastasis [89]. Furthermore, like uPAR, elevated uPA expression is associated with many cancer types [90]. uPA and uPAR exert overlapping functions for two main reasons: 1) uPA interacts with other components such as integrins, PAI-1 or LRP-1 [4, 28, 29]; 2) uPAR is a multi-ligand receptor binding to factor XII another serine protease, and interacting with other receptors, i.e. integrins, RTKs or GPCRs [1,2,3,4,5,6]. Inhibiting uPA activity decreases tumour growth and metastasis but increases apoptosis [91,92,93]. In both uPA−/− and uPAR−/− mice, tumour volume is reduced but diminution is more pronounced in uPA−/− mice with significantly less macrophage infiltration [94]. In uPA−/− mice, tumoural cells do not progress into melanoma showing that uPA participates in tumourigenesis [95]. Here again, uPA released by stroma surrounding the tumour plays an important role [92]. In another model of transgenic breast cancer mice, uPA deficiency did not alter the growth of the primary tumour but reduced metastasis [96]. On the other hand, uPA null mice were shown to exhibit more colon adenoma than their wild-type counterparts [97]. In addition, uPAR-dependent sustained ERK activation and concomitant apparition of cancer stem cell-like properties appear to be vitronectin (VN)-dependent and uPA-independent [98, 99]. VN is both a ligand of integrins and uPAR [1,2,3,4,5,6]. However, VN- and uPA-dependent signalling are different [46], and VN induces cell migration via integrins not uPAR [15, 100]. Although an important role has been attributed to VN-uPAR interaction [101], stem cell differentiation and tumour formation occur via a VN/αvβ3 integrin-dependent mechanism [102]. VN knockout mice are viable but display delayed blood vessel growth and wound healing, reduced macrophage infiltration and leaky blood vessels [103]. The role of VN in angiogenesis and tumour formation is further supported by conditional αv integrins knockout mice, which showed that αv is essential for vascular remodelling [104]. In β3 knockout mice, the formation of larger tumours goes along with pathological angiogenesis [105, 106]. Another mouse model revealed that DiYF (two Y to F mutations) β3 knock-in mice with defective β3 phosphorylation have highly impaired angiogenesis due to altered cell adhesion and spreading, cell migration and formation of capillary tubes [107]. β3 integrins knockout mice also exhibit vascular leak [108], but reduced osteolytic bone metastasis [109]. These mice have poorly functioning osteoclasts and increased bone mass [109]. Osteoclasts are also affected in EGFR−/− mice, which show delayed ossification due to impaired recruitment of both osteoclasts and osteoblasts, and decreased osteoblast proliferation [110, 111]. Interestingly, bone homeostasis is likewise disturbed in uPAR−/− mice [112]. Bone mass is augmented with altered osteoclasts and highly proliferative osteoblasts [112].
In this report, we also investigated the correlation between the structure of uPAR and its function as signalling receptor promoting cell growth. We identified the mitogenic epitope of uPAR as the D2A sequence present in its domain 2, and we precisely demonstrated that its mitogenic activity is associated with the GEEG motif harboured by this D2A sequence. The growth-promoting activity of D2A is comparable to that of EGF promoting the phosphorylation of EGFR, and subsequently triggering similar intracellular signalling as shown by ERK phosphorylation. Polyclonal antibodies raised against the D2A epitope inhibited both D2A and EGF-induced cell proliferation. The reverse relationship is also true as a blocking antibody against EGFR blocked D2A- and EGF-promoted cell growth. Furthermore, the EGFR tyrosine kinase inhibitor, AG 1478, and MEK inhibitor, PD98059, equally blocked uPA- and EGF-induced effects on cell proliferation and EGFR phosphorylation. Lastly, both D2A and EGF failed to induce proliferation of NR6 cells expressing EGFR-K721A, a kinase-defective mutant of EGFR, whereas the same promoted the growth of NR6 cells expressing wild-type EGFR.
The data showing that D2A has a particular 3D structure are especially encouraging. Most of the time, peptides have no special 3D structure and assume the correct conformation only when they bind to their corresponding site. D2A peptide stands apart and has a specific 3D structure. By using this structure template, it will be possible to design new agonistic and antagonistic molecules of integrins for the treatment of diseases where cell proliferation and migration play a leading role including inflammation, vascular diseases and cancers.
Conclusion
We have shown in this study that uPAR promotes cell proliferation through transactivation of EGFR, which mediates uPAR-induced cell proliferation. The region responsible of the growth-promoting activity of uPAR is sequence D2A located in domain 2. The derived synthetic D2A peptide matches the effects of EGF on cell proliferation and possesses a particular 3D structure. Moreover, uPAR expression and activity are required for EGFR functioning. EGF failed to induce cell proliferation and EGFR phosphorylation when uPAR is not expressed or its activity blocked.
Abbreviations
- 3D:
-
Three-dimensional
- EGF:
-
Epidermal growth factor
- EGFR:
-
Epidermal growth factor receptor
- ERK:
-
Extracellular signal-regulated kinase
- GPCR:
-
G protein-coupled receptor
- MAP:
-
Mitogen-activated protein
- MEK:
-
Mitogen-activated protein kinase kinase
- RTK:
-
Receptor tyrosine kinase
- uPA:
-
Urokinase
- uPAR:
-
Urokinase receptor
References
Blasi F, Sidenius N (2009) The urokinase receptor: focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett 584:1923–1930. https://doi.org/10.1016/j.febslet.2009.12.039
Archinti M, Britto M, Eden G, Furlan F, Murphy R, Degryse B (2011) The urokinase receptor in the central nervous system. CNS Neurol Disord Drug Targets 10:271–294
Del Rosso M, Margheri F, Serratì S, Chillà A, Laurenzana A, Fibbi G (2011) The urokinase receptor system, a key regulator at the intersection between inflammation, immunity, and coagulation. Curr Pharm Des 17:1924–1943
Eden G, Archinti M, Furlan F, Murphy R, Degryse B (2011) The urokinase receptor interactome. Curr Pharm Des 17:1874–1889
Montuori N, Ragno P (2014) Role of uPA/uPAR in the modulation of angiogenesis. Chem Immunol Allergy 99:105–122
Gonias SL, Hu J (2015) Urokinase receptor and resistance to targeted anticancer agents. Front Pharmacol 6:154. https://doi.org/10.3389/fphar.2015.00154
Degryse B, Britto M, Shan CX, Wallace RG, Rochfort KD, Cummins PM, Meade G, Murphy RP (2017) Moesin and merlin regulate urokinase receptor-dependent endothelial cell migration, adhesion and angiogenesis. Int J Biochem Cell Biol 88:14–22. https://doi.org/10.1016/j.biocel.2017.04.012
Degryse B (2008) The urokinase receptor (uPAR) and integrins constitute a cell migration signalosome. In: Edwards D, Hoyer-Hansen G, Blasi F, Sloane BF (eds) The cancer degradome—proteases and cancer biology. Springer, New York, pp 451–474
Archinti M, Eden G, Murphy R, Degryse B (2010) Follow the leader: when the urokinase receptor coordinates cell adhesion, motility and proliferation with cytoskeleton organization. In: Lansing S, Rousseau T (eds) Cytoskeleton: cell movement, cytokinesis and organelles organization. Nova Publishers, New York, pp 1–36
Eden G, Archinti M, Furlan F, Fitzpatrick P, Murphy R, Degryse B (2011) The cell migration signalosome. In: Williams TC (ed) Chemotaxis: types, clinical significance, and mathematical models. Nova Publishers, New York, pp 1–52
Llinas P, Le Du MH, Gårdsvoll H, Danø K, Ploug M, Gilquin B, Stura EA, Ménez A (2005) Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide. EMBO J 24:1655–1663
Huang M, Mazar AP, Parry G, Higazi AA, Kuo A, Cines DB (2005) Crystallization of soluble urokinase receptor (suPAR) in complex with urokinase amino-terminal fragment (1–143). Acta Crystallogr D Biol Crystallogr 61:697–700
Barinka C, Parry G, Callahan J, Shaw DE, Kuo A, Bdeir K, Cines DB, Mazar A, Lubkowski J (2006) Structural basis of interaction between urokinase-type plasminogen activator and its receptor. J Mol Biol 363:482–495
Huai Q, Zhou A, Lin L, Mazar AP, Parry GC, Callahan J, Shaw DE, Furie B, Furie BC, Huang M (2008) Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes. Nat Struct Mol Biol 15:422–423. https://doi.org/10.1038/nsmb.1404
Degryse B, Resnati M, Czekay RP, Loskutoff DJ, Blasi F (2005) Domain 2 of the urokinase receptor contains an integrin-interacting epitope with intrinsic signaling activity: generation of a new integrin inhibitor. J Biol Chem 280:24792–24803
Chaurasia P, Aguirre-Ghiso JA, Liang OD, Gardsvoll H, Ploug M, Ossowski L (2006) A region in urokinase plasminogen receptor domain III controlling a functional association with alpha5beta1 integrin and tumor growth. J Biol Chem 281:14852–14863
Wei Y, Tang CH, Kim Y, Robillard L, Zhang F, Kugler MC, Chapman HA (2007) Urokinase receptors are required for alpha5 beta1 integrin-mediated signaling in tumor cells. J Biol Chem 282:3929–3939
LaRusch GA, Mahdi F, Shariat-Madar Z, Adams G, Sitrin RG, Zhang WM, McCrae KR, Schmaier AH (2010) Factor XII stimulates ERK1/2 and Akt through uPAR, integrins, and the EGFR to initiate angiogenesis. Blood 115:5111–5120. https://doi.org/10.1182/blood-2009-08-236430
Larusch GA, Merkulova A, Mahdi F, Shariat-Madar Z, Sitrin RG, Cines DB, Schmaier AH (2013) Domain 2 of uPAR regulates single-chain urokinase-mediated angiogenesis through β1-integrin and VEGFR2. Am J Physiol Heart Circ Physiol 305:H305–H320. https://doi.org/10.1152/ajpheart.00110.2013
Kirchheimer JC, Wojta J, Christ G, Binder BR (1987) Proliferation of a human epidermal tumor cell line stimulated by urokinase. FASEB J 1:125–128
Rabbani SA, Desjardins J, Bell AW, Banville D, Mazar A, Henkin J, Goltzman D (1990) An amino-terminal fragment of urokinase isolated from a prostate cancer cell line (PC-3) is mitogenic for osteoblast-like cells. Biochem Biophys Res Commun 173:1058–1064
He CJ, Rebibou JM, Peraldi MN, Meulders Q, Rondeau E (1991) Growth factor-like effect of urokinase type plasminogen activator in human renal cells. Biochem Biophys Res Commun 176:1408–1416
Fischer K, Lutz V, Wilhelm O, Schmitt M, Graeff H, Heiss P, Nishiguchi T, Harbeck N, Kessler H, Luther T, Magdolen V, Reuning U (1998) Urokinase induces proliferation of human ovarian cancer cells: characterization of structural elements required for growth factor function. FEBS Lett 438:101–105
Naldini L, Tamagnone L, Vigna E, Sachs M, Hartmann G, Birchmeir W, Daikuhara Y, Tsubouchi H, Blasi F, Comoglio PM (1992) Extracellular proteolytic cleavage by urokinase is required for activation of hepatocyte growth factor/scatter factor. EMBO J 11:4825–4833
Odekon LE, Blasi F, Rifkin DB (1994) Requirement for receptor-bound urokinase in plasmin-dependent cellular conversion of latent TGF-beta to TGF-beta. J Cell Physiol 158:398–407
Ossowski L, Aguirre-Ghiso JA (2000) Urokinase receptor and integrin partnership: coordination of signaling for cell adhesion, migration and growth. Curr Opin Cell Biol 12:613–620
Schwartz MA, Assoian RK (2001) Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J Cell Sci 114:2553–2560
Tarui T, Akakura N, Majumdar M, Andronicos N, Takagi J, Mazar AP, Bdeir K, Kuo A, Yarovoi SV, Cines DB, Takada Y (2006) Direct interaction of the kringle domain of urokinase-type plasminogen activator (uPA) and integrin αvβ3 induces signal transduction and enhances plasminogen activation. Thromb Haemost 95:524–534
Degryse B, Fernandez-Recio J, Citro V, Blasi F, Cubellis MV (2008) In silico docking of urokinase plasminogen activator and integrins. BMC Bioinform 9(Suppl 2):S8. https://doi.org/10.1186/1471-2105-9-S2-S8
Liu D, Aguirre Ghiso J, Estrada Y, Ossowski L (2002) EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell 1:445–457
Jo M, Thomas KS, Marozkina N, Amin TJ, Silva CM, Parsons SJ, Gonias SL (2005) Dynamic assembly of the urokinase-type plasminogen activator signaling receptor complex determines the mitogenic activity of urokinase-type plasminogen activator. J Biol Chem 280:17449–17457
Kiyan J, Kiyan R, Haller H, Dumler I (2005) Urokinase-induced signaling in human vascular smooth muscle cells is mediated by PDGFR-beta. EMBO J 24:787–1797
Schmeidler-Sapiro KT, Ratnoff OD, Gordon EM (1991) Mitogenic effects of coagulation factor XII and factor XIIa on HepG2 cells. Proc Natl Acad Sci USA 88:4382–4385
Gordon EM, Venkatesan N, Salazar R, Tang H, Schmeidler-Sapiro K, Buckley S, Warburton D, Hall FL (1996) Factor XII-induced mitogenesis is mediated via a distinct signal transduction pathway that activates a mitogen-activated protein kinase. Proc Natl Acad Sci USA 93:2174–2179
Reuning U, Bang NU (1992) Regulation of the urokinase-type plasminogen activator receptor on vascular smooth muscle cells is under the control of thrombin and other mitogens. Arterioscler Thromb 12:1161–1170
Reuning U, Little SP, Dixon EP, Bang NU (1994) Effect of thrombin, the thrombin receptor activation peptide, and other mitogens on vascular smooth muscle cell urokinase receptor mRNA levels. Blood 84:3700–3708
Ueshima S, Fukao H, Okada K, Matsuo O (2004) Growth inhibition of vascular smooth muscle cells derived from urokinase receptor (u-PAR)-deficient mice in the presence of carcinoma cells. Thromb Res 113:41–49
Jo M, Thomas KS, Takimoto S, Gaultier A, Hsieh EH, Lester RD, Gonias SL (2007) Urokinase receptor primes cells to proliferate in response to epidermal growth factor. Oncogene 26:2585–2594
Rønne E, Behrendt N, Ellis V, Ploug M, Danø K, Høyer-Hansen G (1991) Cell-induced potentiation of the plasminogen activation system is abolished by a monoclonal antibody that recognizes the NH2-terminal domain of the urokinase receptor. FEBS Lett 288:233–236
Resnati M, Pallavicini I, Daverio R, Sidenius N, Bonini P, Blasi F (2006) Specific immunofluorimetric assay detecting the chemotactic epitope of the urokinase receptor (uPAR). J Immunol Methods 308:192–202
Degryse B, Resnati M, Rabbani SA, Villa A, Fazioli F, Blasi F (1999) Src-dependence and pertussis-toxin sensitivity of urokinase receptor-dependent chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. Blood 94:649–662
Masucci MT, Pedersen N, Blasi F (1991) A soluble, ligand binding mutant of the human urokinase plasminogen activator receptor. J Biol Chem 266:8655–8658
Pruss RM, Herschman HR (1977) Variants of 3T3 cells lacking mitogenic response to epidermal growth factor. Proc Natl Acad Sci USA 74:3918–3921
Honegger AM, Dull TJ, Felder S, Van Obberghen E, Bellot F, Szapary D, Schmidt A, Ullrich A, Schlessinger J (1987) Point mutation at the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell 51:199–209
Honegger AM, Szapary D, Schmidt A, Lyall R, Van Obberghen E, Dull TJ, Ullrich A, Schlessinger J (1987) A mutant epidermal growth factor receptor with defective protein tyrosine kinase is unable to stimulate proto-oncogene expression and DNA synthesis. Mol Cell Biol 7:4568–4571
Degryse B, Orlando S, Resnati M, Rabbani SA, Blasi F (2001) Urokinase/urokinase receptor and vitronectin/αvβ3 integrin induce chemotaxis and cytoskeleton reorganization through different signaling pathways. Oncogene 20:2032–2043
Moro L, Dolce L, Cabodi S, Bergatto E, Boeri Erba E, Smeriglio M, Turco E, Retta SF, Giuffrida MG, Venturino M, Godovac-Zimmermann J, Conti A, Schaefer E, Beguinot L, Tacchetti C, Gaggini P, Silengo L, Tarone G, Defilippi P (2002) Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and p130Cas and leads to phosphorylation of specific EGF receptor tyrosines. J Biol Chem 277:9405–9414
Mazzieri R, D’Alessio S, Kenmoe RK, Ossowski L, Blasi F (2006) An uncleavable uPAR mutant allows dissection of signaling pathways in uPA-dependent cell migration. Mol Biol Cell 17:367–378
Wüthrich K (1986) NMR with proteins and nucleic acids. Europhys News 17:11–13
Bohuslav J, Horejsí V, Hansmann C, Stöckl J, Weidle UH, Majdic O, Bartke I, Knapp W, Stockinger H (1995) Urokinase plasminogen activator receptor, beta 2-integrins, and Src-kinases within a single receptor complex of human monocytes. J Exp Med 181:1381–1390
Simon DI, Rao NK, Xu H, Wei Y, Majdic O, Rønne E, Kobzik L, Chapman HA (1996) Mac-1 (CD11b/CD18) and the urokinase receptor (CD87) form a functional unit on monocytic cells. Blood 88:3185–3194
Liu Y, Pixley R, Fusaro M, Godoy G, Kim E, Bromberg ME, Colman RW (2009) Cleaved high-molecular-weight kininogen and its domain 5 inhibit migration and invasion of human prostate cancer cells through the epidermal growth factor receptor pathway. Oncogene 28:2756–2765. https://doi.org/10.1038/onc.2009.132
D’Alessio S, Gerasi L, Blasi F (2008) uPAR-deficient mouse keratinocytes fail to produce EGFR-dependent laminin-5, affecting migration in vivo and in vitro. J Cell Sci 121:3922–3932. https://doi.org/10.1242/jcs.037549
Wei Y, Waltz DA, Rao N, Drummond RJ, Rosenberg S, Chapman HA (1994) Identification of the urokinase receptor as an adhesion receptor for vitronectin. J Biol Chem 269:32380–32388
Carter RE, Sorkin A (1998) Endocytosis of functional epidermal growth factor receptor-green fluorescent protein chimera. J Biol Chem 273:35000–35007
Abu-Ali S, Fotovati A, Shirasuna K (2008) Tyrosine-kinase inhibition results in EGFR clustering at focal adhesions and consequent exocytosis in uPAR down-regulated cells of head and neck cancers. Mol Cancer 7:47. https://doi.org/10.1186/1476-4598-7-47
Hu J, Jo M, Cavenee WK, Furnari F, VandenBerg SR, Gonias SL (2011) Crosstalk between the urokinase-type plasminogen activator receptor and EGF receptor variant III supports survival and growth of glioblastoma cells. Proc Natl Acad Sci USA 108:15984–15989. https://doi.org/10.1073/pnas.1113416108
Wykosky J, Hu J, Gomez GG, Taylor T, Villa GR, Pizzo D, VandenBerg SR, Thorne AH, Chen CC, Mischel PS, Gonias SL, Cavenee WK, Furnari FB (2015) A urokinase receptor-Bim signaling axis emerges during EGFR inhibitor resistance in mutant EGFR glioblastoma. Cancer Res 75:394–404. https://doi.org/10.1158/0008-5472.CAN-14-2004
Mazzieri R, Furlan F, D’Alessio S, Zonari E, Talotta F, Verde P, Blasi F (2007) A direct link between expression of urokinase plasminogen activator receptor, growth rate and oncogenic transformation in mouse embryonic fibroblasts. Oncogene 26:725–732
Montuori N, Rossi G, Ragno P (1999) Cleavage of urokinase receptor regulates its interaction with integrins in thyroid cells. FEBS Lett 460:32–36
Lee EJ, Whang JH, Jeon NK, Kim J (2007) The epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 (Iressa) suppresses proliferation and invasion of human oral squamous carcinoma cells via p53 independent and MMP, uPAR dependent mechanism. Ann N Y Acad Sci 1095:113–128
Miyamoto S, Teramoto H, Gutkind JS, Yamada KM (1996) Integrins can collaborate with growth factors for phosphorylation of receptor tyrosine kinases and MAP kinase activation: roles of integrin aggregation and occupancy of receptors. J Cell Biol 135:1633–1642
Yu X, Miyamoto S, Mekada E (2000) Integrin alpha 2 beta 1-dependent EGF receptor activation at cell-cell contact sites. J Cell Sci 113:2139–2147
Kuwada SK, Li X (2000) Integrin alpha5/beta1 mediates fibronectin-dependent epithelial cell proliferation through epidermal growth factor receptor activation. Mol Biol Cell 11:2485–2496
Streuli CH, Akhtar N (2009) Signal co-operation between integrins and other receptor systems. Biochem J 418:491–506. https://doi.org/10.1042/BJ20081948
Schlessinger J (2002) Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110:669–672
Irwin ME, Bohin N, Boerner JL (2011) Src family kinases mediate epidermal growth factor receptor signaling from lipid rafts in breast cancer cells. Cancer Biol Ther 12:718–726. https://doi.org/10.4161/cbt.12.8.16907
Wu W, Graves LM, Gill GN, Parsons SJ, Samet JM (2002) Src-dependent phosphorylation of the epidermal growth factor receptor on tyrosine 845 is required for zinc-induced Ras activation. J Biol Chem 277:24252–24257
Jo M, Thomas KS, O’Donnell DM, Gonias SL (2003) Epidermal growth factor receptor-dependent and -independent cell-signaling pathways originating from the urokinase receptor. J Biol Chem 278:1642–1646
Stork PJ (2002) ERK signaling: duration, duration, duration. Cell Cycle 1:315–317
Boonstra MC, Verspaget HW, Ganesh S, Kubben FJ, Vahrmeijer AL, van de Velde CJ, Kuppen PJ, Quax PH, Sier CF (2011) Clinical applications of the urokinase receptor (uPAR) for cancer patients. Curr Pharm Des 17:1890–1910
Yu W, Kim J, Ossowski L (1997) Reduction in surface urokinase receptor forces malignant cells into a protracted state of dormancy. J Cell Biol 137:767–777
Alfano D, Iaccarino I, Stoppelli MP (2006) Urokinase signaling through its receptor protects against anoikis by increasing BCL-xL expression levels. J Biol Chem 30:17758–17767
Tkachuk N, Kiyan J, Tkachuk S, Kiyan R, Shushakova N, Haller H, Dumler I (2008) Urokinase induces survival or pro-apoptotic signals in human mesangial cells depending on the apoptotic stimulus. Biochem J 415:265–273. https://doi.org/10.1042/BJ20071652
Kook YH, Adamski J, Zelent A, Ossowski L (1994) The effect of antisense inhibition of urokinase receptor in human squamous cell carcinoma on malignancy. EMBO J 13:3983–3991
Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D, Russell R, DePinho RA, Lenz J, van Lohuizen M (2002) Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet 32:160–165
Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C, Regl G, Kroismayr R, Moriggl R, Sibilia M, Aberger F (2009) Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res 69:1284–1292. https://doi.org/10.1158/0008-5472.CAN-08-2331
Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R (1995) Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376:337–341
Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M (2007) The epidermal growth factor receptor: from development to tumorigenesis. Differentiation 75:770–787
Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC (1999) Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development 126:2739–2750
Bugge TH, Suh TT, Flick MJ, Daugherty CC, Rømer J, Solberg H, Ellis V, Danø K, Degen JL (1995) The receptor for urokinase-type plasminogen activator is not essential for mouse development or fertility. J Biol Chem 270:16886–16894
Dewerchin M, Nuffelen AV, Wallays G, Bouché A, Moons L, Carmeliet P, Mulligan RC, Collen D (1996) Generation and characterization of urokinase receptor-deficient mice. J Clin Investig 97:870–878
Jo M, Takimoto S, Montel V, Gonias SL (2009) The urokinase receptor promotes cancer metastasis independently of urokinase-type plasminogen activator in mice. Am J Pathol 175:190–200. https://doi.org/10.2353/ajpath.2009.081053
Almholt K, Lærum OD, Nielsen BS, Lund IK, Lund LR, Rømer J, Jögi A (2015) Spontaneous lung and lymph node metastasis in transgenic breast cancer is independent of the urokinase receptor uPAR. Clin Exp Metastasis 32:543–554. https://doi.org/10.1007/s10585-015-9726-1
Al-Hassan NN, Behzadian A, Caldwell R, Ivanova VS, Syed V, Motamed K, Said NA (2012) Differential roles of uPAR in peritoneal ovarian carcinomatosis. Neoplasia 14:259–270
Srivatsa S, Paul MC, Cardone C, Holcmann M, Amberg N, Pathria P, Diamanti MA, Linder M, Timelthaler G, Dienes HP, Kenner L, Wrba F, Prager GW, Rose-John S, Eferl R, Liguori G, Botti G, Martinelli E, Greten FR, Ciardiello F, Sibilia M (2017) EGFR in tumor-associated myeloid cells promotes development of colorectal cancer in mice and associates with outcomes of patients. Gastroenterology 153(178–190):e10. https://doi.org/10.1053/j.gastro.2017.03.053
Almholt K, Green KA, Juncker-Jensen A, Nielsen BS, Lund LR, Rømer J (2007) Extracellular proteolysis in transgenic mouse models of breast cancer. J Mammary Gland Biol Neoplasia 12:83–97
Carmeliet P, Moons L, Dewerchin M, Rosenberg S, Herbert JM, Lupu F, Collen D (1998) Receptor-independent role of urokinase-type plasminogen activator in pericellular plasmin and matrix metalloproteinase proteolysis during vascular wound healing in mice. J Cell Biol 140:233–245
Ossowski L, Reich E (1983) Antibodies to plasminogen activator inhibit human tumor metastasis. Cell 35:611–619
Duffy MJ (2004) The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des 10:39–49
Gutierrez LS, Schulman A, Brito-Robinson T, Noria F, Ploplis VA, Castellino FJ (2000) Tumor development is retarded in mice lacking the gene for urokinase-type plasminogen activator or its inhibitor, plasminogen activator inhibitor-1. Cancer Res 60:5839–5847
Frandsen TL, Holst-Hansen C, Nielsen BS, Christensen IJ, Nyengaard JR, Carmeliet P, Brünner N (2001) Direct evidence of the importance of stromal urokinase plasminogen activator (uPA) in the growth of an experimental human breast cancer using a combined uPA gene-disrupted and immunodeficient xenograft model. Cancer Res 61:532–537
Henneke I, Greschus S, Savai R, Korfei M, Markart P, Mahavadi P, Schermuly RT, Wygrecka M, Stürzebecher J, Seeger W, Günther A, Ruppert C (2010) Inhibition of urokinase activity reduces primary tumor growth and metastasis formation in a murine lung carcinoma model. Am J Respir Crit Care Med 181:611–619. https://doi.org/10.1164/rccm.200903-0342OC
Zhang J, Sud S, Mizutani K, Gyetko MR, Pienta KJ (2011) Activation of urokinase plasminogen activator and its receptor axis is essential for macrophage infiltration in a prostate cancer mouse model. Neoplasia 13:23–30
Shapiro RL, Duquette JG, Roses DF, Nunes I, Harris MN, Kamino H, Wilson EL, Rifkin DB (1996) Induction of primary cutaneous melanocytic neoplasms in urokinase-type plasminogen activator (uPA)-deficient and wild-type mice: cellular blue nevi invade but do not progress to malignant melanoma in uPA-deficient animals. Cancer Res 56:3597–3604
Almholt K, Lund LR, Rygaard J, Nielsen BS, Danø K, Rømer J, Johnsen M (2005) Reduced metastasis of transgenic mammary cancer in urokinase-deficient mice. Int J Cancer 113:525–532
Karamanavi E, Angelopoulou K, Lavrentiadou S, Tsingotjidou A, Abas Z, Taitzoglou I, Vlemmas I, Erdman SE, Poutahidis T (2014) Urokinase-type plasminogen activator deficiency promotes neoplasmatogenesis in the colon of mice. Transl Oncol 7(174–187):e5. https://doi.org/10.1016/j.tranon.2014.02.002
Eastman BM, Jo M, Webb DL, Takimoto S, Gonias SL (2012) A transformation in the mechanism by which the urokinase receptor signals provides a selection advantage for estrogen receptor-expressing breast cancer cells in the absence of estrogen. Cell Signal 24:1847–1855. https://doi.org/10.1016/j.cellsig.2012.05.011
Jo M, Eastman BM, Webb DL, Stoletov K, Klemke R, Gonias SL (2010) Cell signaling by urokinase-type plasminogen activator receptor induces stem cell-like properties in breast cancer cells. Cancer Res 70:8948–8958. https://doi.org/10.1158/0008-5472.CAN-10-1936
Bartsch JE, Staren ED, Appert HE (2003) Adhesion and migration of extracellular matrix-stimulated breast cancer. J Surg Res 110:287–294
Madsen CD, Ferraris GM, Andolfo A, Cunningham O, Sidenius N (2007) uPAR-induced cell adhesion and migration: vitronectin provides the key. J Cell Biol 177:927–939
Hurt EM, Chan K, Serrat MA, Thomas SB, Veenstra TD, Farrar WL (2010) Identification of vitronectin as an extrinsic inducer of cancer stem cell differentiation and tumor formation. Stem Cells 28:390–398. https://doi.org/10.1002/stem.271
Leavesley DI, Kashyap AS, Croll T, Sivaramakrishnan M, Shokoohmand A, Hollier BG, Upton Z (2013) Vitronectin—master controller or micromanager? IUBMB Life 65:807–818. https://doi.org/10.1002/iub.1203
Carmeliet P (2002) Integrin indecision. Nat Med 8:14–16
Reynolds LE, Wyder L, Lively JC, Taverna D, Robinson SD, Huang X, Sheppard D, Hynes RO, Hodivala-Dilke KM (2002) Enhanced pathological angiogenesis in mice lacking beta3 integrin or beta3 and beta5 integrins. Nat Med 8:27–34
Taverna D, Moher H, Crowley D, Borsig L, Varki A, Hynes RO (2004) Increased primary tumor growth in mice null for beta3- or beta3/beta5-integrins or selectins. Proc Natl Acad Sci USA 101:763–768
Mahabeleshwar GH, Feng W, Phillips DR, Byzova TV (2006) Integrin signaling is critical for pathological angiogenesis. J Exp Med 203:2495–2507
Su G, Atakilit A, Li JT, Wu N, Bhattacharya M, Zhu J, Shieh JE, Li E, Chen R, Sun S, Su CP, Sheppard D (2012) Absence of integrin αvβ3 enhances vascular leak in mice by inhibiting endothelial cortical actin formation. Am J Respir Crit Care Med 185:58–66. https://doi.org/10.1164/rccm.201108-1381OC
Bakewell SJ, Nestor P, Prasad S, Tomasson MH, Dowland N, Mehrotra M, Scarborough R, Kanter J, Abe K, Phillips D, Weilbaecher KN (2003) Platelet and osteoclast beta3 integrins are critical for bone metastasis. Proc Natl Acad Sci USA 100:14205–14210
Wang K, Yamamoto H, Chin JR, Werb Z, Vu TH (2004) Epidermal growth factor receptor-deficient mice have delayed primary endochondral ossification because of defective osteoclast recruitment. J Biol Chem 279:53848–53856
Sibilia M, Wagner B, Hoebertz A, Elliott C, Marino S, Jochum W, Wagner EF (2003) Mice humanised for the EGF receptor display hypomorphic phenotypes in skin, bone and heart. Development 130:4515–4525
Furlan F, Galbiati C, Jorgensen NR, Jensen JE, Mrak E, Rubinacci A, Talotta F, Verde P, Blasi F (2007) Urokinase plasminogen activator receptor affects bone homeostasis by regulating osteoblast and osteoclast function. J Bone Miner Res 22:1387–1396
Acknowledgements
We thank Drs. Simona Polo and Sara Sigismund (IFOM-IEO campus, Milan, Italy) for providing NR6 cell lines and helpful suggestions. We are grateful to Prof. Francesco Blasi (IFOM-IEO campus, Milan, Italy) for scientific support, valuable advices and critically review of the manuscript. This work was supported by grant from Science Park RAF SpA, Milan, Italy. The NMR spectrometers were funded by Istituto di Chimica Biomolecolare, Consiglio Nazionale delle Ricerche, Pozzuoli (Naples), Italy.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Eden, G., Archinti, M., Arnaudova, R. et al. D2A sequence of the urokinase receptor induces cell growth through αvβ3 integrin and EGFR. Cell. Mol. Life Sci. 75, 1889–1907 (2018). https://doi.org/10.1007/s00018-017-2718-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2718-3