Abstract
Objective
Ulcerative colitis (UC) and Crohn’s disease (CD) result from an interaction between genetic and environmental factors. Though several polymorphisms have been identified in PTPN2, their roles in the incidence of UC and CD are conflicting. This meta-analysis was aimed to clarify the impact of these polymorphisms on UC and CD risk.
Method
PubMed, EMBASE, Cochrane Library and CBM were searched until 23 July 2013 for eligible studies on three PTPN2 polymorphisms: rs2542151, rs1893217 and rs7234029. Data were extracted, and pooled odd ratios (ORs) as well as 95 % confidence intervals (95 % CIs) were calculated.
Conclusion
The meta-analysis indicated that rs2542151, rs1893217 and rs1893217 were associated with increased CD risk, while the former was associated with increased UC risk. The differences in age of onset and ethnic groups may influence the associations. Gene–gene and gene–environment interactions should be investigated in the future.
Results
Seventeen studies with 18,308 cases and 20,406 controls were included. Significant associations were found between rs2542151 polymorphism and CD susceptibility (OR = 1.22, 95 % CI, 1.15–1.30, I 2 = 32 %), as well as between rs2542151 and UC susceptibility (OR = 1.16, 95 % CI, 1.07–1.25, I 2 = 39 %). A similar result was found in Caucasians, but not in Asians. Moreover, a significant increase in CD risk for all carriers of the minor allele of rs1893217 (OR = 1.45, 95 % CI, 1.23–1.70, I 2 = 0 %) and rs7234029 (OR = 1.36, 95 % CI, 1.16–1.59, I 2 = 0 %) were found. For children, the rs1893217 polymorphism appeared to confer susceptibility to CD (OR = 1.56, 95 % CI, 1.28–1.89, I 2 = 0 %).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The major forms of inflammatory bowel disease (IBD), ulcerative colitis (UC) and Crohn’s disease (CD) are chronic inflammatory disorders of the gastrointestinal tract that have been empirically defined by clinical, pathological, endoscopic and radiological features [1]. UC is characterized by inflammation that is limited to the colon: it begins in the rectum, spreads proximally in a continuous fashion and frequently involves the periappendiceal region; being different from UC, CD involves any part of the gastrointestinal tract (most commonly the terminal ileum or the perianal region) in a non-continuous fashion, and is commonly associated with complications such as strictures, abscesses and fistulas [2, 3]. Yet, other than the fact that several genes, gut microbiota and unknown environmental factors contribute to it, little is known about its cause [4, 5].
Genome-wide association studies (GWAS) for IBD susceptibility loci performed in the last few years have been highly successful in identifying genes that contribute to disease susceptibility, identifying 99 non-overlapping genetic risk loci, including 28 that are shared between UC and CD [6, 7]. Despite distinct clinical features, approximately 30 % of IBD-related genetic loci are shared between UC and CD, indicating that these diseases engage common pathways and may be part of a mechanistic continuum [8]. Though many susceptibility loci associated with UC and CD, including ECM1, CDH1, NOD2, IBD5, IL23R, ATG16L1 and so on, have been identified, their roles in the incidence of CD are conflicting [9–16].
The family of protein tyrosine phosphatases (PTPs) plays a critical role in regulating fundamental cellular signaling events, including cell proliferation, differentiation and survival [17]. As a member of this superfamily, PTPN2 (protein tyrosine phosphatase, nonreceptor type 2), which is located at chromosome 18p11.2, is expressed in intestinal epithelial cells and helps to maintain barrier function [18, 19], and the encoded protein (T cell protein tyrosine phosphatase) is involved in the regulation of the immune system. In intestinal cells, PTPN2 regulates epithelial permeability and is a key negative regulator of important immune mediators STAT1 and STAT3, as well as p38 and ERK1/2 phosphorylation by modulating cytokine secretion and by restricting epithelial barrier defects [20, 21]. Consequently, it attenuates both IFNγ-induced and TNFα-induced signaling and release of proinflammatory cytokines. Evidence from PTPN2−/− mouse knockouts suggests that PTPN2 is a multiple regulator of the immune system, affecting hematopoiesis in several lineages and controlling systemic inflammatory responses [22, 23]. Three common polymorphisms recognized by restriction enzymes have been reported within the region of PTPN2: rs2542151 which is located 5.5 kb upstream of the PTPN2 gene; rs1893217 which is located in intron 7; and rs7234029 which are located within an intronic region.
Recently, a couple of case–control studies across multiethnic groups have assessed the associations between PTPN2 polymorphisms with UC and CD. However, the results have been proposed for the disparity, including small sample sizes, low statistical power and subjects from different ethnic backgrounds. Therefore, in order to overcome the limitations of these individual studies, resolve inconsistencies, and reduce the likelihood that random errors are responsible for false-positive or false-negative associations, we conducted a meta-analysis of the previously published studies involving the associations between PTPN2 polymorphisms with UC and CD.
Materials and methods
Search strategy
Pubmed, Embase, Cochrane Library and CBM were searched for studies focused on the relationships between the three common polymorphisms of PTPN2 and the risk of UC and CD. The latest search was done on July 23, 2013 by two independent reviewers (J.X.Z. and J.W.). The key words and subject terms used are as follows: “Crohn’s disease” or ‘‘CD’’, “ulcerative colitis” or “UC”, “inflammatory bowel disease” or “IBD”, “protein tyrosine phosphatase nonreceptor type 2” or “PTPN2”, and “genetic polymorphism” or “polymorphism” or “variant”. The reference lists in the retrieved studies were also further screened for additional eligible publications. When data were unclear or incomplete, the corresponding author was contacted to clarify data extraction.
Inclusion and exclusion criteria
Studies were included into the meta-analysis if they met the following criteria: (1) the study assessed the associations between UC or CD with at least one of the three polymorphisms; (2) controls of included studies were from a healthy population or were subjects without diseases related to IBD; (3) the study offered sufficient and complete data to calculate the number of each allele identified. Studies were excluded if: (1) the research did not study any of the three polymorphisms; (2) the design was based on family data; (3) the genotype frequency was not reported; (4) the article was a review; (5) there was insufficient information to support integrity of the data upon extraction; (6) the study was without control group.
Data extraction
The following information regarding each eligible trial was recorded: year of publication; ethnicity of subjects; sample sizes; and Hardy–Weinberg equilibrium (HWE) of control. Baseline information and data were extracted by two reviewers (J.X.Z. and J.W.) independently, using the same standard. The results were compared and disagreements were resolved by consensus.
Statistical analysis
Variant genotype frequencies were compared between cases and controls. The odds ratio (OR) and 95 % confidence interval (CI) for the heterozygote and variant homozygote were calculated as compared with the wildtype homozygote. In addition to overall comparisons, a subgroup analysis was performed based on ethnicity when adequate data were available. Between-study heterogeneity was estimated using the χ 2-based Q statistic and I 2 test. When I 2 > 50 % and P < 0.1, heterogeneity was considered statistically significant, and random effects model was used to analyze the data subsequently. On the contrary, fixed effects model was chosen. Egger’s test was used to assess the publication bias. A χ 2 test was performed to examine HWE, and P < 0.05 was considered statistically significant.
All analyses were performed by the Review Manager 5.1 and Stata 12 (Stata Corporation, College Station, Texas) software packages.
Results
Studies included in the meta-analysis
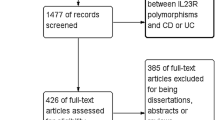
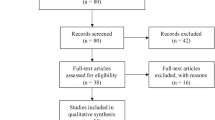
Following the searching strategy, 31 potentially relevant studies were retrieved. According to the inclusion criteria, 17 studies [24–40] with full-text were included into this meta-analysis and 14 studies were excluded (Fig. 1). Eight studies [24, 26–28, 30, 31, 33, 37] examined the association between rs2542151 polymorphisms and UC risk; 13 [25–37] studied the association between rs2542151 polymorphisms and CD risk; three [38–40] involved the association between rs1893217 polymorphisms and CD and two [30, 39] studied the association between rs7234029 and CD risk. In these studies, seven examined the association between PTPN2 polymorphisms and CD as well as UC (Table 1). The distribution of genotypes in the controls was consistent with HWE for all selected studies, except for two studies [26, 39] for rs2542151.
Associations between PTPN2 polymorphisms and CD
A summary of the meta-analysis findings concerning associations between PTPN2 polymorphisms and CD is shown in Tables 2 and 3.
Thirteen studies [25–37], which were comprised of 9,804 cases and 10,645 controls, reported the rs2542151 polymorphisms. It was found that rs2542151 polymorphism was associated with CD risk (TG vs. TT: OR = 1.19, 95 % CI, 1.12–1.27, I 2 = 25 %; GG vs. TT: OR = 1.60, 95 % CI, 1.35–1.91, I 2 = 17 %; TG + GG vs. TT: OR = 1.22, 95 % CI, 1.15–1.30, I 2 = 32 %; GG vs. TG + TT: OR = 1.53, 95 % CI, 1.28–1.82, I 2 = 8 %; G vs. T: OR = 1.22, 95 % CI, 1.15–1.28, I 2 = 30 %) (Table 2; Fig. 2). According to the subgroup analysis by ethnicity, the patients of ten studies [25, 27–33, 35, 36] were Caucasian and that of the other three [26, 34, 37] were Asian. Interestingly, rs2542151 polymorphism was associated with CD risk in Caucasians, but not in Asians (Table 3).
Three studies [38–40] including 1,296 cases and 1,668 controls examined rs1893217 polymorphisms. Significant association between rs1893217 polymorphisms and CD risk was found (TC vs. TT: OR = 1.38, 95 % CI, 1.17–1.63, I 2 = 13 %; CC vs. TT: OR = 2.39, 95 % CI, 1.49–3.82, I 2 = 0 %; TC + CC vs. TT: OR = 1.45, 95 % CI, 1.23–1.70, I 2 = 0 %; TT vs. TC + CC: OR = 2.18, 95 % CI, 1.37–3.48, I 2 = 0 %; C vs. T: OR = 1.43, 95 % CI, 1.24–1.65, I 2 = 0 %) (Table 2; Fig. 3). According to the stratified analysis by patients’ onset age, rs1893217 polymorphism was associated with CD risk for patients diagnosed during their childhood (Table 3).
There were two studies [29, 30], containing 1,452 cases and 1,496 controls, that discussed rs7234029 polymorphisms. Significant association was found between rs7234029 polymorphisms and CD risk (AG vs. AA: OR = 1.33, 95 % CI, 1.13–1.56, I 2 = 0 %; GG vs. AA: OR = 1.75, 95 % CI, 1.11–2.78, I 2 = 0 %; AG + GG vs. AA: OR = 1.36, 95 % CI, 1.16–1.59, I 2 = 0 %; GG vs. AG + AA: OR = 1.62, 95 % CI, 1.03–2.56, I 2 = 0 %; G vs. A: OR = 1.33, 95 % CI, 1.15–1.52, I 2 = 0 %) (Table 2; Fig. 4).
Associations between PTPN2 polymorphisms and UC
A summary of the meta-analysis findings concerning associations between PTPN2 polymorphisms and UC is shown in Tables 4 and 5.
Eight studies [24, 26–28, 30, 31, 33, 37], which were comprised of 5,720 cases and 6,597 controls, reported the rs2542151 polymorphisms. It was found that rs2542151 polymorphism was associated with CD risk (TG vs. TT: OR = 1.14, 95 % CI, 1.05–1.24, I 2 = 34 %; GG vs. TT: OR = 1.32, 95 % CI, 1.05–1.67, I 2 = 0 %; TG + GG vs. TT: OR = 1.16, 95 % CI, 1.07–1.25, I 2 = 39 %; GG vs. TG + TT: OR = 1.28, 95 % CI, 1.01–1.61, I 2 = 0 %; G vs. T: OR = 1.14, 95 % CI, 1.07–1.23, I 2 = 32 %) (Table 4; Fig. 5). According to the subgroup analysis by ethnicity, interestingly, rs2542151 polymorphism was associated with UC risk in Caucasians, but not in Asians (Table 5).
Test of heterogeneity and publication bias
Heterogeneity of the included studies pertaining to each polymorphism is presented in Tables 2, 3, 4 and 5. During the meta-analysis, heterogeneity was found in the subgroup analysis for Asian, but sensitivity analysis found these studies that contributed to heterogeneity did not influence the results.
For the results indicated by Egger’s Test in Tables 2 and 4, potential publication bias was only found for GG vs. TT (Egger’s Test P = 0.024) and GG vs. TG + TT (Egger’s Test P = 0.024).
Discussion
The pathogenesis of IBD relates, at least in part, to a dysregulated host response to intestinal microbiota in subjects with an underlying genetic predisposition. There is increasing evidence of global variation of susceptibility genes for IBD, and of clinically important differences in polymorphism frequency between racial and ethnic groups in the same region [41]. To date, 99 IBD susceptibility loci have been identified: 71 associated with CD, 47 with UC, and 28 with both CD and UC [15]. Among these IBD-related genes, PTPN2 is a key negative regulator of important immune mediators STAT1 and STAT3, as well as p38 and ERK1/2 phosphorylation, and helps to maintain barrier function [20, 21]. So far, several loci located in PTPN2 have been identified and the associations between them and IBD susceptibility have been evaluated in some studies. But because of the differences, including sample sizes, statistical power and ethnic backgrounds, all the conclusions are not unanimous. Therefore, performing a meta-analysis to evaluate the associations between PTPN2 polymorphisms and IBD is necessary. This meta-analysis, including 17 studies with 18,308 cases and 20,406 controls concerning rs2542151, rs1893217 and rs7234029, is the first quantitative evaluation with respect to the associations between PTPN2 polymorphisms with UC and CD risk.
Rs2542151, located 5.5 kb upstream of the PTPN2 gene, was first reported in the GWAS [30, 39]. It has been demonstrated that the presence of PTPN2 variant rs2542151 causes impaired autophagosome formation, and dysfunctional autophagy resulted in increased levels of autophagosome formation in colonic lamina propria fibroblasts derived from CD patients [19]. Our meta-analysis contained 13 studies [25–37] with 9,804 cases and 10,645 controls, and reported the association between rs2542151 polymorphism with CD; eight studies [24, 26–28, 30, 31, 33, 37] with 5,720 cases and 6,597 controls paid attention to the association between it and UC. We found that rs2542151 polymorphism was associated with UC and CD susceptibility. In other word, the risk “G” allele increased the UC and CD risk. Interesting, similar results were found in Caucasians but not in Asians. But the result for Asians should be interpreted cautiously, because only two studies (for UC) and three studies (for CD) whose patients were Asian were included in the meta-analysis.
In addition, the second major finding of this meta-analysis is that the minor allele “C” of rs1893217 was associated with increased CD risk. And in the stratified analysis, significant association between rs1893217 polymorphisms and pediatric CD was found. But due to the insufficient number of studies, the association between it and UC susceptibility was not evaluated. Rs1893217, which is located in intron 7 of PTPN2, is a potential binding site for transcription factors such as HFN4-α, PPAR-γ, and STAT6 are pertinent. HFN4-α plays a crucial role in immune homeostasis in the gastrointestinal tract [42, 43], and knockdown of HFN4-α in cells results in oxidative stress and accentuation of cellular inflammatory activation [39]. Intronic variation at single-nucleotide polymorphism (SNP) rs1893217 may hamper the binding of HFN4-α and may hinder transcription of the PTPN2 gene, which could in turn induce cellular changes that contribute to the inflammatory cascade. The presence of the rs1893217 variation also results in elevated MAPK signaling in response to the bacterial wall component, MDP, but doesn’t affect the activation of the NF-κB pathway. Moreover, elevated MAPK signaling results in elevated mRNA expression of T-bet transcription factor and increased IFN-γ secretion, which contributes to an aberrant, out-of-control inflammatory state in the intestine.
In this meta-analysis, two studies [26, 27] containing 1,452 cases and 1,496 controls discussed the association between rs7234029 polymorphisms and CD. The result indicated that the risk allele “G” of rs7234029 was associated with increased CD susceptibility. Regretfully, because of insufficient number of studies, the association between it and UC susceptibility was not evaluated. Like rs1893217, intronic variation at SNP rs7234029 also hampers the binding of important transcription factors, such as OCT-1, NFIL3, CEBP-b, and EVI1, independently and then contributes to the inflammatory cascade.
Though we attempted to minimize the likelihood of bias by developing a detailed protocol before initiating the study, some insurmountable limitations of this meta-analysis may affect the results and even the subsequent conclusions. First, because of incomplete raw data or publication limitations, several relevant studies could not be included in this meta-analysis. Second, the numbers of subjects and studies included in the meta-analysis were small, and may not have been sufficient to reveal the associations between rs1893217 and rs7234029 with UC and CD. Third, most of the included studies were hospital-based rather than population-based, which makes results more prone to potential selection bias. Fourth, because not all necessary information could be obtained from most included studies, relevant stratifications could not be made for many studies.
In conclusion, this meta-analysis summarized the associations between PTPN2 polymorphisms and UC and CD susceptibility. We found the minor allele “G” of rs2542151 was associated with both increased UC and CD risk, especially in Caucasians, but not in Asians. Moreover, a significant increase in CD risk for all carriers of the minor allele of rs1893217 and rs7234029 were found. For children, the rs1893217 polymorphism appeared to confer susceptibility to CD. Because IBD-related genetic factors may interfere with non-genetic risk factors, such as environment and diet, future studies should therefore be stratified in these ways. In addition, gene–gene and gene–environment interactions should be investigated in the future.
References
Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347(6):417–29.
Leone V, Chang EB, Devkota S. Diet, microbes, and host genetics: the perfect storm in inflammatory bowel diseases. J Gastroenterol. 2013;48(3):315–21.
Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, et al. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307(5710):734–8.
Hammer HF. Gut microbiota and inflammatory bowel disease. Dig Dis. 2011;29(6):550–3.
Simmons A. Crohn’s disease: Genes, viruses and microbes. Nature. 2010;466(7307):699–700.
Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–25.
Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–52.
Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307–17.
Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411(6837):603–6.
Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–3.
Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39(2):207–11.
Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3(4):e58.
Silverberg MS, Duerr RH, Brant SR, Bromfield G, Datta LW, Jani N, et al. Refined genomic localization and ethnic differences observed for the IBD5 association with Crohn’s disease. Eur J Hum Genet. 2007;15(3):328–35.
Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet. 2005;14(22):3499–506.
Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60(12):1739–53.
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78.
Tonks NK, Neel BG. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr Opin Cell Biol. 2001;13(2):182–95.
Scharl M, Paul G, Weber A, Jung BC, Docherty MJ, Hausmann M, et al. Protection of epithelial barrier function by the Crohn’s disease associated gene protein tyrosine phosphatase n2. Gastroenterology. 2009;137(6):2030–40.
Scharl M, Wojtal KA, Becker HM, Fischbeck A, Frei P, Arikkat J, et al. Protein tyrosine phosphatase nonreceptor type 2 regulates autophagosome formation in human intestinal cells. Inflamm Bowel Dis. 2012;18(7):1287–302.
Scharl M, McCole DF, Weber A, Vavricka SR, Frei P, Kellermeier S, et al. Protein tyrosine phosphatase N2 regulates TNFα-induced signalling and cytokine secretion in human intestinal epithelial cells. Gut. 2011;60(2):189–97.
Scharl M, Hruz P, McCole DF. Protein tyrosine phosphatase non-receptor Type 2 regulates IFN-γ-induced cytokine signaling in THP-1 monocytes. Inflamm Bowel Dis. 2010;16(12):2055–64.
You-Ten KE, Muise ES, Itié A, Michaliszyn E, Wagner J, Jothy S, et al. Impaired bone marrow microenvironment and immune function in T cell protein tyrosine phosphatase-deficient mice. J Exp Med. 1997;186(5):683–93.
Heinonen KM, Nestel FP, Newell EW, Charette G, Seemayer TA, Tremblay ML, et al. T-cell protein tyrosine phosphatase deletion results in progressive systemic inflammatory disease. Blood. 2004;103(9):3457–64.
Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat Genet. 2008;40(6):710–2.
Jung C, Colombel JF, Lemann M, Beaugerie L, Allez M, Cosnes J, et al. Genotype/phenotype analyses for 53 Crohn’s disease associated genetic polymorphisms. PLoS ONE. 2012;7(12):e52223.
Lv C, Yang X, Zhang Y, Zhao X, Chen Z, Long J, et al. Confirmation of three inflammatory bowel disease susceptibility loci in a Chinese cohort. Int J Colorectal Dis. 2012;27(11):1465–72.
Waterman M, Xu W, Stempak JM, Milgrom R, Bernstein CN, Griffiths AM, et al. Distinct and overlapping genetic loci in Crohn’s disease and ulcerative colitis: correlations with pathogenesis. Inflamm Bowel Dis. 2011;17(9):1936–42.
Yu W, Hegarty JP, Berg A, Kelly AA, Wang Y, Poritz LS, et al. PTPN2 is associated with Crohn’s disease and its expression is regulated by NKX2-3. Dis Markers. 2012;32(2):83–91.
Morgan AR, Han DY, Huebner C, Lam WJ, Fraser AG, Ferguson LR, et al. PTPN2 but not PTPN22 is associated with Crohn’s disease in a New Zealand population. Tissue Antigens. 2010;76(2):119–25.
Glas J, Wagner J, Seiderer J, Olszak T, Wetzke M, Beigel F, et al. PTPN2 gene variants are associated with susceptibility to both Crohn’s disease and ulcerative colitis supporting a common genetic disease background. PLoS ONE. 2012;7(3):e33682.
Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40(6):713–5.
Peter I, Mitchell AA, Ozelius L, Erazo M, Hu J, Doheny D, et al. Evaluation of 22 genetic variants with Crohn’s disease risk in the Ashkenazi Jewish population: a case-control study. BMC Med Genet. 2011;12:63.
Latiano A, Palmieri O, Latiano T, Corritore G, Bossa F, Martino G, et al. Investigation of multiple susceptibility loci for inflammatory bowel disease in an Italian cohort of patients. PLoS ONE. 2011;6(7):e22688.
Yamazaki K, Takahashi A, Takazoe M, Kubo M, Onouchi Y, Fujino A, et al. Positive association of genetic variants in the upstream region of NKX2-3 with Crohn’s disease in Japanese patients. Gut. 2009;58(2):228–32.
Weersma RK, Stokkers PC, Cleynen I, Wolfkamp SC, Henckaerts L, Schreiber S, et al. Confirmation of multiple Crohn’s disease susceptibility loci in a large Dutch-Belgian cohort. Am J Gastroenterol. 2009;104(3):630–8.
Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39(7):830–2.
Chen ZY. Correlation between gene polymorphism of IL23R, PTPN2, 10q and inflammatory bowel disease. Guangzhou: Southern Medical University; 2008.
Amre DK, Mack DR, Morgan K, Israel D, Deslandres C, Seidman EG, et al. Susceptibility loci reported in genome-wide association studies are associated with Crohn’s disease in Canadian children. Aliment Pharmacol Ther. 2010;31(11):1186–91.
Marcil V, Mack DR, Kumar V, Faure C, Carlson CS, Beaulieu P, et al. Association between the PTPN2 gene and Crohn’s disease: dissection of potential causal variants. Inflamm Bowel Dis. 2013;19(6):1149–55.
Scharl M, Mwinyi J, Fischbeck A, Leucht K, Eloranta JJ, Arikkat J, et al. Crohn’s disease-associated polymorphism within the PTPN2 gene affects muramyl-dipeptide-induced cytokine secretion and autophagy. Inflamm Bowel Dis. 2012;18(5):900–12.
Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, et al. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–5.
Marcil V, Seidman E, Sinnett D, Boudreau F, Gendron FP, Beaulieu JF, et al. Modification in oxidative stress, inflammation, and lipoprotein assembly in response to hepatocyte nuclear factor 4alpha knockdown in intestinal epithelial cells. J Biol Chem. 2010;285(52):40448–60.
UK IBD Genetics Consortium, Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41(12):1330–4.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Ian Ahnfelt-Rønne.
J. X. Zhang and J. H. He contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, JX., He, JH., Wang, J. et al. Associations between PTPN2 polymorphisms and susceptibility to ulcerative colitis and Crohn’s disease: a meta-analysis. Inflamm. Res. 63, 71–79 (2014). https://doi.org/10.1007/s00011-013-0673-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-013-0673-5