Abstract
Objective
Published studies on the association between NCF4 rs4821544T/C polymorphism and inflammatory bowel disease (IBD) risk in Caucasian have yielded conflicting results. The present study aimed to provide more reliable conclusions by conducting a meta-analysis.
Methods
All eligible studies were extracted from Wiley Online Library, Chinese National Knowledge Infrastructure and PubMed databases. Odds ratios (ORs) with 95 % confidence intervals (CIs) were used to assess the associations between rs4821544T/C polymorphism and IBD risk in Caucasian.
Results
A total of 13 case–control studies comprising 7441 Crohn’s disease (CD) patients, 2565 ulcerative colitis (UC) patients and 8315 controls were included in this meta-analysis. Significant associations were found between CD and the rs4821544T/C polymorphism in three genetic models (C vs T: OR = 1.11, 95 % CI: 1.06, 1.16, P = 0.000; CC vs TT: OR = 1.31, 95 % CI: 1.18, 1.45, P = 0.000; CC/TC vs TT: OR = 1.07, 95 % CI: 1.01, 1.13, P = 0.014; CC vs TC/TT: OR = 1.28, 95 % CI: 1.16, 1.42, P = 0.000). However, significant associations were not found in UC under any genetic models (C vs T: OR = 1.04, 95 % CI: 0.97, 1.11, P = 0.264; CC vs TT: OR = 1.10, 95 % CI: 0.93, 1.30, P = 0.284; TC vs TT: OR = 1.04, 95 % CI: 0.95, 1.13, P = 0.429; CC/TC vs TT: OR = 1.04, 95 % CI: 0.95, 1.13, P = 0.390; CC vs TC/TT: OR = 1.07, 95 % CI: 0.91, 1.26, P = 0.409).
Conclusion
This meta-analysis suggested that the rs4821544T/C polymorphism was associated with CD, but not UC in Caucasian.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inflammatory bowel disease (IBD), a chronic non-specific gastrointestinal inflammatory disease, is typically classified into two clinical forms: Crohn’s disease (CD) and ulcerative colitis (UC). Recently, the incidence of UC and CD has increased overall in Europe from 6.0/100,000 person-years in UC and 1.0 per 100,000 person-years in CD in 1962 to 9.8/100,000 person-years and 6.3 per 100,000 person-years in 2010, respectively [1]. Of note, there is wide geographic variability in the incidence and prevalence of IBD [1]. IBD seriously affects quality of life by causing abdominal pain, vomiting, diarrhea, and other extra-intestinal symptoms [2]. It has been established that IBD is associated with both an increased risk of colorectal cancer and cardiovascular disease [3, 4]. Therefore, patients with IBD carry a slightly higher risk of dying than the general population [3–5].
Epidemiological data obtained from previous studies supports the underlying etiology of IBD is thought to be multifactorial [1]. Environmental factors including the composition of the gut microbiota, dietary fiber, saturated fats, depression and even impaired sleep act as an essential player in IBD [1]. Since CARD15 was initially identified as a candidate gene for CD in 2001 [6], people come to realize that IBD also arises as a result of a genetic predisposition [7]. Oxidative stress resulted from excessive reactive oxygen species (ROS) in the intestinal tract is also regarded as another major factor contributing to the pathogenesis and progression of gastrointestinal inflammation in IBD [8]. Nicotinamide adenine dinucleotide phosphate oxidase (NADPH oxidase) enzyme family often acts as catalyzer of the production ROS [8]. Genes encoding the NADPH complex consisted of CYBB, CYBA, NCF4, NCF2 and RAC2. The NCF4, encoding for the p40-phox protein, located on chromosome 22 [9]. It has been found that genetic polymorphisms of NADPH oxidase including NCF4 modulate subunit expression and enzyme activity [10]. Previous studies indicated that variation in NCF4 was associated with colorectal cancer and rheumatoid arthritis risk [11, 12]. Therefore, variation in NCF4 may influence susceptibility to IBD. Recently, a North American genome-wide association study identified variant in NCF4 as being associated with CD [13]. Considerable efforts have been devoted to investigating the relationship between NCF4 polymorphism and IBD risk in Caucasian [13–21]. However, results from previous studies have been inconsistent. Therefore, we designed a meta-analysis to better clarify the relationship between NCF4 polymorphism and IBD risk in Caucasian.
Materials and methods
Search strategy
Genetic association studies regarding associations between rs4821544T/C polymorphism and IBD risk were searched in Wiley Online Library, PubMed and Chinese National Knowledge Infrastructure (CNKI) databases. Different combinations of the search terms were as follows: “Inflammatory bowel disease”, “IBD”, “Crohn’s disease”, “ulcerative colitis”, ‘‘CD’’, ‘‘UC’’, and “NCF4”, “rs4821544”, and “polymorphism” or “variant” or “mutation”. Search results were restricted to human populations and articles written in English or Chinese (up to May 20, 2015). Moreover, additional studies were identified by a full manual search from the reference of selected papers on this topic.
Criteria for inclusion and exclusion
The studies eligible should meet the following inclusion criteria: (1) case–control studies; (2) studies should be related to association between rs4821544T/C polymorphism and IBD; (3) studies that clearly describe IBD diagnoses and the sources of cases and controls; (4) enough information to calculate the odds ratio (OR) with 95 % confidence interval (CI); (5) studies in which the genotype distribution of the control population was in Hardy–Weinberg equilibrium (HWE); (6) subjects in all studies included must be Caucasians. Accordingly, the exclusion criteria were as follows: (1) duplicated studies; (2) not case–control study or family-based case–control study; (3) studies containing overlapping data; (4) investigations of the associations of other genes with IBD or the relationships between NCF4 gene polymorphisms and other diseases; (5) subjects in all studies included are not Caucasians; (6) studies classified as review, case reports, animal or cell studies; (7) insufficient information to calculate OR and 95 % CI.
Data extraction
Two investigators independently extracted all the following data from the eligible studies: the name of first author, country of study, year of publication, the number of cases and controls, minor allele frequencies (MAF) in cases and controls.
Statistical analysis
To assess the association between rs4821544T/C polymorphism and IBD risk, ORs and its 95 % CI were calculated in four distinct genetic models: homozygote comparison, heterozygote comparison, dominant model and recessive model. Heterogeneity between studies was calculated by χ2-based Q test and I 2 test. If the data showed no heterogeneity (P > 0.10, I 2 < 50 %), Mantel–Haenszel fixed effect model was used [22], or else DerSimonian–Laird random effect model was used [22, 23]. The significance of the pooled OR was determined by the Z test. Publication bias was assessed by the shapes of the Begg’s funnel plots and Egger’s test [24]. The HWE in control group was assessed by χ2 goodness of fit. When heterogeneity between studies existed, subgroup analysis according to study characteristics should be performed [25]. All reported P values were two sided with P < 0.05 being considered as significant. STATA package version 11.0 (Stata Corporation, College Station, Texas) was used in our study.
Results
Main characteristics of eligible studies
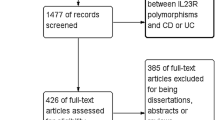
Figure 1 shows the flowchart of study selection for this meta-analysis. In total, 24 relevant records were identified. After initial examination of titles and abstracts, 11 records were excluded because they were not related to NCF4 (N = 6) or other unrelated disease (N = 5). After further screening full records, 4 records were excluded because they were not case–control studies (N = 3) or review (N = 1). Thus, a total of 9 records comprising 13 case–control studies were identified [13–21].
The studies were published from 2007 to 2013. 7441 CD patients, 2565 UC patients and 8315 controls were included in 9 and 4 case–control studies in 9 records. The characteristics of studies included in the meta-analysis are shown in Table 1. The genotype distribution of the control group in all studies was consistent with HWE (Table 1).
Associations between rs4821544T/C and CD
For rs4821544T/C polymorphism, 9 case–control studies with 7441 cases and 8 315 controls were identified. Significant associations were found under three genetic models (C vs T: OR = 1.11, 95 % CI: 1.06, 1.16, P = 0.000; CC vs TT: OR = 1.31, 95 % CI: 1.18, 1.45, P = 0.000 (Fig. 2 ); CC/TC vs TT: OR = 1.07, 95 % CI: 1.01, 1.13, P = 0.014 (Fig. 3 ); CC vs TC/TT: OR = 1.28, 95 % CI: 1.16, 1.42, P = 0.000 (Fig. 4). However, no significant associations was found under the genetic model CC vs TT (OR = 1.05, 95 % CI: 0.95, 1.12, P = 0.085).
Associations between rs4821544T/C and UC
For rs4821544T/C polymorphism, 4 case–control studies with 2565 cases and 4624 controls were identified. No significant association was found in any of the genetic models (C vs T: OR = 1.04, 95 % CI: 0.97, 1.11, P = 0.264; CC vs TT: OR = 1.10, 95 % CI: 0.93, 1.30, P = 0.284; TC vs TT: OR = 1.04, 95 % CI: 0.95, 1.13, P = 0.429; CC/TC vs TT: OR = 1.04, 95 % CI: 0.95, 1.13, P = 0.390; CC vs TC/TT: OR = 1.07, 95 % CI: 0.91, 1.26, P = 0.409).
Publication bias
Begg’s funnel plot and Egger’s test were applied to assess the publication bias of the included studies. The results of Egger’s test and the shapes of the funnel plots for all of the polymorphism indicated that there was no publication bias in the meta-analysis for the association between the NCF4 polymorphism and CD (Fig. 5; Table 2).
Discussion
Although the pathogenesis of IBD was not fully elucidated, IBD is widely regarded to be product of interaction between intestinal microbial flora and genetic predisposition(s) [26]. As we know, phagocytic leukocytes (especially neutrophil) are critical for bacterial killing by ROS produced from NOX2 NADPH oxidase complex [27]. Interestingly, a very recent study indicates patients with chronic granulomatous disease, an inflammatory colitis indistinguishable from CD, have a mutated NADPH complex and are therefore deficient in ROS production [28]. NADPH complex was comprised with many subunits [28], of which p40-phox encoded by NCF4 was widely studied [29, 30]. Neutrophils collected from a p40phox-/− mice exhibit severe defects in NADPH oxidase regulation and oxidant-dependent bacterial killing [27]. The studies mentioned above indicated p40(phox) may act as an essential player in IBD.
From a genetic prospective, variant in NCF4 may lead to functional alterations in granulocyte ROS production [31]. Recently, several reported genetic markers of NCF4 have been suggested to have effects on rheumatoid arthritis identifies [12] whose genetic predisposition is partly similar to IBD [32]. To date, considerable efforts have been devoted to investigating the relationship between NCF4 polymorphism and IBD risk. However, the results of existing studies are inconsistent. Given that subjects in all studies included are Caucasian, we designed a meta-analysis to better clarify the relationship between rs4821544T/C polymorphism and IBD risk in Caucasian.
To our knowledge, this is the first meta-analysis to comprehensively assess the associations between rs4821544T/C polymorphism and IBD risk in Caucasian. A total of 13 case–control studies comprising 7441 CD patients, 2565 UC patients and 8315 controls were included in this meta-analysis. Our results suggest that the rs4821544T/C polymorphism is associated with CD, but not UC in Caucasian. To date, there are limited data on how NCF4 may functionally influence IBD susceptibility. A possible mechanism is that rs4821544 influences ROS production [14] and consequently impairs bacterial-killing capacity [27], which leads to alteration of intestinal flora.
Of note, there are several genome-wide association studies (GWASs) included in our meta-analysis [13, 17, 21]. GWASs, aimed at increasing g the power of studies by combining the results from different study populations, have led to the identification of novel associations that would not otherwise have been identified in individual studies with small sample. Most results of GWASs also were consistent with our meta-analysis.
Meta-analysis has been considered an effective tool to comprehensively estimate the effect of selected genetic polymorphism on disease risk [33, 34]. Publication bias, also known as the “file-drawer problem”, can be considered a major drawback of meta-analyses by compromising their validity [35]. No significant publication bias was detected in all analyses. Heterogeneity, another important issue when performing a meta-analysis, was also not found in also analyses. Furthermore, sample size in each study is relatively larger. Based on the above analyses, our results are relatively scientific and reliable.
As in other systematic reviews and meta-analyses, some limitations require careful consideration. First, there was a lack data available, and we did not analyze the association between rs4821544T/C polymorphism and different histological subtypes, which requires further investigation. Second, only genetic factors were under consideration. A more precise analysis should be based on additional factors including smoking, drinking and other environmental factors. Last but not least, our studies only involved Caucasian. Therefore, representativeness bias of the study was unavoidable.
In conclusion, our meta-analysis suggested that the rs4821544T/C polymorphism was associated with CD, but not UC in Caucasian. Considering there were some limitations in our study, further well-designed case–control studies are necessary to validate the results of our present meta-analysis.
References
Burisch J, Munkholm P. The epidemiology of inflammatory bowel disease. Scand J Gastroenterol. 2015;50(8):942–51. doi:10.3109/00365521.2015.1014407.
Singh S, Blanchard A, Walker JR, Graff LA, Miller N, Bernstein CN. Common symptoms and stressors among individuals with inflammatory bowel diseases. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2011;9(9):769–75. doi:10.1016/j.cgh.2011.05.016.
Romberg-Camps M, Kuiper E, Schouten L, Kester A, Hesselink-van de Kruijs M, Limonard C, et al. Mortality in inflammatory bowel disease in the Netherlands 1991–2002: results of a population-based study: the IBD South-Limburg cohort. Inflamm Bowel Dis. 2010;16(8):1397–410. doi:10.1002/ibd.21189.
Manninen P, Karvonen AL, Huhtala H, Rasmussen M, Salo M, Mustaniemi L, et al. Mortality in ulcerative colitis and Crohn’s disease. A population-based study in Finland. J Crohn’s Colitis. 2012;6(5):524–8. doi:10.1016/j.crohns.2011.10.009.
Ekbom A, Helmick CG, Zack M, Holmberg L, Adami HO. Survival and causes of death in patients with inflammatory bowel disease: a population-based study. Gastroenterology. 1992;103(3):954–60.
McGovern DP, van Heel DA, Ahmad T, Jewell DP. NOD2 (CARD15), the first susceptibility gene for Crohn’s disease. Gut. 2001;49(6):752–4.
Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369(9573):1627–40. doi:10.1016/s0140-6736(07)60750-8.
Lam G, Apostolopoulos V, Zulli A, Nurgali K. NADPH oxidases and inflammatory bowel disease. Curr Med Chem. 2015;22(17):2100–9.
Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40(8):955–62. doi:10.1038/ng.175.
Schirmer M, Hoffmann M, Kaya E, Tzvetkov M, Brockmoller J. Genetic polymorphisms of NAD(P)H oxidase: variation in subunit expression and enzyme activity. Pharmacogenomics J. 2008;8(4):297–304. doi:10.1038/sj.tpj.500467.
Ryan BM, Zanetti KA, Robles AI, Schetter AJ, Goodman J, Hayes RB, et al. Germline variation in NCF4, an innate immunity gene, is associated with an increased risk of colorectal cancer. In J Cancer. 2014;134(6):1399–407. doi:10.1002/ijc.28457.
Olsson LM, Lindqvist AK, Kallberg H, Padyukov L, Burkhardt H, Alfredsson L, et al. A case–control study of rheumatoid arthritis identifies an associated single nucleotide polymorphism in the NCF4 gene, supporting a role for the NADPH-oxidase complex in autoimmunity. Arthritis Res Ther. 2007;9(5):R98. doi:10.1186/ar2299.
Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi:10.1038/ng2032.
Amre DK, Mack DR, Israel D, Krupoves A, Costea I, Lambrette P, et al. NELL1, NCF4, and FAM92B genes are not major susceptibility genes for Crohn’s disease in Canadian children and young adults. Inflamm Bowel Dis. 2012;18(3):529–35. doi:10.1002/ibd.21708.
Eglinton TW, Roberts R, Pearson J, Barclay M, Merriman TR, Frizelle FA, et al. Clinical and genetic risk factors for perianal Crohn’s disease in a population-based cohort. Am J Gastroenterol. 2012;107(4):589–96. doi:10.1038/ajg.2011.437.
Glas J, Seiderer J, Pasciuto G, Tillack C, Diegelmann J, Pfennig S, et al. Rs224136 on chromosome 10q21.1 and variants in PHOX2B, NCF4, and FAM92B are not major genetic risk factors for susceptibility to Crohn’s disease in the German population. Am J Gastroenterol. 2009;104(3):665–72. doi:10.1038/ajg.2008.65.
Muise AM, Xu W, Guo CH, Walters TD, Wolters VM, Fattouh R, et al. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut. 2012;61(7):1028–35. doi:10.1136/gutjnl-2011-300078.
Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn’s disease in a population-based cohort. Genes Immun. 2008;9(6):561–5. doi:10.1038/gene.2008.49.
Jung C, Colombel JF, Lemann M, Beaugerie L, Allez M, Cosnes J, et al. Genotype/phenotype analyses for 53 Crohn’s disease associated genetic polymorphisms. PLoS One. 2012;7(12):e52223. doi:10.1371/journal.pone.0052223.
Nasir BF, Griffiths LR, Nasir A, Roberts R, Barclay M, Gearry RB, et al. An envirogenomic signature is associated with risk of IBD-related surgery in a population-based Crohn’s disease cohort. J Gastrointest Surg Off J Soc Surg Aliment Tract. 2013;17(9):1643–50. doi:10.1007/s11605-013-2250-1.
Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40(6):713–5. doi:10.1038/ng.148.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical Research ed). 1997;315(7109):629–34.
Scholten RJ, Assendelft WJ, Kostense PJ, Bouter LM. The practice of systematic reviews. V. Heterogeneity between studies and subgroup analysis. Ned Tijdschr Geneeskd. 1999;143(16):843–8.
Gkouskou KK, Deligianni C, Tsatsanis C, Eliopoulos AG. The gut microbiota in mouse models of inflammatory bowel disease. Front Cell Infect Microbiol. 2014;4:28. doi:10.3389/fcimb.2014.00028.
Ellson CD, Davidson K, Ferguson GJ, O’Connor R, Stephens LR, Hawkins PT. Neutrophils from p40phox-/- mice exhibit severe defects in NADPH oxidase regulation and oxidant-dependent bacterial killing. J Exp Med. 2006;203(8):1927–37. doi:10.1084/jem.20052069.
van de Veerdonk FL, Dinarello CA. Deficient autophagy unravels the ROS paradox in chronic granulomatous disease. Autophagy. 2014;10(6):1141–2. doi:10.4161/auto.28638.
Li ZY, Jiang WY, Cui ZJ. An essential role of NAD(P)H oxidase 2 in UVA-induced calcium oscillations in mast cells. Photochem Photobiol Sci Off J Eur Photochem Assoc Eur Soc Photobiol. 2015;14(2):414–28. doi:10.1039/c4pp00304g.
Munnamalai V, Weaver CJ, Weisheit CE, Venkatraman P, Agim ZS, Quinn MT, et al. Bidirectional interactions between NOX2-type NADPH oxidase and the F-actin cytoskeleton in neuronal growth cones. J Neurochem. 2014;130(4):526–40. doi:10.1111/jnc.12734.
Somasundaram R, Deuring JJ, van der Woude CJ. Peppelenbosch MP, Fuhler GM. Linking risk conferring mutations in NCF4 to functional consequences in Crohn’s disease. Gut 2012;61(7):1097; author reply -8. doi:10.1136/gutjnl-2011-301344.
Wellcome Trust Case Control Consortium. Genome-wide association study of 14000 cases of seven common diseases and 3000 shared controls. Nature 2007;447(7145):661–78.
Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45(2):202–7. doi:10.1038/ng.2520.
Lu XC, Tao Y, Wu C, Zhao PL, Li K, Zheng JY, et al. Association between variants of the autophagy related gene—IRGM and susceptibility to Crohn’s disease and ulcerative colitis: a meta-analysis. PLoS One. 2013;8(11):e80602. doi:10.1371/journal.pone.0080602.
Siddiqi N. Publication bias in epidemiological studies. Cent Eur J Public Health. 2011;19(2):118–20.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: John Di Battista.
P.-B. Wu, J.-F. Dai and Q. Wang contribute equally to this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, PB., Dai, JF., Wang, Q. et al. Association between NCF4 rs4821544T/C polymorphism and inflammatory bowel disease risk in Caucasian: a meta-analysis. Inflamm. Res. 64, 825–831 (2015). https://doi.org/10.1007/s00011-015-0866-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-015-0866-1