Abstract
Programmable DNA nucleases such as TALENs and CRISPR/Cas9 are emerging as powerful tools for genome editing. Dual-fluorescent surrogate systems have been demonstrated by several studies to recapitulate DNA nuclease activity and enrich for genetically edited cells. In this study, we created a single-strand annealing-directed, dual-fluorescent surrogate reporter system, referred to as C-Check. We opted for the Golden Gate Cloning strategy to simplify C-Check construction. To demonstrate the utility of the C-Check system, we used the C-Check in combination with TALENs or CRISPR/Cas9 in different scenarios of gene editing experiments. First, we disrupted the endogenous pIAPP gene (3.0 % efficiency) by C-Check-validated TALENs in primary porcine fibroblasts (PPFs). Next, we achieved gene-editing efficiencies of 9.0–20.3 and 4.9 % when performing single- and double-gene targeting (MAPT and SORL1), respectively, in PPFs using C-Check-validated CRISPR/Cas9 vectors. Third, fluorescent tagging of endogenous genes (MYH6 and COL2A1, up to 10.0 % frequency) was achieved in human fibroblasts with C-Check-validated CRISPR/Cas9 vectors. We further demonstrated that the C-Check system could be applied to enrich for IGF1R null HEK293T cells and CBX5 null MCF-7 cells with frequencies of nearly 100.0 and 86.9 %, respectively. Most importantly, we further showed that the C-Check system is compatible with multiplexing and for studying CRISPR/Cas9 sgRNA specificity. The C-Check system may serve as an alternative dual-fluorescent surrogate tool for measuring DNA nuclease activity and enrichment of gene-edited cells, and may thereby aid in streamlining programmable DNA nuclease-mediated genome editing and biological research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Programmable DNA nucleases are efficient tools for precision genome editing. Zinc finger nucleases (ZFNs) [1–3], the transcription activator-like effector nucleases (TALENs) [4–6], and especially, the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated nuclease (Cas) or CRISPR/Cas [7–9] are emerging as the most promising tools to introduce site-specific modifications in endogenous genomic loci of living cells and organisms.
Engineered TALEN proteins have three characteristic domains; a nuclear localization domain, a nuclease domain derived from the FokI endonuclease, and a DNA binding domain consisting of various numbers of tandem 34-aa repeats. Each repeat in the TALEN tandem array is identical except for the two residues at position 12 and 13, known as the repeat-variable di-residue (RVD), which defines the DNA binding specificity using an “RVD-DNA” codon [6]. The RVDs NI, NG, HD, and NN/NK preferentially recognize adenine (A), thymine (T), cytosine (C), and guanine (G), respectively [6, 10]. The adaptation of golden gate cloning for construction of custom TALENs has greatly promoted the utility of TALENs for gene editing in a variety of cell types and organisms such as human pluripotent stem cells [4], zebrafish [11], rats [12], pigs [13], and rice [14]. Furthermore, the development of the cost-effective fast ligation-based automatable solid-phase high-throughput (FLASH) system for large-scale assembly of TALENs has made efficient genome-scale editing procedures possible [15].
The Cas9 nuclease derived from the type II bacterial CRISPR system of Streptococcus pyogenes can, by usage of a small guide RNA (sgRNA), introduce position-specific double-strand breaks at endogenous genomic loci [8, 16]. Unlike TALENs, the specific DNA binding of the CRISPR/Cas9 system is mediated by the complementarity between the 20-nucleotide sgRNA spacer and the target DNA sequences (protospacer) preceding an NGG trinucleotide, which is known as the protospacer-adjacent motif (PAM) [17]. This simple RNA–DNA binding principle of the CRISPR/Cas9 system has greatly simplified its design and construction, and has thus rapidly revolutionized biological research during the last 3 years [18–23]. Genome editing using CRISPR/Cas9 has been applied in various cell types and organisms such as plants [19, 24], bacteria [25, 26], C. elegans [27, 28], zebrafish [29–31], mice [8, 21], rats [22, 23], pigs [32, 33], primates [34], and human cells [7, 8, 16], including human pluripotent stem cells [35, 36] and human hematopoietic stem cells [20].
The targeted genomic loci, number of the tandem repeats (TRs), and the spacer length between the TALEN pairs can affect the TALEN nuclease activity [13, 37, 38]. Similarly, a dependence of the activity on the sgRNA sequence has been reported in many studies of CRISPR/Cas9-mediated genome editing [7, 8, 16, 28, 39, 40]. Thus, a cost-effective, and sensitive method for activity quantification and selection of the most efficient TALENs and sgRNAs would increase the utility of these tools. Another challenge that can hamper the utility of these programmable DNA nucleases is the selection of cells with the required genetic modifications, a problem encountered especially with cell types that are difficult to transfect. Several approaches have been applied to enrich genetically modified cells, including fusion of TALENs and Cas9 to a fluorescent or antibiotic protein [18, 41], or co-transfection with a fluorescent or antibiotic resistance encoding marker gene [13, 33, 36, 42]. However, such protein fusions or co-transfections only reflect the transfection efficiency but not the actual nuclease activity. Previous studies have reported frequent synergistic biallelic gene modifications at the single-cell level using ZFNs, TALENs, or CRISPR/Cas9 [13, 31, 43]. To recapitulate nuclease activity, episomal surrogate reporter vectors, comprising the same targeting sequence as the endogenous genomic sequence to be modified, have been developed to enrich for gene-edited cells by flow cytometry, magnetic separation, or antibiotic selection [44–46]. The episomal surrogate reporter system functions through the DNA double-strand break (DSB) repair pathways non-homologous end joining (NHEJ) and single-strand annealing (SSA) [44–48]. Although NHEJ is the preferential pathway for DSB repair in cells, the exclusion of an in-frame stop codon in the targeting sequence of the NHEJ-based surrogate reporter system has limited its broad application [44, 49]. Ren et al. have recently shown that the SSA-based surrogate reporter system with homology arms of more than 200 bp is more sensitive in detecting nuclease activity than the NHEJ-based system [44].
Construction of surrogate reporter vectors can be a laborious and time-consuming procedure and the surrogate systems described are all based on the cloning of the target sequence into a multiple cloning site using type II restriction enzymes [44, 45, 49, 50]. We sought to develop a simple approach for reporter vector construction that is easy in design and construction, compatible with multiplex target sites, and independent of targeting sequence. The Golden Gate cloning method is a robust method for assembly of multiple DNA fragments into a plasmid vector in a single reaction. This method has facilitated the generation of TALENs and sgRNA expression vectors [18, 51]. Previously, we have also shown that rAAV-mediated gene targeting vectors can be generated in one step using the Golden Gate cloning method [52].
In the present study, we took advantage of the Golden Gate Cloning method to generate a dual-fluorescent vector system for CHECKing programable DNA nuclease-mediated Cleavage activity, hereafter called C-Check. We have demonstrated the application of the C-Check system in several genome editing settings. First, we showed that C-Check reporter could be used for in vitro functional assay and selection of TALENs and CRISPR/Cas9 with functional active nuclease activity. Second, we used donor plasmids and C-Check-validated CRISPR/Cas9 vectors to modify two porcine neurodegeneration-associated genes (MAPT and SORL1) in primary porcine fibroblasts. Third, we used donor plasmids and C-Check-validated CRISPR/Cas9 vectors to introduce an in-frame EGFP domain into the C-terminus of the human genes COL2A1 and MYH6 gene in human fibroblasts. Fourth, using the C-Check surrogate vector and Fluorescence-Activated Cell Sorting (FACS), we achieved a knockout efficiency >85 % by CRISPR/Cas9 in two human cell lines (HEK293T and MCF-7). Finally, we also demonstrated that the C-Check system could be used to test the mismatch tolerance of CRISPR/Cas9 when introducing mismatches between the sgRNA guide sequence and the protospacer.
Results
Generation of a modified dual-fluorescent reporter vector, C-Check, for assaying TALENs activity in vitro
Apart from the simplicity in vector design, construction, and screening, one of the essential requirements for a dual-fluorescent reporter vector for DNA nuclease activity measurements is its general compatibility with all types and sequences of target sites. We surveyed the previously reported dual-fluorescent reporter systems and selected the SSA-based system, which, unlike the NHEJ-based system, does not require the exclusion of an in-frame stop codon in the target sites [44, 46, 49, 53, 54]. The modified dual-fluorescent reporter vector (C-Check) we have generated is composed of two expression cassettes: a truncated EGFP expression cassette for detecting DSB-induced SSA events and an AsRED expression cassette for measuring transfection efficiency and for normalization purposes (Fig. 1a). Two truncated EGFP fragments: EGFP (1–600) and EGFP (100–720) were generated with complete disruption of fluorescence encoded from either fragment while retaining a maximum length of homology sequences (500 bp) thereby facilitating recombination mediated generation of one functional EGFP encoding sequence from the two fragments [55]. To facilitate the cloning and screening of the target site of interest in the C-Check vector, a Golden Gate cloning site comprising a lacZ selection cassette was inserted between the two truncated EGFP genes. Insertion of the target sequences is mediated by BsaI (Eco31I)-based Golden Gate cloning (Additional File 1). Two in-frame stop codons were inserted to flank the target sequences to prevent any possible read through of the truncated EGFP gene resulting from nuclease-induced indels. Once TALENs or CRISPR/Cas9 have induced DBSs at the target sites in the C-Check vector, the reporter cells will express both AsRED and EGFP if repaired by SSA, whereas only an AsRED signal will be observed if there is no nuclease activity or the DBSs are repaired via the NHEJ pathway (Fig. 1a). To facilitate the utilization of the C-Check vector for gene editing, this system has been deposited to the non-profit global plasmid repository Addgene (ID: 66817).
Generation of the C-Check system and validation for functional assay of TALEN-mediated DNA cleavage activity. a Schematic illustration of the C-Check reporter system. PGK phosphoglycerate kinase 1 promoter; the coding sequence of the EGFP gene is indicated with codon numbering from 5′ to 3′; the homology arms within the two truncated EGFP fragments (trEGFP1 and trEGFP2) are indicated by the yellow boxes; different poly A terminal signals were used to avoid recombination as indicated in different color. Binding of TALENs and CRISPR/Cas9 to the target sites in the C-Check reporter vector is illustrated. After cleavage of the episomal C-Check reporter vector in cells, the C-Check vector can be repaired through two pathways: single-strand annealing (SSA) or non-homologous end joining (NHEJ). Two stop codons were included to flank the target sites in the C-Check vector. The first stop codon was pre-built in the 5′-end and the second stop codon at the 3′-end is introduced by Golden Gate insertion of the target site sequence. b Schematic representation of the porcine IAPP locus and IAPP target site. Exons are indicated with gray boxes and the target site sequences are highlighted in blue. NLS nuclear localization signal, TRs tandem repeats, L and R denote the TALEN monomer protein that binds to the target site at the coding and non-coding strand. Figures are not drawn to scale. c Representative fluorescence imaging of the C-Check assaying of IAPP TALENs activity. Scr scrambled TALENs that do not target IAPP. d Representative flow cytometry diagram of the nuclease activity quantification by C-Check. Weak transmission from the AsRED spectrum to the EGFP detector was observed. The indicated gating (P1 and P2) was applied to avoid any false positive results. Efficiency was calculated as the percentage of cells in P2 out of the total number of cells in P1 and P2. This gating and quantification strategy was applied to all C-Check nuclease quantification assays throughout the study. e Quantification of IAPP TALEN activity. Asterisk (*) indicates a p value less than 0.05 compared to the remaining groups. f, g Representative fluorescence images and quantification of dose-dependent TALEN nuclease activities determined by C-Check analysis. Asterisk (*) indicates a p value less than 0.05 between the compared groups. h T7E1 assay of IAPP TALEN-induced indels in primary porcine fibroblasts. i Identification of TALEN-induced indels by Sanger sequencing. Three out of 96 clones analyzed carried 1, 7, and 6 bp deletions at the TALEN spacer sites, respectively. TALEN target sites are underlined
To validate the C-Check reporter system, we generated one pair of TALEN proteins targeting intron 2 of the porcine islet amyloid polypeptide (pIAPP) gene (Fig. 1b, Additional File 2) by Golden Gate cloning [13, 51]. The reason for choosing a porcine gene in validating the C-Check system is related to our long-term work of generating genetically modified pig models of human diseases [56, 57]. Humans can be predisposed to type 2 diabetes by aggregation of the IAPP protein as a consequence of either altered expression or mutations, whereas the wild-type porcine IAPP fragments are refractory to aggregation [58]. To generate an IAPP-based porcine model of type 2 diabetes, we aimed at replacing the endogenous porcine IAPP gene with a mutant human-derived IAPP gene by TALEN-mediated homologous recombination. We generated a pIAPP C-Check vector and transfected HEK293T cells with the pIAPP C-Check vector alone and in different combinations with pIAPP TALENs and scrambled TALEN vectors (Fig. 1c). Only HEK293T cells transfected with the C-Check-pIAPP and a pair of functional pIAPP TALENs expressed EGFP 24 h post transfection, whereas EGFP expression was not observed in cells transfected with either a single pIAPP TALEN protein encoding vector or a pair of scrambled TALEN vectors. Maximum EGFP expression was detected at 48–72 h post transfection (Fig. 1c). Microscopically, we noticed a weak transmission of the AsRED signal through the EGFP filter, which was also confirmed by flow cytometry analysis (Fig. 1d). This may be due to both overlap in the spectra of the two fluorescent molecules and due to low levels of leakage of EGFP expression from the unrepaired C-Check vector. This highlights the importance of including control transfections such as transfection with C-Check reporter vector only, or the C-Check reporter vector together with a TALEN scrambled control when performing the C-Check assay. Quantification of nuclease activity by flow cytometry was calculated as the percentage of EGFP positive cells (P2) out of the total number of AsRED positive cells (P1 + P2) and this calculation was applied to all nuclease activity assays throughout the study (Fig. 1d). A significant increase in the population of EGFP positive cells was detected in HEK293T cells transfected with the C-Check-pIAPP vector and functionally active pIAPP TALENs (Fig. 1e). Previous reports have observed that TALEN activity is dose dependent [13, 59, 60] and this is further supported by our C-Check reporter system (Fig. 1f, g). However, the dose-dependent increase of TALEN activity was not linear. A 12-fold increase in TALENs plasmid resulted in only 20 % increase of efficiency (approx. 50 % efficiency using 30 ng TALENs in contrast to 70 % efficiency using 360 ng TALENs) (Fig. 1f, g).
To further prove that the C-Check reporter vector actually reflects the nuclease activity at the endogenous gene level, we next examined the C-Check-validated pIAPP TALENs in mediating pIAPP gene disruption in primary pig fibroblasts. We chose primary fibroblasts established from newborn Göttingen minipigs. Efficient transfection of the primary fibroblasts is a critical step for successful delivery of TALENs and subsequent analysis of generated indels by T7E1 digestion, an assay that allows for discrimination between homoduplex and heteroduplex double-stranded DNA. We optimized the transfection efficiency of porcine fibroblasts by nucleofection by testing a combination of 5 nucleofection reagents and 15 different nucleofection programs using a 4D-Nucleofector (Additional File 3). Using the optimized nucleofection protocol (reagent P1, program CA137: transfection efficiency >50 %; viability >25 %), we transfected the porcine fibroblasts with the pIAPP TALENs, extracted genomic DNA from the cells 48 h post transfection, and amplified the target region by PCR. A nuclease activity of 2.99 % was revealed by the T7E1 assay (Fig. 1h). We also cloned the PCR product into competent bacterial cells. Three out of 96 clones (3.125 %) analyzed by Sanger sequencing carried different deletions at the target site (Fig. 1i). These results suggested that the C-Check-validated TALENs are also functional active at the endogenous genomic target locus.
CRISPR/Cas9-mediated double-gene targeting by homologous recombination in primary porcine fibroblasts using C-Check-validated CRISPR/Cas9 vectors
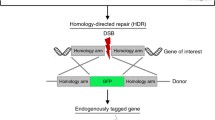
We next tested whether the C-Check system is also useful as a nuclease activity assay for CRISPR/Cas9. Gene targeting by homology-directed repair (HDR) in primary porcine cells is an important application in generating porcine model of human diseases. In order to develop porcine models of neurodegeneration [57], we selected two porcine genes (MAPT and SORL1) that are involved in the pathogenesis of Alzheimer’s disease. Two gRNA vectors were generated for each gene (referred to as pMAPT-T1, pMAPT-T2, pSORL1-T1, and pSORL1-T2) as well as a C-Check vector for each gene comprising either the two SORL1 or the two MAPT gRNA target sites. The efficiencies of the pMAPT and pSORL1 gRNA vectors were tested using the relevant C-Check vector. HEK293T cells were transfected with equal amounts of gRNA vector, Cas9 vector, and C-Check vector, and flow cytometry was performed 48 h post transfection. Cells transfected with the pSORL1 or pMAPT gRNAs yielded efficiencies ranging from 11.7 to 18.1 %, whereas cells transfected with the C-Check vectors alone showed only background EGFP expression (2.37–2.91 %) (Fig. 2a, b). Based on the C-Check assay, the pMAPT-T2 and the pSORL1-T1 gRNA vectors were chosen for genome editing in primary Göttingen fibroblasts.
Double-gene targeting in primary porcine fibroblasts with C-Check-validated CRISPR/Cas9. Quantification of MAPT (a) and SORL1 (b) CRISPR/Cas9 sgRNA activity by C-Check. HEK293T cells were co-transfected with the C-Check vector alone (control) or in different combinations of each sgRNA and the Cas9 vector. Cells were harvested 48 h post transfection and subjected to flow cytometry analysis. Schematic illustration of CRISPR/Cas9-mediated porcine MAPT (P301L) knockin (KI) by HDR (c) and CRISPR/Cas9-mediated pSORL1 knockout (KO) by HDR (d). Exons for each gene are indicated by black boxes. The CRISPR sgRNA target sites (pMAPT-T1, pMAPT-T2, pSORL1-T1, and pSORL1-T2) are indicated by a red, light blue, green, dark blue box, respectively. An asterisk (*) indicates the MAPT (P301L) mutation in the targeting vector and the targeted locus. Blue triangles denote LoxP sites. ITR inverted terminal repeats in the rAAV targeting plasmid, LHA and RHA left and right homology arm, respectively, Hygr hygromycin antibiotic resistance gene driven by a PGK promoter, Neo neomycin antibiotic resistance gene driven by a PGK promoter. Primers for PCR screening are indicated by arrows. Figures are not drawn to scale. e Summary of single- and double-gene targeting frequency using pMAPT-T2 and pSORL1-T1 in primary porcine fibroblasts (PPF)
In this experiment, we investigated if CRISPR/Cas9 was capable of inducing simultaneous double-gene targeting by knocking out the porcine SORL1 gene while at the same time knocking in the P301L mutation in the porcine MAPT gene. Two donor plasmids, pSORL1-KO-Neo and pMAPT-KI-Hygr, were generated using our previously established Golden Gate cloning approach (Fig. 2c, d) [61] and used in combination with the CRISPR/Cas9 system to introduce the intended genomic changes. The two donor plasmids also comprised an antibiotic resistance gene (neomycin and hygromycin, respectively) allowing for selection of targeted cells (Fig. 2c, d).
Two gene targeting experiments were conducted using fibroblasts established from either male or female Göttingen minipigs. In both cases, 1.5 × 106 cells were co-transfected with the pMAPT-T2 gRNA and pSORL1-T1gRNA vectors (both 1200 ng), and the Cas9-encoding plasmid (1200 ng) together with the rAAV-based donor plasmids pMAPT-KI-Hygr (1500 ng) and pSORL1-KO-Neo (2900 ng). The ratio of the two donor plasmids was adjusted to the double amount of the pSORL1-KO-Neo donor compared to the pMAPT-KI-Hygr donor (since we in a pilot study using equal amounts of the two donors obtained only pMAPT KI clones and no pMAPT KI/SORL1 KO clones). The cells were trypsinized 48 h post transfection and half of the cell suspension was seeded in either 15 (for male cells) or 5 (for female cells) 96-well dishes. Selection with 0.8 mg/ml neomycin and 0.14 mg/ml hygromycin was initiated the next day and maintained throughout the experiment. Neo+/Hygr+ cell clones were screened by PCR using primers located within the selection cassettes and both 5′ and 3′ to the targeted region of the pMAPT and pSORL1 genes (Additional File 4). As shown in Fig. 2e, a total of 225 female cell clones were selected and analyzed by PCR. Of these, nine comprised both the intended MAPT KI and SORL1 KO yielding a double-targeting efficiency of 4 % (% cell clones with MAPT KI and SORL1 KO/% Neo+ and Hygr+ cell clones). For the male cells, 184 clones were selected and analyzed by PCR. Eleven of these comprised both MAPT KI and SORL1 KO resulting in a double-targeting frequency of 6 % (Fig. 2e). As expected, the individual gene targeting frequencies for each gene alone was higher than for the double targeting. Thus, in male cells the targeting efficiency was 7.6 % for SORL1 and 13.6 % for MAPT, whereas in female cells the SORL1 and MAPT targeting efficiencies were found to be 10.2 and 25.8 %, respectively (Fig. 2e). Thus, though with somewhat varying efficiency in the two cell types, this demonstrates that it is possible to conduct dual gene targeting with the CRISPR/Cas9 system in primary Göttingen fibroblasts. In both cell types, however, all the double targeted clones also had some random integration of the donor plasmid (data not shown). Thus, for animal production purposes, further adjustment of the amounts of the donor plasmids will be warranted in order to eliminate random integration of the donor plasmid.
HDR-mediated gene editing in human fibroblasts using C-Check-validated CRISPR/Cas9 vectors
Fluorescent tagging of endogenous genes is a powerful tool in stem cell and biological research [62, 63]. Also, it is evident, as well as proven by this study (Fig. 2), that HDR-mediated gene editing is enhanced with CRISPR/Cas9 [64–67]. Thus, we attempted to generate a versatile system for fluorescent tagging of endogenous genes (Fig. 3a). By usage of CRISPR/Cas9-induced DSBs, we aimed at inserting the EGFP coding sequence preceding the stop codon of the target gene upon HDR between the endogenous target locus and the targeting fluorescence tagging vector (Fig. 3a). To facilitate the construction of the targeting vector, we further developed our Golden Gate cloning toolkit by introducing three extra Golden Gate cloning modules: pGolden-EGFP, pGolden-PGK-Neo-1A, and pGolden-1A-TK (Fig. 3b) [52]. We next selected two human genes MYH6 and COL2A1, which are lineage specific for cardiomyocyte and chondrocyte differentiation, respectively. We generated one and two sgRNAs targeting the 3′ untranslated region (3′UTR) in the MYH6 (MYH6-T1) and COL2A1 (COL2A1-T1 and COL2A1-T2) genes, respectively (Fig. 3a). To avoid the potential alteration in RNA stability resulting from CRISPR/Cas9-induced 3′UTR disruption [68], all sgRNAs target sites were designed downstream and as close to the stop codon as possible, which results in minimal deletion of the 3′UTR after homologous recombination. We also generated two targeting fluorescence tagging vectors (pGolden-MYH6-EGFP-tagging and pGolden-COL2A1-EGFP-tagging) using the Golden Gate cloning method (Fig. 3a, b). Forty-eight hours after co-transfecting the MYH6 and COL2A1 CRISPR/Cas9 vectors (CRISPR/Cas9-MYH6-T1, CRISPR/Cas9-COL2A1-T1, and CRISPR/Cas9-COL2A1-T2) and the corresponding C-Check vector, 33.9, 7.5, and 19.8 %, respectively, of the transfected HEK293T cells were EGFP/AsRED positive (Fig. 3c, d). We further co-transfected human dermal fibroblasts with CRISPR/Cas9-MYH6-T1 and the pGolden-MYH6-EGFP-tagging vectors, or CRISPR/Cas9-COL2A1-T2 and pGolden-COL2A1-EGFP-tagging vectors. Following selection of G418-resistant cell clones in 96-well plates for 2–3 weeks, we isolated 10 and 13 resistant clones, respectively, for MYH6 and COL2A1 and analyzed the gene targeting by PCR. Screening PCR revealed that 10 % (1/10) and 7.69 % (1/13) of the MYH6 and COL2A1 G418-resistant clones were targeted, respectively (Fig. 3e, f). The correct fusion of EGFP into the endogenous gene was further validated by subjecting the PCR product to Sanger sequencing (data not shown). These two MYH6 and COL2A1 EGFP-tagged fibroblast cell lines will be important tools for studying differentiation of cardiomyocytes and chondrocytes directly or after prior dedifferentiation into iPS cells [69].
Fluorescence tagging of MYH6 and COL2A1 in human fibroblasts. a Schematic representation of the CRISPR/Cas9-mediated C-terminal fluorescence tagging of endogenous genes by homologous recombination. One and two sgRNAs were generated for MYH6 and COL2A1, respectively, indicated with a correspondingly colored box. UTR, TS, LHA, RHA, and 2A denote untranslated region, target site, left homology arm, right homology arm, and 2A peptide, respectively. TK thymidine kinase cassette for Cre-mediated excision screening, PGK-Neo PGK promoter-driven neomycin expression cassette for gene targeting selection; two LoxP sites were included for excision of the antibiotic markers by Cre recombinase. b Schematic illustration of the Golden Gate assembly of the C-terminal EGFP-tagging system. “Kan” and “amp” denote bacterial kanamycin and ampicillin selection cassettes. The 2A peptide sequences are generated upon the correct assembly of the PGK-Neo-1A fragment and the 1A-TK fragment. c, d Quantitative analysis of MYH6 and COL2A1 sgRNA activity by C-Check assays. Asterisk (*) indicates statistical significance compared to the control (C-Check only); hash symbol indicates statistical significance compared to the Cas9 + COL2A1-T1. e, f Screening PCR of MYH6 and COL2A1 knockin of 10 and 13 G418+ clones. Symbols (P, +, −) indicate targeted knockin clones, positive control, and negative control templates, respectively
Efficient generation of null knockout human embryonic kidney cells (HEK293T) using the C-Check surrogate reporter system
The aforementioned studies have demonstrated that the C-Check system can recapitulate the nuclease activity of TALENs and CRISPR/Cas9. Another important application of the dual-fluorescent reporter system is to use it to enrich for gene-edited cells [45, 49]. To determine whether the C-Check system could serve as a surrogate reporter, three CRISPR/Cas9 vectors and one C-Check vector were generated for targeting the human insulin-like growth factor I receptor (IGF1R) gene, which plays a crucial role in cell proliferation [70]. We first generated three sgRNAs targeting exon 2 (common coding exon in all IGF1R isoforms) of IGF1R (Fig. 4a). The IGF1R target sites were amplified by PCR and inserted into the C-Check vector by Golden Gate Cloning (Fig. 4a). Two days after the co-transfection, a significant increase in EGFP and AsRED double-positive HEK293T cells was detected exclusively in the co-transfections comprising the IGF1R C-Check vector, CRISPR/Cas9, and the target sgRNA. Efficiencies of 31.9, 42.8 and 22.7 % for IGF1R T1, T2, and T3 were observed, respectively, (Fig. 4b, c) indicating that all three designed IGF1R sgRNA were functionally active.
Enrichment of IGF1R null-modified HEK293T cells with the C-Check surrogate reporter vector. a Schematic illustration of the endogenous IGF1R locus and the IGF1R C-Check vector. All sgRNAs target sites (T1–T3) were on the coding strand of exon 2. Primers for generating the IGF1R C-Check vector (F1 + R1) and for screening of IGF1R knockout (F2 + R2) are indicated with black arrows. b, c Representative fluorescence imaging and quantification of sgRNAs activity by C-Check. Asterisk (*) indicates statistical significance between the comparisons; hash symbol indicates statistical significance compared to CC (transfected with the IGF1R C-Check plasmid only). d C-Check surrogate reporter-based FACS (upper panel) and PCR quantification of targeted IGF1R deletion frequency (KO%) in the indicated population of cells. The HEK93T cells were transfected with the IGF1R CRISPR/Cas9 (T2 and T3) and IGF1R C-Check (lower panel). Representative plot of AsRED-based (e) or EGFP-based (f) FACS sorting of HEK293T cells co-transfected with the IGF1R CRISPR/Cas9 (T2 and T3) and either a scrambled C-Check vecor (e) or the IGF1R C-Check vector (f). Gatings are illustrated with numbers (3–18). g Quantification of targeted IGF1R deletion efficiency based on PCR screening and Image J. Groups (3–18) are the corresponding sorted cells. Group 1 and 2 are unsorted cells co-transfected with the IGF1R CRISPR/Cas9 (T2 and T3) and either a scrambled C-Check vector or the IGF1R C-Check vector, respectively. Wild-type (WT) cells were used as control. h The HEK293T cells co-transfected with the IGF1R CRISPR/Cas9 (T2 and T3) and the IGF1R C-Check vector and sorted into six populations based on both AsRED and EGFP signal. The IGF1R knockout efficiency was quantified by PCR and image J (h, lower panel). i, j The cell populations (h, P1, P3, and P6) were also single-cell sorted into a 96-well plate for clonogenic cell growth followed by IGF1R knockout PCR screening of clonogenic cell clones. Letters o, e, and w represent homozygous, heterozygous, and “wild type” clones, respectively, based on PCR. Note that small indels could not be distinguished by PCR. The “wild type” bands appearing in the heterozygous and wild-type clones might therefore actually be mutated. This was further validated by Sanger sequencing (Additional File 6). k Western blot analysis of IGF1R in three IGF1R knockout cell clones. Wild-type (WT) parental HEK293T cells were used as control. Beta-actin was used as loading control
We next tested whether the C-Check system in combination with FACS could be used to enrich for CRISPR/Cas9-induced mutations in HEK293T cells. Detection of indels induced by a single CRISPR/Cas9 vector, using T7E1 or Surveyor Nuclease assays, is often laborious and expensive. To circumvent this problem, we used a pair of sgRNAs for which indels (mostly deletions) are expected and easily detected by PCR (Fig. 4a) [18, 71]. In this experiment, to enable IGF1R knockout screening by PCR, we selected the IGF1R sgRNAs T2 and T3 for co-transfection of HEK293T cells together with the IGF1R C-Check vector. Seventy-two hours post transfection, we sorted four populations of cells based on the fluorescence intensity of EGFP and AsRED cells (M1–M4 in Fig. 4d, upper panel). A clear increase in indel frequency was detected by PCR-based screening yielding 8.7 % in the EGFP and AsRED negative cells and 97.9 % in the EGFP and AsRED double-positive cells of highest fluorescence intensity (Fig. 4d, lower panel) indicating efficient enrichment of CRISPR/Cas9-induced IGF1R mutations.
Two effects have been reported that could additively contribute to the enrichment of programmable DNA nuclease-induced mutated cells: a co-transfection effect and a surrogate reporter effect [45]. To distinguish between these effects, we co-transfected the HEK293T cells with IGF1R CRISPR/Cas9 (sgRNAs T2 and T3) and either a scrambled C-Check vector (Fig. 4e) or the IGF1R C-Check vector (Fig. 4f), and sorted the transfected cells based on AsRED signal (Fig. 4e) or EGFP signal (Fig. 4f). As the NHEJ-based surrogate reporter vector [45], the C-Check surrogate reporter system can be used to efficiently enrich for the gene-edited cells (Fig. 4g). Most importantly, the C-Check-based enrichment did not concordantly enrich for off-target events (Additional File 5).
We further sorted the cells into six populations (P1–P6) based on both EGFP and AsRED fluorescence intensity (Fig. 4h, upper panel). We observed a clear increase in targeted deletion efficiency associated with the EGFP intensity (13 % increase in P3 vs. P2, and 5.4 % increase in P3 vs. P4), whereas only 1.6 % increase was observed while comparing cells which only differed in AsRED intensity (P3 vs. P5) (Fig. 4h, lower panel). Apart from the advantage in enrichment of cells with desired mutations, the surrogate reporter system could facilitate the antibiotic-selection-free establishment of clonogenic cells [45, 49]. To determine whether the C-Check surrogate system is compatible with clonogenic cells establishment, we co-transfected HEK293T cells with the IGF1R C-Check and the IGF1R CRISPR/Cas9 vectors (T2 + T3), and sorted single cells based on three gatings (P1, P3, and P6) into 96-well plates (3, 1, and 1 plates each for P1, P3, and P6, respectively) 72 h after transfection. Single-cell-derived clonogenic cell clones (56, 40, and 40 clones for P1, P3, and P6, respectively) were selected for analyses of IGF1R knockout by PCR screening (Fig. 4i). Of the clonogenic cell clones sorted from P1 (AsRED++EGFP++), 36 and 61 % were null and heterozygous knockout, respectively. A clear decrease in null frequency was detected in the clonogenic cell clones sorted from P3 (AsRED+EGFP+) (Fig. 4j). Since small indels could not be distinguished by the PCR-based gel electrophoresis, we speculated that the mutation rate might be underestimated in these cell clones. All the clones identified as heterozygous and wild type might be compound heterozygous and mutated. To test this, we randomly selected a few clonogenic cell clones established from P1, P3, and P6 and further validated these by Sanger sequencing (Additional File 6). All eight clones established from P1 were null modified, including the single clone that appeared to be “wild type” based on the PCR screening (Fig. 4i), indicating that all clones established from P1 were null mutated. We further confirmed that the out-of-frame null mutations in exon 2 of IGF1R led to complete loss of IGF1R at the protein level in three clones by Western blot analysis (Fig. 4k). In summary, this experiment indicates that the C-Check system could be used as a surrogate reporter system to enrich for CRISPR/Cas9-mediated gene-edited cells.
Efficient generation of CBX5 null human breast cancer cells using the C-Check surrogate reporter system
To further demonstrate the application of the C-Check surrogate system in enriching cell clones with desired mutations, we tested the C-Check system using another gene in another cell line. MCF-7 is a human breast cancer cell line widely used for studies of tumor biology and hormone responsiveness [72]. Using the same approach as established in the C-Check/CRISPR/Cas9-mediated IGF1R knockout in HEK293T cells, we designed three sgRNAs and one C-Check vector targeting exon 2 (common coding exon in all isoforms) of the Chromobox Homolog 5 (CBX5) gene in MCF-7 cells (Fig. 5a). CBX5 encodes the Heterochromatin Protein 1α (HP1α), which has been shown to be important for DNA packing and maintaining heterochromatin and gene silencing as well as playing an important role in breast cancer cell metastasis [73, 74]. All three CBX5 CRISPR/Cas9 vectors (CBX5 T1–T3) were functionally active as measured by the C-Check assays in HEK293T cells (Fig. 5b, c). We next transfected MCF7 breast cancer cells with the CBX5 C-Check vector alone or together with three different combinations of CBX5 CRISPR/Cas9 vectors: CRISPR/Cas9-CBX5-T1, CRISPR/Cas9-CBX5-T1 + T2, and CRISPR/Cas9-CBX5-T1 + T3. Compared to HEK293T cells, the percentage of EGFP and AsRED positive cells was much lower in the MCF7 cells—most likely due to differences in transfection efficiency and the SSA-mediated DSB repair efficiency between the two cell lines [75] (Fig. 5d). Seventy-two hours after co-transfection with CBX5-C-Check and CRISPR/Cas9 CBX5 vectors, single cells were sorted from the EGFP+AsRED+ cells into 96-well plates (Fig. 5d). Twenty out of 23 clones (86.9 %) were modified in all alleles as genotyped by PCR screening and Sanger sequencing (Fig. 5e, Additional File 7). Most CRISPR/Cas9-induced CBX5 indels in these CBX5 knockout clones were nonsense mutations that caused a decrease in mRNA level and complete loss of CBX protein (Fig. 5f, g). Taken together, these results corroborated that the C-Check surrogate reporter system facilitates efficient generation and enrichment of selection-free genetically modified cells.
Enrichment of CBX5 null MCF-7 cells with the C-Check surrogate reporter vector. a Schematic illustration of the endogenous CBX5 locus and the CBX5 C-Check vector. All sgRNA target sites (T1–T3) were on the coding strand of CBX5 exon 2. Primers for generating the CBX5 C-Check vector (F1 + R1) and for screening of CBX5 knockout (F2 + R2) are denoted with black arrows. b, c Representative fluorescence imaging and quantification of sgRNAs activity by C-Check. Asterisk (*) indicates statistical significance between the corresponding comparisons; hash symbol indicates statistical significance compared to C-Check control (transfected with CBX5 C-Check plasmid only). d Illustration of FACS diagram and C-Check based gating for single-cells sorting. G1, G2, and G3 samples were population sorted into 96-well plates as described in the method section. e Genotyping by PCR and Sanger sequencing of CBX5 knockout clonogenic MCF-7 cells resulting from single-cell sorting. A summary of all clones genotyped by PCR and Sanger Sequencing is provided in the lower panel (Additional File 7). Note that small indels could not be distinguished by PCR screening. null targeted modified in all alleles, he. heterozygously modified. f qPCR analysis of 9 biallelic CBX5 knockout clones. Asterisk (*) indicates statistical significance compared to wild-type cells (WT); ns not significant. g Western blot analysis of CBX5 in five CBX5 knockout cell clones. Wild-type (WT) parental MCF-7 cells were used as control. Beta-actin was used as loading control
The C-Check system is compatible with testing of multiplex target sites
To investigate whether a single C-Check vector system is compatible with multiple target sites, we generated two C-Check vectors: One C-Check vector comprising target sites from exon 5 of the porcine HPRT gene (referred to as C-Check-HPRT), and another C-Check vector containing a synthetic DNA fragment comprising an array of 10 CRIRSPR/Cas9 targeting sites (referred to as C-Check-M10) (Fig. 6a). The C-Check-HPRT vector was co-transfected with a combination of five left and five right TALEN monomer encoding vectors into HEK293T cells, and nuclease activity was quantified by flow cytometry 48 h post transfection. Based on the results from the C-Check system, the pHPRT TALENs pair (L3 + R3) with the highest activity (>60 %) could easily be selected (Fig. 6b). We next transfected the HEK293T cells with the C-Check-M10 alone or in combination with each of ten different sgRNAs for CRISPR/Cas9 nuclease. Significant, but variable, CRISPR/Cas9 nuclease activity was observed for all ten CRISPR sgRNAs with an efficiency ranging from 15 to 42 % compared to the controls demonstrating that the C-Check system is compatible with multiplexing analyses of gRNAs activities (Fig. 6c).
C-Check system is compatible with multiplexing nuclease analyses. a Schematic representation of the porcine HPRT locus (exon 5 and partial flanking introns), and the HPRT TALEN target sites (left panel) and the multiplex C-Check vector containing ten CRISPR/Cas9 targeting sites (right panel). Numbers of tandem repeats for each TALEN monomer protein is given with numbers in red. b Heatmap presentation of the TALEN nuclease activities for 25 pairs of HPRT TALENs. c Quantification of activity of ten CRISPR/Cas9 vectors by flow cytometry using a single multiplexing C-Check vector. Asterisk (*) indicates statistical significance compared to control (C-Check only); ns not significant compared to control; hash symbol indicates statistical significance compared to all other sgRNAs
The C-Check system can be used for studying CRISPR/Cas9 sgRNA specificity
One of the major concerns in CRISPR/Cas9-mediated gene editing is the potential off-target events resulting from unspecific binding of sgRNAs to similar protospacer sites [7, 8]. Several approaches to avoid sgRNAs with potential off-target sites, such as in silico design, including mismatches [76, 77], using truncated sgRNAs (at either the 3′ end or the spacer sequences) [78–80], and CRISPR/Cas9 nickase, have been described [81, 82]. It has been reported that CRISPR/Cas9 sgRNAs are more sensitive to mismatches at the seed region of the protospacer (1–12 bp preceding the PAM, Fig. 7a) [8, 83]. To investigate whether the C-Check system could recapitulate sgRNA specificity of CRISPR/Cas9, we generated a C-Check vector to determine CRISPR/Cas9 mismatch tolerance (Fig. 7a). Two sgRNA target sites were inserted into the Golden Gate cloning site of the C-Check vector. In this experiment, we generated ten sgRNAs for each of the two target sites harboring mismatches at positions 1–3, 10–12, and 17–19 nt preceding the PAM (Additional file 8). For CRISPR/Cas9 target site 1 (T1), one mismatch at position 1, 10, and 19 decreases the activity from 25.1 % (on-target activity) to 4.4, 8.3, and 15.2 %, respectively. Introduction of two or more mismatches at the T1 seed region completely abolished cleavage by CRISPR/Cas9 (Fig. 7b) consistent with previous reports suggesting CRISPR/Cas9 to be more sensitive to mismatches at the 3′-end of sgRNAs (seed region) [7, 8, 16, 29, 83]. For the CRISPR/Cas9 sgRNA2 (T2), one mismatch introduced at position 1 or 10 decreased the activity from 34.5 % (on-target activity) to 9.5 % and 32.0 %, respectively, whereas one mismatch introduced at position 19 retained higher CRISPR/Cas9-mediated cleavage activity (43.7 %, Fig. 7c). Similar observations have been reported for some sgRNAs that retain robust CRISPR/Cas9-mediated on-target cleavage activity with mismatches or truncation at the 5′-end [7, 17, 78, 83]. Similar to T1, introduction of three mismatches at target site T2 completely abolished the CRISPR/Cas9-mediated DNA cleavage (Fig. 7c) further validating the utility of the C-Check system as a versatile tool for studying CRISPR/Cas9 functions.
Quantification of CRISPR/Cas9 sgRNA specificity with C-Check. a Illustration of the C-Check CRISPR OFF vector. Two sgRNA target sites were cloned into the C-Check vector. Positions for each nucleotide, represented with an individual box, in the protospacer sequence are annotated as 1–20 from the 3′-end to the 5′-end. Position 1 is the nucleotide preceding the PAM. The seed region of the target site (TS, 1–12) is colored yellow. b, c Quantification of CRISPR/Cas9 nuclease activity of one on-target (ON) sgRNA and nine off-target (OFF) sgRNAs for target site T1 (b) and T2 (c). The filled boxes in the lower panels represent mismatches between the sgRNA and the target site. Asterisk (*) indicates statistical significance compared to both controls; hash symbol, represents statistical significance compared to all remaining groups
Discussion
Systems for accurate and sensitive detection of programmable DNA nuclease activity and enrichment of cells with desired genetic modification are essential tools to facilitate genome editing in living cells [44–46, 49, 54, 84]. In this study, we demonstrated in different scenarios that the modified HDR-directed dual-fluorescent C-Check system could be used for assaying TALENs and CRISPR/Cas9 activity in vitro. Several similar dual-fluorescent reporter systems, either based on the SSA or NHEJ pathways, have been developed for in vitro functional analysis of programmable DNA nuclease activity and as surrogate reporters to enrich for genetically modified cells [44, 45, 49]. The SSA-based C-Check system developed in this study offers an alternative tool in the current surrogate-reporter-vector toolbox to facilitate genome editing. Consistent with previous surrogate reporter vectors, the C-Check vector could reflect nuclease activity and enrich for genetically modified cells with desired mutations [45, 49]. Using the Golden Gate Cloning approach, the C-Check system simplified the cloning procedure. In addition, unlike the NHEJ-based reporter system, exclusion of in-frame stop codons at the target sequences is not required for the SSA-based C-Check reporter system, thus simplifying the in silico design and broadening its utility. Furthermore, although DBSs are predominantly repaired by NHEJ [85], the HDR-based C-Check system exhibits high sensitivity in detecting nuclease activity in HEK293T cells. Although comparison between the C-Check system and other surrogate systems was not conducted in this study, a previous study by Ren et al. demonstrated that the SSA-based system with homology arm lengths >200 bp is more efficient and sensitive than the NHEJ-based system [44].
In this study, we have demonstrated the usefulness of C-Check system for functional in vitro assays of TALENs and CRISPR/Cas9 vector activity. Since the C-Check system is functioning via programmable DNA nuclease-induced double-strand repair by SSA, the C-Check system should also be compatible for in vitro functional assays of other programmable DNA nucleases such as ZFNs [3], CRIPSR/Cas9 nickase [86], and dimeric CRISPR/dCas9-FokI nuclease [87] although this was not addressed in this study.
One major advantage offered by the dual-fluorescent surrogate system is to enhance the generation of clonogenic cells with desired genetic modifications [45, 49]. With the C-Check system, we generated clonogenic HEK293T and MCF-7-null-mutated cells with a targeted modification rate of 86.9 % in MCF-7 cells and nearly 100 % in HEK293T cells. Although only two genes and two cell lines were tested in this study, the C-Check surrogate reporter system could in principle be applied to any transfectable cell type that is compatible with clonogenic formation from single cells. Furthermore, since the C-Check system is based on SSA, the C-Check system may serve as a real-time indicator of the endogenous cellular machinery for homologous recombination [88]. Thus, the C-Check surrogate reporter system might be compatible with enhancing antibiotic-selection-free gene targeting by programmable DNA nuclease-mediated homologous recombination. This will be addressed in future studies.
Off-target effects of CRISPR/Cas9 have been reported by many CRISPR/Cas9-mediated gene editing studies [7, 8, 16, 17, 78–80, 83, 89]. Although off-target events were not examined for all the CRISPR/Cas9 sgRNAs used in this study, previous studies have suggested that both NHEJ- and HDR-based surrogate reporter systems did not exacerbate off-target effects in the enriched cells [44, 45]. Furthermore, we demonstrated that the C-Check system could be used for quantifying CRISPR/Cas9 sgRNA specificity. Our findings based on the C-Check CRISPR OFF system further confirmed that CRISPR/Cas9 is more sensitive to mismatches close to PAM as two or more mismatches at this position completely abolished the CRISPR/Cas9 nuclease activity [7, 8, 17] suggesting that sgRNAs with potential off-target sites should comprise more than three mismatches in the seed region to avoid targeting of these sites. The C-Check system provides a versatile tool for studying optimizing approaches, such as usage of truncated sgRNAs and other modifications to both the sgRNA and Cas9 nuclease, that could improve CRISPR/Cas9 specificity in future studies [78].
In this study, we also demonstrated that CRISPR/Cas9 could mediate efficient gene targeting by homologous recombination in primary Göttingen porcine and human fibroblasts. We demonstrated for the first time that double-gene targeting in primary porcine fibroblasts could be efficiently achieved by CRISPR/Cas9-mediated homologous recombination. Furthermore, we provided two examples on generating fluorescently tagged human fibroblasts that could be used for generating induced pluripotent stem cells and provide the capacity of subsequent real-time monitoring of lineage-specific differentiation [90]. Although the gene targeting experiments were not conducted for cells without CRISPR/Cas9 vectors, many studies have reported that the targeting frequency with only one gene targeting plasmid, even with homology arms larger than 10 kb, is lower than 1 % in primary cells [91–93]. The length of the homology arms in our targeting vectors was approximately 1 kb each, suggesting that the high targeting frequency was enhanced by CRISPR/Cas9. This is expected to facilitate the generation of genetically modified pig models for human diseases by somatic cell nuclear transfer in the future [33, 94].
Conclusions
In summary, our C-Check system provides an alternative dual-fluorescent surrogate reporter system to monitor programmable DNA nuclease activity, enrich for genetically modified cells at desired genomic loci, establish antibiotic-selection-free clonogenic cells with desired genetic modifications, and for studying CRISPR/Cas9 specificity. Thus, the C-Check system provides an attractive alternative to other similar dual-fluorescent surrogate reporter systems and a useful tool in genome editing.
Materials and methods
All DNA oligonucleotide syntheses and Sanger sequencing in this study were performed by Eurofins Genomics, Germany. All Fast Digest restriction enzymes were purchased from Thermo Scientific, Life Technologies, Denmark.
Cells
Human embryonic kidney 239T (HEK293T) cells and human breast cancer MCF7 cells were cultured in DMEM medium supplemented with 10 % FBS, 1× GlutaMAX, and 1× P/S in a 2-gas tissue culture incubator (5 % CO2, 37 °C). Normal human dermal fibroblasts (NHDF) and primary porcine fibroblasts (PPF, established from Göttingen minipigs) were cultured in DMEM medium supplemented with 15 % FBS, 1× GlutaMAX, and 1× P/S in a tri-gas tissue culture incubator (5 % CO2, 5 % O2, 37 °C). During selection, basic human fibroblast growth factor (5 ng/ml, Life technologies) was supplemented to the media for NHDF and PPF.
Construction of the C-Check vector
The C-Check vector was constructed by a modular cloning strategy. Four DNA fragments, including the PGK-EGFP1–600 (the first 600 bp coding sequences of EGFP driven by the PGK promoter), B-lacZ-B (the bacterial lacZ expression cassette flanked by two BsaI (Eco31I) restriction enzyme sites), EGFP100–720-SV40pA (the 100–720 bp coding sequence of EGFP and SV40 poly A signal), and CMV-AsRED-BGHpA (the AsRED expression cassette driven by the CMV promoter), were amplified by PCR and digested with BsaI, BsmBI, BsmBI, and BsmBI, respectively. These four PCR fragments were ligated with the BsaI digested pFUS-A plasmid backbone (plasmid from the Golden Gate TALEN and TAL Effector Kit, Addgene ID 1000000024). The C-Check vector was validated by restriction enzyme digestion and Sanger sequencing, and has been deposited at Addgene (plasmid ID 66817).
Construction of gene-specific C-Check reporter vectors
A more detailed protocol on how to generate and validate C-Check reporter vectors is provided in Additional Files 9 and 10. Complementary oligonucleotide annealing (COA) or PCR-based protocols were used for the C-Check reporter vector construction.
For the COA approach, two complementary oligonucleotides were synthesized:
-
C-Check-COA-F: 5′-GTCGGAt(SS-TS)ataGGT,
-
C-Check-COA-R: 5′-CGGTACCtat(AS-TS)aTC.
Sequences in brackets denote the sense strand (SS) and antisense strand (AS) of the target site (TS) sequences recognized by programmable DNA nucleases such as TALENs and CRISPR/Cas9. Upon annealing, the two complementary oligonucleotides form a double-strand DNA fragment with a 5′-GTCG overhang in the sense strand and a TGGC-5′ in the antisense strand. The annealed oligonucleotides are subsequently cloned into the BsaI-digested C-Check vector, transformed into competent bacterial cells, and plated on LB agar plates containing spectinomycin (50 µg/ml) and 8 µl IPTG (0.5 M) and 8 µl X-gal (100 mg/µl). Spectinomycin/X-gal positive bacterial clones were selected for plasmid DNA purification. To facilitate the bacterial clone screening of the C-Check reporter vector, a KpnI restriction enzyme site will be incorporated into the C-Check vector upon correct ligation. Thus, the C-Check reporter vector can be further screened by co-digestion with BamH1 and KpnI (Additional File 9).
As errors and cost in DNA oligonucleotide synthesis will increase accordingly with the oligonucleotide length, an alternative PCR-based approach is established to generate the C-Check vector (Additional File 9). First, the targeted regions of the gene-of-interest were analyzed for the presence of three popular type IIS restriction enzymes which cleave DNA outside their recognition sequences: BsaI, BsmBI, and BbsI. The targeted region was then amplified (less than 300 bp) by PCR with the C-Check PCR primers. Linkers containing one of the aforementioned type IIS restriction enzyme absent in the targeted region were chosen. The PCR products were then digested with the corresponding restriction enzyme and ligated into the C-Check vector. When using the BsaI restriction enzyme, digestion and ligation can be performed together. All C-Check reporter vectors used in this study were validated by Sanger sequencing.
Generation of TALEN vectors
All TALEN vectors in this study were generated using the TAL Effector Kit (Addgene ID 1000000024) and the GoldyTALEN (Addgene ID 38143). TALEN vectors were generated by Golden Gate Assembling according to the protocols previously described by us and other groups [51, 95]. TALEN target regions and the TALEN modulars are listed in Additional File 2.
Generation of CRISPR/Cas9 sgRNAs
CRISPR/Cas9 sgRNAs were designed using an online sgRNA designing tool (http://crispr.mit.edu/). Guide RNA sequences with more than three mismatches were chosen to minimize potential off-target events. Two CRISPR/Cas9 systems were used in this study. A two-vector CRISPR/Cas9 system was chosen for the porcine and the human fibroblasts to avoid the integration of CRISPR/Cas9 vector into the targeted cells. The human codon-optimized Cas9 [a gift from George Church (Addgene plasmid # 41815)] and sgRNA (pFUS-U6-sgRNA, generated by us) were expressed in two separate plasmids. An all-in-one CRISPR system (pSpCas9(BB)-2A-Puro (PX459), a gift from Feng Zhang (Addgene plasmid # 48139), was used for the rest of CRISPR experiments described in this study.
To generate CRISPR sgRNA vectors, two complementary guide oligonucleotides (100 pmol each) were first denatured in 1× NEB buffer 2 (in a total volume of 20 µl) at 95 °C for 5 min using a heating block followed by slow annealing by turning off the heating block. For sgRNA ligation, one microliter of the annealed oligonucleotides and 100 ng of the sgRNA scaffold plasmid (pFUS-U6-sgRNA) or the all-in-one PX459 plasmid were mixed with 1 µl BsaI (for pFUS-U6-sgRNA) or BbsI (for PX459) restriction enzyme, 1 µl T4 ligase (Thermo Scientific), and 2 µl T4 ligase buffer (10×) in a total volume of 20 µl. Digestion and ligation were performed in a thermal cycler using the following program: ten cycles of 37 °C for 5 min and 22 °C for 10 min; one cycle of 37 °C for 30 min; and one cycle of 75 °C for 15 min. The ligation product was stored at 4 °C or used directly (2 µl ligation product) to transform competent bacterial cells. Using this protocol, we have experienced that over 95 % of the bacterial clones are positive. Bacterial colony screening was also performed by PCR using a U6 forward primer (Additional File 4) and the antisense guide oligonucleotide (template strand of the sgRNA spacer). All sgRNA vectors used in this study have been validated by Sanger sequencing. Target sites and oligonucleotides for construction of all sgRNAs are listed in Additional Files 1 and 4, respectively.
Transfection
Three transfection methods have been used in this study. Both nucleofection (Amaxa™ 4D-Nucleofector) and Lipofectamine LTX Plus transfection (Life technologies) were used to transfect primary porcine fibroblasts. Transfection of PPF with TALENs was carried out by nucleofection. The Primary Cell Optimization 4D-Nucleofector™ X Kit was used to optimize the nucleofection program in primary porcine fibroblasts (Additional File 3). The optimized nucleofection program (reagent P1, program CA137) was used for delivering the TALENs into PPF. Lipofectamine LTX Plus reagent was used to deliver CRISPR/Cas9 vectors into both NHDF and PPF. Transfection was performed according to the manufacturer’s instruction. To minimize cell toxicity, we reduced the amount of DNA used by a factor of 0.6. A ratio of 1:3 was chosen for the use of DNA (µg) and Lipofectamine LTX (µl) reagent. All transfections of the HEK393T cells and MCF7 cells were performed using the X-tremeGENE 9 reagent (Roche) exactly following the manufacturer’s instruction. The following principle was applied for the plasmid DNA mixture used in co-transfections: For co-transfection of C-Check with a TALENs pair or co-transfection of C-Check with separated Cas9 and sgRNA vectors, a ratio of 1:1:1 in DNA amount was used. For co-transfection of C-Check with all-in-one CRISPR/Cas9 vector, a ratio of 1:1 in DNA amount was used. For control transfections, TALENs, Cas9, or sgRNA plasmids were replaced with equal amounts of a control plasmid pUC19.
Selection and PCR screening of gene knockout and knockin NHDF and PPF
NHDF or PPF (1.5 × 106 cells) were seeded onto a gelatin-coated 10-cm cell culture dish the day before transfection. The C-Check-validated CRISPR/Cas9 vectors were transfected with the donor plasmid using Lipofectamine LTX Plus transfection reagent. One day after transfection, cells were trypsinized and seeded into gelatin-coated 96-well plates at a density of about 400–500 cells per well. Three days after transfection (2 days after splitting the cells into 96-well plates), cells were selected with G418 (500 μg/ml for NHDF, and 800 µg/ml for PPF) for 2 weeks with the medium changed every 3–4 days. Basic fibroblast growth factor (bFGF) (5 ng/ml) was supplemented to the growth medium. For pMAPT and pSORL1 double-gene targeting, selection was carried out using both G418 (800 µg/ml) and hygromycin (140 µg/ml). Following selection, G418-resistant, or (for double targeting) G418-resistant and hygromycin-resistant, cell clones were trypsinized and 1/3 of the cells were transferred to 96-well PCR plates for PCR screening, while the remaining 2/3 of the cells were cultured in gelatin-coated 96-well plates and further expanded for downstream applications.
Fluorescent imaging and flow cytometry analysis
The day before transfection, HEK293T cells were seeded into a 24-well plate (1 × 105 cells per well). At least three independent transfection experiments were carried out for each C-Check transfection. Fluorescence microscopy and photographing were performed 48 h post transfection. Exposure times were adjusted to the control C-Check transfection group, that was transfected with C-Check only or C-Check with a scrambled gRNA vectors, to avoid overexposure of the EGFP signal. The same adjusted exposure time was applied to all transfection groups. At least three random regions were analyzed by fluorescence imaging. Following fluorescence imaging, the transfected cells were harvested by trypsinization (0.05 % Trypsin–EDTA), washed twice in PBS, re-suspended in 250 µl 5 % FBS-PBS, and analyzed with a BD LSRFortessa Analyzer (FACS CORE facility at the Department of Biomedicine, Aarhus University). At least 10,000 events were analyzed per sample. All flow cytometry results were analyzed with FlowJo version 10.
Fluorescence-activated cell sorting (FACS)
Fluorescence-activated cell sorting was performed using a four-laser FACSAria III cell sorter (FACS CORE facility, Department of Biomedicine, Aarhus University). Cells (HEK293T and MCF7) were transfected with X-tremeGENE 9 in 6-well plates. Briefly, the transfected cells were harvested by trypsinization 72 h post transfection, washed twice with PBS, and re-suspended in ice cold 2 % FBS-PBS. Cells were kept on ice until FACS analysis. For population sorting, the corresponding populations of cells (10,000 cells per population) were sorted into a 1.5-ml tube, followed by cell lysis and genotyping by PCR. For single-cell sorting of the C-Check and IGF1R CRISPR/Cas9 transfected cells, transfected cells in gates P1, P3, and P6 were sorted into three, one, and one 96-well plates, respectively, containing 100 µl complete culture medium supplemented with 0.005 M HEPES per well. For single-cell sorting of the C-Check and CBX5 CRISPR/Cas9 transfected cells, the corresponding populations (G1, G2, G3) of cells were single-cell sorted into one 96-well plate each. Medium was changed every 3–4 days. Cell colonies formed from single cells were ready for screening and passaging 2–3 weeks after sorting for HEK293T cells (3–4 weeks for MCF7 cells).
PCR screening of gene knockout and knockin cell clones
PCR-based screening of gene knockout and knockin cell clones in 96-well plates using cell lysates was performed as described previously [96]. Briefly, cell colonies at >60 % confluence per well in 96-well plates were washed twice with PBS, and incubated with 30 µl 0.05 % trypsin–EDTA at 37 °C for 4 min. 90 µl complete cell culture medium was added to the cells to stop trypsinization. One-third of the cells (40 µl) were transferred to a 200-µl PCR tube (or 96-well PCR plate if many clones were to be analyzed). The remaining two-thirds of the cells were seeded into two new wells of a 96-well plate, supplemented with 60 µl complete cell culture medium. The cells in the PCR tube or PCR plate were spun down at 2000 rpm for 10 min. Then, 30 µl of the supernatant was carefully removed with a transfer pipette or multichannel pipette without disturbing the cell pellet. The cell pellet was lysed by adding 30 µl cell lysis buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–Cl, pH 8.5, 0.5 % Nonidet P40, 0.5 % Tween, 400 µg/ml proteinase K) to each PCR tube. The cells were lysed at 65 °C for 30 min followed by inactivation of proteinase K at 95 °C for 10 min in a thermal cycler. One microliter of cell lysate was used for PCR-based screenings in a 25 µl PCR reaction volume.
T7E1 assay and quantification of gel
The T7 endonuclease 1 (T7E1) assay was performed as described previously [46]. Briefly, genomic DNA was isolated from primary porcine fibroblasts 48 h after nucleofection with the pIAPP TALENs using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol. PCR was carried using the Platinum® Pfx DNA Polymerase kit (Life Technologies) using 50 ng genomic DNA as template according to the manufacturer’s method. The amplicons were checked by 1 % agarose gel electrophoresis and the PCR products were extracted from the gel using a NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel). For each sample, 200 ng amplicon was diluted in 30 µl TE buffer prior to denaturation at 95 °C for 5 min and slow annealing to form heteroduplex DNA. Two-thirds of the annealed amplicon volume were treated with 5 units of T7 endonuclease 1 (NEB) at 37 °C for 20 min. The remaining 1/3 of the annealed amplicon volume was used as untreated controls. Untreated and T7E1-treated samples were analyzed by agarose gel electrophoresis (2 %). Semi-quantification of indels was performed with ImageJ.
Quantitative PCR (qPCR)
Total RNA was isolated from freshly cultured cells using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s protocol. The integrity and quantity of isolated RNA was assessed by gel electrophoresis and ND-1000 UV spectrophotometer (Nanodrop), respectively. cDNA was synthesized from 500 ng total RNA per sample with the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR (qPCR) assays were performed with the LightCycler 480 SYBR Green I Master Mix (Roche) using a LightCycler® 480 Instrument (Roche). Each qPCR reaction mix contained 2 µl of 5× diluted cDNA and a primer concentration of 500 nM. The following qPCR program was used for both CBX5 and GAPDH; one cycle of denaturation at 95 °C for 5 min followed by 45 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s, and extension at 72 °C for 10 s. At the end of the PCR assay, a melting curve was recorded with continuous acquisition of fluorescence intensity from 65 to 95 °C. The qPCR assay was performed in triplicate for all samples. Relative gene expression of CBX5 was calculated using the \(2^{{ - \Delta \Delta C_{\text{t}} }}\) method [97]. Briefly, the triplicate CBX5 Ct values in each sample were subtracted the mean Ct value of GAPDH of this sample depicted as Δ C t. The ΔΔ C t values were then calculated by subtracting the Δ C t value in each sample by the Δ C t value of the wild-type parental MCF7 cells. Fold changes in relative gene expression was calculated by \(2^{{ - \Delta \Delta C_{\text{t}} }}\). The qPCR primer efficiencies used for CBX5 and GAPDH were validated by standard curve assays. The primer sequences are provided in Additional File 4.
Western blot analysis
Western blot was performed as described previously [61]. Briefly, cells were grown to confluency in a 6-well plate and lysed with 200 µl RTK Lysis buffer containing a proteinase inhibitor cocktail. Protein concentration was measured using a NanoDrop instrument and similar protein amounts were used for further analysis. Proteins were resolved on NuPAGE®Novex® Tris–acetate protein gels and immunoblotted onto a PVDF membrane using the following antibodies: anti-IGF1R (Cell Signaling, #3027), anti-bActin (Sigma, #A5316), anti-HP1α (Millipore MAB3446, 2G9), polyclonal goat anti-rabbit HRP (DAKO, P0448), and polyclonal goat anti-mouse HRP (DAKO, P0447).
Statistical analysis
All data were represented as mean ± standard deviation. Unless stated elsewhere, one-way analysis of variance (ANOVA) with Bonferroni correction for multiple comparisons was used for all statistical analysis in this study. All statistical analyses were conducted using Stata (version 10). p values less than 0.05 were considered statistically significant.
References
Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF 3rd (2012) Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods 9:805–807
Kim YG, Chandrasegaran S (1994) Chimeric restriction endonuclease. Proc Natl Acad Sci USA 91:883–887
Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B et al (2009) Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27:851–857
Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC et al (2011) Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29:731–734
Bogdanove AJ, Voytas DF (2011) TAL effectors: customizable proteins for DNA targeting. Science 333:1843–1846
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512
Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA (2010) Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ et al (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29:143–148
Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG 2nd, Tan W, Penheiter SG, Ma AC, Leung AY et al (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491:114–118
Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L et al (2011) Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 29:695–696
Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC (2012) Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA 109:17382–17387
Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392
Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30:460–465
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. eLife 2:e00471
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L et al (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111:4632–4637
Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, Vrbanac V, Garrison BS, Stortchevoi A, Bryder D et al (2014) Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15:643–652
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M et al (2014) CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159:440–455
Shao Y, Guan Y, Wang L, Qiu Z, Liu M, Chen Y, Wu L, Li Y, Ma X, Liu M, Li D (2014) CRISPR/Cas-mediated genome editing in the rat via direct injection of one-cell embryos. Nat Protoc 9:2493–2512
Chapman KM, Medrano GA, Jaichander P, Chaudhary J, Waits AE, Nobrega MA, Hotaling JM, Ober C, Hamra FK (2015) Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Rep 10:1828–1835
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23:1233–1236
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239
Yosef I, Manor M, Kiro R, Qimron U (2015) Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA 112:7267–7272
Dickinson DJ, Ward JD, Reiner DJ, Goldstein B (2013) Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 10:1028–1034
Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10:741–743
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Jao LE, Wente SR, Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA 110:13904–13909
Hai T, Teng F, Guo R, Li W, Zhou Q (2014) One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24:372–375
Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S et al (2015) Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci 72:1175–1184
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W et al (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156:836–843
Wakayama S, Kohda T, Obokata H, Tokoro M, Li C, Terashita Y, Mizutani E, Nguyen VT, Kishigami S, Ishino F, Wakayama T (2013) Successful serial recloning in the mouse over multiple generations. Cell Stem Cell 12:293–297
Merkle FT, Neuhausser WM, Santos D, Valen E, Gagnon JA, Maas K, Sandoe J, Schier AF, Eggan K (2015) Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep 11:875–883
Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E et al (2013) A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31:251–258
Christian ML, Demorest ZL, Starker CG, Osborn MJ, Nyquist MD, Zhang Y, Carlson DF, Bradley P, Bogdanove AJ, Voytas DF (2012) Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues. PLoS One 7:e45383
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Cho SW, Kim S, Kim JM, Kim JS (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232
Duda K, Lonowski LA, Kofoed-Nielsen M, Ibarra A, Delay CM, Kang Q, Yang Z, Pruett-Miller SM, Bennett EP, Wandall HH et al (2014) High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res 42:e84
Feng Y, Zhang S, Huang X (2014) A robust TALENs system for highly efficient mammalian genome editing. Sci Rep 4:3632
Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL et al (2008) Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26:808–816
Ren C, Xu K, Liu Z, Shen J, Han F, Chen Z, Zhang Z (2015) Dual-reporter surrogate systems for efficient enrichment of genetically modified cells. Cell Mol Life Sci 72:2763–2772
Ramakrishna S, Cho SW, Kim S, Song M, Gopalappa R, Kim JS, Kim H (2014) Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat Commun 5:3378
Lim S, Wang Y, Yu X, Huang Y, Featherstone MS, Sampath K (2013) A simple strategy for heritable chromosomal deletions in zebrafish via the combinatorial action of targeting nucleases. Genome Biol 14:R69
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211
Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18:99–113
Kim H, Um E, Cho SR, Jung C, Kim H, Kim JS (2011) Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods 8:941–943
Liu Y, Lv X, Tan R, Liu T, Chen T, Li M, Liu Y, Nie F, Wang X, Zhou P et al (2014) A modified TALEN-based strategy for rapidly and efficiently generating knockout mice for kidney development studies. PLoS One 9:e84893
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82
Luo Y, Lin L, Bolund L, Sorensen CB (2014) Efficient construction of rAAV-based gene targeting vectors by Golden Gate cloning. Biotechniques 56:263–268
Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T (2007) Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25:786–793
Ochiai H, Fujita K, Suzuki K, Nishikawa M, Shibata T, Sakamoto N, Yamamoto T (2010) Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells 15:875–885
Kim HK, Kaang BK (1998) Truncated green fluorescent protein mutants and their expression in Aplysia neurons. Brain Res Bull 47:35–41
Luo Y, Lin L, Bolund L, Jensen TG, Sorensen CB (2012) Genetically modified pigs for biomedical research. J Inherit Metab Dis 35:695–713
Holm IE, Alstrup AK, Luo Y (2015) Genetically modified pig models for neurodegenerative disorders. J Pathol. doi:10.1002/path.4654
Zhang X, Cheng B, Gong H, Li C, Chen H, Zheng L, Huang K (2011) Porcine islet amyloid polypeptide fragments are refractory to amyloid formation. FEBS Lett 585:71–77
Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW (2013) Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 31:23–24
Xin J, Yang H, Fan N, Zhao B, Ouyang Z, Liu Z, Zhao Y, Li X, Song J, Yang Y et al (2013) Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS One 8:e84250
Blechingberg J, Luo Y, Bolund L, Damgaard CK, Nielsen AL (2012) Gene expression responses to FUS, EWS, and TAF15 reduction and stress granule sequestration analyses identifies FET-protein non-redundant functions. PLoS One 7:e46251
Sigal A, Danon T, Cohen A, Milo R, Geva-Zatorsky N, Lustig G, Liron Y, Alon U, Perzov N (2007) Generation of a fluorescently labeled endogenous protein library in living human cells. Nat Protoc 2:1515–1527
Zhou D, Ren JX, Ryan TM, Higgins NP, Townes TM (2004) Rapid tagging of endogenous mouse genes by recombineering and ES cell complementation of tetraploid blastocysts. Nucleic Acids Res 32:e128
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL (2015) Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33:538–542
Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196:961–971
Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24:142–153
Port F, Chen HM, Lee T, Bullock SL (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA 111:E2967–E2976
Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3(REVIEWS000):4
Zou L, Luo Y, Chen M, Wang G, Ding M, Petersen CC, Kang R, Dagnaes-Hansen F, Zeng Y, Lv N et al (2013) A simple method for deriving functional MSCs and applied for osteogenesis in 3D scaffolds. Sci Rep 3:2243
Siddle K (2011) Signalling by insulin and IGF receptors: supporting acts and new players. J Mol Endocrinol 47:R1–R10
Brandl C, Ortiz O, Rottig B, Wefers B, Wurst W, Kuhn R (2015) Creation of targeted genomic deletions using TALEN or CRISPR/Cas nuclease pairs in one-cell mouse embryos. FEBS Open Bio 5:26–35
Levenson AS, Jordan VC (1997) MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res 57:3071–3078
Norwood LE, Grade SK, Cryderman DE, Hines KA, Furiasse N, Toro R, Li Y, Dhasarathy A, Kladde MP, Hendrix MJ et al (2004) Conserved properties of HP1(Hsalpha). Gene 336:37–46
Kirschmann DA, Lininger RA, Gardner LM, Seftor EA, Odero VA, Ainsztein AM, Earnshaw WC, Wallrath LL, Hendrix MJ (2000) Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res 60:3359–3363
Ceccaldi R, Rondinelli B, D’Andrea AD (2015) Repair pathway choices and consequences at the double-strand break. Trends Cell Biol S0962–8924(15):00142–00147
Bae S, Park J, Kim JS (2014) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30:1473–1475
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42:W401–W407
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31:839–843
Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24:132–141
Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS (2012) Precision genome engineering with programmable DNA-nicking enzymes. Genome Res 22:1327–1333
Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Li Y, Park AI, Mou H, Colpan C, Bizhanova A, Akama-Garren E, Joshi N, Hendrickson EA, Feldser D, Yin H et al (2015) A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol 16:111
Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18:134–147
Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, Skarnes WC (2014) Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 11:399–402
Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32:569–576
Sung P, Klein H (2006) Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 7:739–750
Voit RA, Hendel A, Pruett-Miller SM, Porteus MH (2014) Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucleic Acids Res 42:1365–1378
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872
Lorson MA, Spate LD, Samuel MS, Murphy CN, Lorson CL, Prather RS, Wells KD (2011) Disruption of the survival motor neuron (SMN) gene in pigs using ssDNA. Transgenic Res 20:1293–1304
Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN et al (2002) Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295:1089–1092
Williams SH, Sahota V, Palmai-Pallag T, Tebbutt SJ, Walker J, Harris A (2003) Evaluation of gene targeting by homologous recombination in ovine somatic cells. Mol Reprod Dev 66:115–125
Du Y, Kragh PM, Zhang Y, Li J, Schmidt M, Bogh IB, Zhang X, Purup S, Jorgensen AL, Pedersen AM et al (2007) Piglets born from handmade cloning, an innovative cloning method without micromanipulation. Theriogenology 68:1104–1110
Luo Y, Lin L, Golas M, Sørensen CB, Bolund L (2015) Targeted porcine genome engineering with TALENs. In: Li X-Q, Jensen TG (eds) Somatic genome manipulation: advances, methods and applications, 1st edn. Springer, Berlin
Luo Y, Bolund L, Sorensen CB (2012) Pig gene knockout by rAAV-mediated homologous recombination: comparison of BRCA1 gene knockout efficiency in Yucatan and Gottingen fibroblasts with slightly different target sequences. Transgenic Res 21:671–676
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Acknowledgments
We are grateful to the FACS CORE facility (with special thanks to Charlotte Christie Petersen) at the Department of Biomedicine, Aarhus University for assistance with flow cytometry and FACS. This work was supported in part by grants from the STAR programme from the R&D Department, Novo Nordisk A/S to YL; and the Danish Research Council for Independent Research (16942) to YL; the Sapere Aude Young Research Talent prize to YL (18382); the Lundbeck Foundation (R173-2014-1105, R151-2013-14439, R126-2012-12448, R100-A9209, R173-2014-993, and R100-A9606) to YL, LB, PB, CBS, and ALN respectively; the China Scholarship Council (CSC) to YZ; the Natural Science Foundation of China (81472126) to ST and GQZ; the Toyota Foundation ALN; and the AUFF AU IDEAS Programme and The Karen Elise Jensen Foundation to CBS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
YL (2012–2014), SGR, and HD were financed by Novo Nordisk A/S. TK was financed by Gubra ApS. A patent claim is declared to the generation of a diabetes pig model based on genetic modification of the porcine IAPP gene. No other competing interests are declared by the authors.
Additional information
Y. Zhou, Y. Liu, D. Hussmann, P. Brøgger contributed equally to the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2015_2128_MOESM3_ESM.pdf
Additional File 3: Optimization of nucleofection in porcine fibroblasts. Five nucleofection reagents (P1-P5) and 15 nucleofection programs were evaluated (PDF 608 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Liu, Y., Hussmann, D. et al. Enhanced genome editing in mammalian cells with a modified dual-fluorescent surrogate system. Cell. Mol. Life Sci. 73, 2543–2563 (2016). https://doi.org/10.1007/s00018-015-2128-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-015-2128-3