Abstract
Using the Raman spectroscopic analysis system that gives the chemical information of the biomaterials, classification is performed through the acquisition of fingerprint signals for each cell line, and the basis of the diagnosis is provided. The origin of diagnosis can be clarified by precise analysis through comparison of local signals and morphology in cells, including measurement at tissue level. In this result, normal breast cell line (MCF-10A) and breast cancer cell lines (MDA-MB-231, MDA-MB-453) were characterized using Raman spectroscopy, atomic force microscopy (AFM), and optical microscopy. These three modalities were combined in order to not only separate cancerous and noncancerous cell lines but to analyze their morphological and optical properties. From the results, the inherent optical properties of cancer cells separated from normal cells in terms of local variation were observed. Bright-field (BF) transmission imaging is also compared to the morphological height difference obtained from AFM and is correlated with surface Raman spectra.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Current cancer detection and diagnosis is accomplished through various processes such as clinical symptoms, medical imaging, biopsy, pathological analysis, and surgery. Histopathologic analysis, especially including immunohistochemical staining, can be relied on at the molecular level, such as structure and concentration, but has disadvantages such as invasiveness, cost, and time-consuming (Rehman et al. 2007). There is a diagnostic method using spectroscopy as an approach to overcome the disadvantages of the pathological analysis method. Such vibration spectroscopy diagnosis is noninvasive and has advantages such as not destroying biological material and requiring no additional labeling (Oshima et al. 2010; Singh et al. 2012; Gautam et al. 2015). Detection of molecular vibrational spectra such as infrared (IR) and Raman spectroscopy has been widely applied in examining chemical and molecular changes in biological samples (Haka et al. 2005, 2009; Saha et al. 2011), and the spectra can fingerprint biomolecule composition without the need for additional labeling or sample separation (Kamemoto et al. 2010; Abramczyk et al. 2012).
Recently, Raman spectroscopy has been reassessed as a measurement method for biological samples of the breast, lungs, stomach, and other organs (Larraona-Puy et al. 2009; Sato et al. 2009; Kirsch et al. 2010; Feng et al. 2011; Kawabata et al. 2011; Larraona-Puy et al. 2011). Furthermore, Raman spectroscopy has been studied for the verification of biological samples of various sizes, such as tissue (Kawabata et al. 2011; Ishigaki et al. 2016), cells (Carvalho et al. 2015; Smith et al. 2016; Talari et al. 2015), bacteria (Liu et al. 2011; Zhou et al. 2014), and protein (Lyng et al. 2007; Maiti et al. 2004; Rygula et al. 2013). The Raman method is based on the inelastic scattering of photons incident on the sample to be measured. When a single energy laser beam is injected into the sample, the energy of a part of the incoming photons is changed by the molecular vibration of the sample. This change is called Stokes shift and the spectral representation of Stokes-shifted photon energy distribution is a Raman spectroscopic signal. Since the initial energy is diverted by various molecular vibration modes, individual peaks in the spectrum include structural information of molecules. Thus, the Raman measurement method that incorporates this chemical information is a great tool for identifying biomaterials such as cancer tissues and cells (Li et al. 2010; Neugebauer et al. 2010; Shapiro et al. 2011; Abramczyk et al. 2011; McQueenie et al. 2012; Marro et al. 2014).
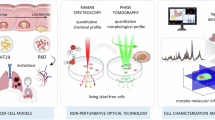
In this study, we compared and analyzed the Raman signal according to the cell surface position through the synchronization between the optical image with high spatial resolution and the measurement result of surface roughness at the nanometer level as shown in Fig. 1. The topography of the cells provided quantitative thickness and quantity indicating the relationship between local Raman spectrum and intensity (Lee et al. 2018). By introducing local RGB intensity in optical measurements, we have identified additional factors that normal cells can use to differentiate cancer cells. The optical microscope shows an image of transmitted light, and the intensity of each pixel in RGB represents an internal event such as absorption and scattering. We compared the optical signals according to the differences in the intracellular heights. As a result, the inelastic scattering Raman and the transmission type optical microscope confirmed that there was rare local variation in the cancer cells, unlike the normal cells.
Schematic diagram showing the examination of cell line using spectroscopy, tomography, and optical signals. Reprinted from Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 205, Lee et al., “Local-dependency of morphological and optical properties between breast cancer cell lines,” p. 132–138, Copyright 2018, with permission from Elsevier
Raman Measurement Mechanism and System Configuration
Raman method is a spectroscopic analysis that measures the inelastic scattering of the photons of an incident single-wavelength laser. In 1928, Raman spectroscopy was carried out to investigate the effect of light, which is different from the energy of light incident upon reflection or scattering. The emission of these other energies is determined by the energy exchange with the molecule vibration that forms between the light and matter, and the difference can be expressed as wavenumber (cm−1) or wavelength (nm) as shown below:
where ΔE is the energy difference, h is Planck’s constant, c is the speed of light, κ is the wavenumber, ν is the frequency, and λ is the wavelength. From this representation of quantum mechanical light, it is possible to explain the difference between the Rayleigh scattering, the Stokes (anti-Stokes) in Raman scattering, and the fluorescence as shown in Fig. 2a. These inelastic scattering and fluorescence signals sometimes interfere with each other (Fig. 2b). Most biomaterials appear as spectra with multiple peaks because of inelastic scattering and emission of fluorescence signals in various forms. On the other hand, these peaks have the advantage of being able to specify a sample like a fingerprint because it provides a parameter, such as the position of the peak and the relative intensity between the various peaks, in a narrow form.
(a) Schematic illustrating the interaction of incident light with materials such as Rayleigh scattering, Raman scattering and fluorescence to show the Raman spectroscopic mechanism. (b) Schematic representation of the spectrum formed by Raman inelastic scattering and fluorescence. (c) Conventional Raman system configuration. © 2020 the authors
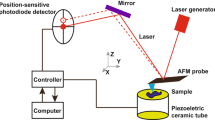
The configuration of a typical Raman system consists of a confocal microscope, a laser source, a monochromator, and a detector as shown in Fig. 2 (c). The Raman signal is obtained by a spectroscope measuring the intensity of an inelastically scattered photon in a sample from a short-wavelength laser (785 nm). 514 nm, 632 nm, and 785 nm, it is possible to improve the specificity of the sample by installing the Raman apparatus of the wavelength variable system because the signal is different depending on the wavelength of the laser used (Synytsya et al. 2014). Since hemoglobin in blood has a large absorption spectrum in the wavelength range of 200–700 nm, it can be seen that the wavelength selection of the incident light is an important issue according to the characteristics of the target sample. By enlarging the scan area through Raman mapping, it is possible to cover a wide range of areas, thereby increasing measurement sensitivity and reliability. In mapping, it is possible to raise the spot size of the beam by reducing the magnification of the lens for a close scan of the area of interest. As a practical experience of our Raman system setting, the spot size is increased from 1.92 um to 3.58 um when switching from ×50 lens (0.5NA) to ×10 lens (0.25NA). Recently, the Raman spectroscopy system has been introduced into the endoscope or made into a portable device to broaden its application range.
Cell Culture and Sample Pretreatment
A variety of cell lines have been used for in vitro models in breast cancer studies at the cellular level. These cell lines have demonstrated advantages such as ease of handling, high homogeneity, and unlimited self-replicating potential (Burdall et al. 2003). Raman spectra have shown not only differences in biological composition between cell lines but also biological processes such as cell differentiation and apoptosis. Therefore, by selecting a few kinds of normal cells and cancer cells related to breast cancer and acquiring the difference between Raman and optical signals, it is possible to secure the appropriateness of Raman and the transmission optical analysis method. Three cells of MCF-10A, MDA-MB-453, and MDA-MB-231 were examined by Raman spectroscopy in order to determine the differences between normal cells and cancer cells, and these cells were selected because they play an important role in cancer metastasis.
For securing these cell lines, one human breast cancer cell line (MDA-MB-453) and two human breast epithelial cell lines (MCF-10 and TNBC) were purchased from ATCC (Manassas, VA), and the same TNBC cell line and human breast cancer cell line (MDA-MB-231) were purchased from Korea Cell Bank. The human breast cancer cell lines were cultured using Dulbecco’s modified Eagle’s Minimal Essential Medium (DMEM; Life Technologies, Inc., Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT, USA) and 1% penicillin–streptomycin solution (Life Technologies, Inc.). The human breast epithelial cell line was grown using mammary epithelium growth medium, MEGM BulletKit (Lonza Group Ltd. Basel, Switzerland), except for GA-1000 (gentamicin–amphotericin B mix), with the addition of 100 ng/ml cholera toxin from Sigma-Aldrich (St. Louis, MO). All of the cell cultures were maintained on a humidified 5% CO2 incubator at 37 °C.
All cells were cultured on gold-coated substrates in Petri dishes until they reached 70 ~ 80% confluency to reduce Raman noise from bare glass. In order to synchronize the results of various measurement devices, it is necessary to fix the cells and measure Raman and optical images. Therefore, phosphate-buffered saline (PBS) was removed after 24 h, and cells were fixed using 4% paraformaldehyde for 20 minutes at 37 °C and 5% humidity in an incubator. Fixed cells were washed three times in PBS for 10 minutes each time. Because dried PBS produces Raman noise signals, the cells were rinsed with distilled water approximately 10 times to remove the PBS before the Raman measurements.
Raman Measurement Conditions and Analysis of Spectral Results
Raman line profiles were measured using a SENTERRA confocal Raman system (Bruker Optics, Billerica, MA, USA), and a 785-nm diode laser with 100 mW power was used for excitation. The laser beam was focused on the fixed cells through a 50 × (0.4 NA) objective with an approximate spot size of 2.4 μm. Raman lines were scanned through a raster scan at approximately 2.5 μm of step which refers to the distance between laser beam centers. The spectral range of each point was measured twice in the range from 400 to 2500 cm−1. A spectral resolution was 5 cm−1 and an integration time was 30 sec at room temperature. Before the Raman measurements, the Raman spectrum was calibrated by measuring a silicon crystal. Following the Raman measurements, the spectrum was preprocessed by a Savitzky–Golay smoothing filter. The autofluorescence background was removed by a third-order polynomial fitting to make the spectrum better suited for an analysis.
The Raman spectra obtained for the normal cell line and the breast cancer cell lines showed major peaks in phenylalanine, amides I and III, CH2 deformation, CH2 wagging/twisting, and S-S bonding vibration regions as shown in Fig. 3. Each spectrum was previously smoothed and baseline corrected, and the spectra obtained from each cell line showed visual differences including standard deviations. The concentrations of amide III, CH2 deformation vibration, CH2 wagging/twisting, amide I, and so on were different in breast cell lines from normal cell lines. The difference in Raman spectra can be distinguished by peak position, relative intensity between peaks, and absolute intensity of a particular peak, and these factors relate to the chemical and quantitative values. The biochemical species such as amide III, CH2 deformation vibration, CH2 wagging/twisting, amide I were labeled from the peak position. The CH2 deformation and amide I regions showed similar intensities in two cancer cell lines, but the MDA-MB-231 spectrum had a higher relative amount of CH2 wagging/twisting. In particular, the Raman spectrum for MDA-MB-231 had higher CH2 wagging/twisting intensity than another cancer cell. This difference is greater than the error bars in situations where the intensities of other peaks, such as S-S bonding and CH2 deformation vibration, are similar. These features seem to indicate the different chemical characteristics between the cancer cells and normal cells, which is a factor in the statistical analysis.
Averaged Raman spectra of the normal breast cell line (a) MCF-10A and the breast cancer cell lines (b) MDA-MB-231 and (c) MDA-MB-453. The standard deviations for each point are represented by shading around the solid line. Reprinted from Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 205, Lee et al., “Local-dependency of morphological and optical properties between breast cancer cell lines,” p. 132–138, Copyright 2018, with permission from Elsevier
Principal Component Analysis (PCA) of Raman Spectroscopic Signals
Analysis of multiple samples can be accessed through statistical analysis. Principal component analysis (PCA) is a method used to analyze a large number of data signals such as individual peaks of Raman signal. It is an algorithm that projects complex data with multiple dimensions into a small number of principal components (PC). Since many data have many dimensions, it is difficult to directly classify the data. The PCA transforms these high-dimensional data into a low-dimensional space and transforms the data so that it can be easily classified.
The remaining Raman spectrum reflects the conformational characteristics of the samples in addition to the background noise. In the Raman spectrum, the x-axis corresponds to the wavelength of light, and the wavelength corresponding to each spectrum can be an independent variable. Since the intensity according to these variables is an input value for analysis, signal processing including smoothing and baseline correction was necessary for accurate data analysis of each sample. We evaluated the spectral differences between the normal and cancer cell lines using PCA. In this study, PCA was conducted as an attempt to group useful information from the preprocessed Raman spectra and to highlight the differences between MCF-10A, MDA-MB-231, and MDA-MB-453. All analyses were conducted using the XLSTAT 2016 software (Microsoft, Redmond, WA, USA).
From the PCA of the spectrum obtained from the normal and cancer cell lines, it became possible to further distinguish between three cell lines as shown in Fig. 4a. It also became possible to distinguish between cancer cells which were difficult to distinguish on the spectrum of Fig. 3. Figure 4a shows a clear separation between the normal breast cells and breast cancer cells in the PCA plot, where the first principle component explained 63.57% of the variance, and the second component explained 20.89%. Additionally, while the breast cancer cells clustered near each other in Fig. 4a, the groups show clear separation from normal cells. Figure 5b shows an additional PC analysis on just the cancer cells, which clearly separates the two cell types, with the first principle component explaining 68.74% of the variance and the second component explaining 23.33%. The Raman spectral variables that were responsible for the discrimination on PC space were the phenylalanine, amide I, amide III, CH2 deformation, CH2 wagging/twisting, and S-S Bonding vibration peaks. Thus, components in the highly confined spectrum including the above chemical constitution alone occupy a large portion as a factor that differentiates each cell line. This statistical analysis method provides the criteria for discrimination between normal and cancer cells. The validity of the diagnosis can be confirmed by peak reference for multiple dimensions.
Principal component analysis results for (a) MCF-10A, MDA-MB-231, and MDA-MB-453 and (b) MDA-MB-231 and MDA-MB-453. Reprinted from Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 205, Lee et al., “Local-dependency of morphological and optical properties between breast cancer cell lines,” p. 132–138, Copyright 2018, with permission from Elsevier
Synchronizing the topographic and optical images of the three cell lines. AFM images of the normal cell (a) MCF-10A and cancer cell lines (b) MDA-MB-231 and (c) MDA-MB-453 on the left column. Transmission optical images are displayed in the right column to match the measured lines, where each black scale bar is 20 μm. Reprinted from Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 205, Lee et al., “Local-dependency of morphological and optical properties between breast cancer cell lines,” p. 132–138, Copyright 2018, with permission from Elsevier
Atomic Force Microscopy for Comparative Analysis of Local Raman Signals and Cellular Morphology
Atomic force microscopy (AFM) has been widely used to study biological phenomena such as the cytotoxicity of cellular systems (Lee et al. 2013) and the mechanical properties of cells and molecules (Haase and Pelling 2015; Bao and Suresh 2003). Furthermore, AFM has been applied to investigate Young’s modulus indicating cellular stiffness, which properties can in turn differentiate between nonmalignant and breast cancer cells (Li et al. 2008; Calzado-Martin et al. 2016). From the AFM results, it is now well-known that cancerous cells are remarkably softer than normal cells, and there is a potential as an analytical tool that can give information on morphology and partial classifications together. In this work, Raman spectroscopy was analyzed by morphological localization through synchronization with AFMs having high spatial resolution and optical microscope. The topography of depending on the in-plane direction of the substrate gives local quantitative thicknesses and volumes that reveal the relationships between local Raman spectra and intensities.
Topography and surface roughness data were acquired using an atomic force microscope (TT-AFM, AFM workshop) in a contact mode at room temperature. Normal and cancer cells fixed on a gold-coated substrate of approximately 15 nm roughness (RMS value) were mounted on the AFM x-y motor stage for scanning. A scan area of 50 μm × 50 μm was constructed using 256 scan lines and a scanning rate of 0.2 Hz. After the samples were prepared to synchronize Raman and AFM, the rinsed cells were dried and alignment markers were printed on them before the loading Raman stage. The individual cells measured by Raman spectroscopy were pre-checked with distance from the alignment marker and magnitude of the objective lens. Prior to approaching and positioning the cantilever, alignment markers on the sample and a crossbar on the AFM optical image were matched to position the cells and rotate the substrate. The magnitudes of the AFM topography and the Raman optical image were synchronized by a scale calibration target on AFM and Raman.
AFM topology and optical images are shown in Fig. 5, indicating matched line profiles of height and Raman spectroscopy. The individual AFM topologies, shown in the left column of (a) MCF-10A, (b) MDA-MB-231, and (c) MDA-MB-453 had 50 μm × 50 μm scan areas covering one or two cells. The minimum data value was shifted such that the height of each cell varied from 0 to 1.5 μm. There were no significant differences in height between cell types or species. The optical image is displayed in the right column corresponding to each AFM topogram column. The optical MCF-10A image showed a whitish cell boundary, a Newton ring with a contour like a rainbow, and blackhead-like points. Most blackheads corresponded to hollows on the AFM topography, but this was not always the case. For the invasive cancer cells MDA-MB-231 and MDA-MB-453, images transmitted from 3e and f showed blackhead-like points, black spots larger than blackheads, and cell boundaries less clear than MCF-10A (See Fig. 3d). The blackheads on the optical images were highly distributed around the hollows shown on the morphology images, but the black spots were at the highest peak in each cell. Note that these spots appear in approximately one out of ten invasive cancer cells. Thus, the whitish boundaries can be a significant indicator of whether or not a cell is invasive, but two cancer cell lines have barely visible boundaries as shown in Fig. 5 on the right column.
To investigate the comparative factors among noninvasive and invasive breast cell lines, line measurements of height and Raman spectroscopy were performed along the white dotted lines on the AFM topography and the red line on the optical image, as shown in Fig. 6.
Comparative results of extracted line morphology (a-c) and Raman line scan (d-f) for matching local variations. Raman line profiles were plotted for phenylalanine, amide I, amide III, CH2 deformation, CH2 wagging/twisting, and S-S bonding vibration. The inset in f shows the corresponding colors. Adapted from Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 205, Lee et al., “Local-dependency of morphological and optical properties between breast cancer cell lines,” p. 132–138, Copyright 2018, with permission from Elsevier
In the AFM results, all cross-sectional intensity height profiles were convex, and thus it was expected that Raman would depend on radial differences in cell morphology. Each profile covered one cell and was 34 μm long for MCF-10A (as shown in Fig. 6a and d), 47 μm long for MDA-MB-231 (as shown in Fig. 6b and e), and 42 μm long for MDA-MB-453 (as shown in Fig. 6c and f). To confirm Raman differences in cell morphology, we plotted the local differences in Raman signal, phenylalanine, amide I, amide III, and CH2 deformation, CH2 wagging/twisting, and S-S bonding vibration peaks for individual cell lines synchronized with AFM. With the normal MDF-10A cells, signals of phenylalanine, CH2 deformation, and amide I looked convex as in AFM morphology, but the signal for amide III was concave. These results indicate that the relative proportions of structural components in normal cells vary radially but not in the invasive cell line. The order of downward peak intensity for the invasive cells was maintained with CH2 wagging/twisting, S-S bonding, CH2 deformation, amide I, and phenylalanine as shown in Figs. 6 e and f.
Comparative Analysis of Raman Signal and RGB Transmission
By introducing local RGB intensity in optical measurements, we further identified factors that can be used to distinguish normal cells from cancer cell lines. An optical microscope shows the image of the transmitted light, and the intensity of each pixel of RGB indicates internal events such as absorption and scattering. Although we found that the difference in height within each cell was about 1.5 μm in the AFM results, the results of the inelastic scattering Raman and transmitted light optical microscope confirmed that there is no local variation in cancer cells, unlike normal cells.
To obtain RGB profile, optical microimages were observed using an optical microscope (BX51, Olympus) with mercury (reflection) and halogen (transmission) white light sources, respectively. A combined 50 × objective lens and a second lens of 2 × magnification were used. To reduce chromatic aberrations and blurred image edges on the reflected image and the transmitted image, an extended focus imaging calculation was performed. The resolution of all images was 1360 × 1024 pixels, and snap images were acquired within 1 sec. For calibration of a white balance, white paper was initially mounted, and subsequently, reflection optical images and RGB profiles were acquired. For acquiring transmission images, white balance was performed on the bare gold surface.
Notable observation from the transmission images was the difference in absorption behavior between the normal and cancer cells. When the transmitted optical images were acquired after light scattering and absorption through the cells, the individual red, green, and blue (RGB) differences gave information associated with scattering and absorption in the cells. At shorter wavelengths in incident white light, Rayleigh scattering is more common, so there is a difference in transmission between RGB. Thus, the RGB sequence on black spots in MDA-MB-231 was as shown in Fig. 7b, where the range from 18 to 22 μm appeared to indicate scattering. In contrast, the relative ratios of RGB from MCF-10A in Fig. 7a showed a fringe with no red-green-blue sequence from 14 to 25 μm; the fringe came from the shape of Newton’s ring as shown in Fig. 5d, which indicated different transmission properties between the normal and cancer cells. By introducing standard deviations for RGB intensity, the intensity was compared quantitatively. The individual deviations in RGB intensity for invasive cell lines were much smaller than those for noninvasive cell lines as shown in Figs. 7 and in Table 1. The standard deviations of RGB in MCF-10A were 28.1, 33.0, and 28.8 where the means were 194.0, 209.8, and 215.9, respectively. For the MDA-MB-231 with absorption information at the black spots removed, the standard deviations for RGB intensity were 10.3, 10.1, and 9.2 with RGB means of 213.2, 215.2, and 214.3. In total, the standard deviations of red, green, and blue intensity in the noninvasive cell lines were nearly triple those of the invasive cells as summarized in Table 1.
RGB intensity for (a) MCF-10A, (b) MDA-MB-231, and (c) MDA-MB-453. Adapted from Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 205, Lee et al., “Local-dependency of morphological and optical properties between breast cancer cell lines,” p. 132–138, Copyright 2018, with permission from Elsevier
Additionally, the local relation between morphology, Raman, and RGB deviation was significant in distinguishing between the normal and cancer cell lines. Since Raman spectroscopy is a technique for the detection of conformational structures, and if the optical transmission is measured with RGB profiling in the radial direction of the cell, the characterization factor that distinguishes the normal and the cancer can be found locally. In the case of the MCF-10A in this Raman study, the position-dependent projected signal difference in the radial direction seemed to be due to the cell organelles. However, in malignant cells of MDA-MB-231 and MDA-MB-453, the spectral difference did not appear to be dependent on the intracellular location. Additionally, since there is little difference in the degree of transmission between the red, green, and blue color components across the cells, it is suggested that RGB and Raman characteristics offer promising data for understanding the mechanism of cancer cell development, diagnosis, and treatment.
Conclusion
In summary, Raman spectra were acquired for the normal cell line (MCF-10A) and cancer cell lines (MDA-MB-231 and MDA-MB-453). The major peaks of Raman spectra are in the regions for phenylalanine, amides I and III, CH2 deformation, CH2 wagging/twisting, and S-S bonding vibration. These peaks were used as the main spectrum markers of cell classification. When those peaks were used as statistical variables, PCA on spectral differences allowed for further discrimination between cancer and normal cells. Although the breast cancer cells of MDA-MB-231 and MDA-MB-453 clustered close to each other when the PCA results were calculated for all three types of cells containing MCF-10A. Each group was well differentiated when only the breast cancer cells were recalculated. The study of local differences of Raman and RGB transmission with the image synchronization in this cell unit suggests the methodology of label-free and nondestructive field study and has potential for further application to other cell lines. When the results obtained in this study are extended to tissue and tumor size, these results will be the basis for establishing clinical diagnostic criteria and endoscopy applications.
Funding
This work was supported by the National Research Foundation of Korea (NRF) (2018R1D1A1B07048562, 2019R1A2C2084122) and MRC grant (2018R1A5A2020732) by the Ministry of Science & ICT (MSIT), and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI18C2391). This study was also supported by a grant (2019-7212) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea.
References
Abramczyk H et al (2011) The label-free Raman imaging of human breast cancer. J Mol Liq 164(1–2):123–131
Abramczyk H et al (2012) Raman 'optical biopsy’ of human breast cancer. Progress in Biophysics & Molecular Biology 108(1–2):74–81
Bao G, Suresh S (2003) Cell and molecular mechanics of biological materials. Nat Mater 2(11):715–725
Burdall SE et al (2003) Breast cancer cell lines: friend or foe? Breast Cancer Res 5(2):89–95
Calzado-Martin A et al (2016) Effect of actin organization on the stiffness of living breast cancer cells revealed by peak-force modulation atomic force microscopy. ACS Nano 10(3):3365–3374
Carvalho LFCS et al (2015) Raman micro-spectroscopy for rapid screening of oral squamous cell carcinoma. Exp Mol Pathol 98(3):502–509
Feng SY et al (2011) Study on gastric cancer blood plasma based on surface-enhanced Raman spectroscopy combined with multivariate analysis. Science China-Life Sciences 54(9):828–834
Gautam R et al (2015) Raman and mid-infrared spectroscopic imaging: applications and advancements. Curr Sci 108(3):341–356
Haase K, Pelling AE (2015) Investigating cell mechanics with atomic force microscopy. J R Soc Interface 12(104)
Haka AS et al (2005) Diagnosing breast cancer by using Raman spectroscopy. Proc Natl Acad Sci U S A 102(35):12371–12376
Haka AS et al (2009) Diagnosing breast cancer using Raman spectroscopy: prospective analysis. J Biomed Opt:14(5)
Ishigaki M et al (2016) Diagnosis of early-stage esophageal cancer by Raman spectroscopy and chemometric techniques. Analyst 141(3):1027–1033
Kamemoto LE et al (2010) Near-infrared micro-Raman spectroscopy for in vitro detection of cervical cancer. Appl Spectrosc 64(3):255–261
Kawabata T et al (2011) Near-infrared multichannel Raman spectroscopy with a 1064 nm excitation wavelength for ex vivo diagnosis of gastric cancer. J Surg Res 169(2):E137–E143
Kirsch M et al (2010) Raman spectroscopic imaging for in vivo detection of cerebral brain metastases. Anal Bioanal Chem 398(4):1707–1713
Larraona-Puy M et al (2009) Development of Raman microspectroscopy for automated detection and imaging of basal cell carcinoma. J Biomed Opt 14(5)
Larraona-Puy M et al (2011) Discrimination between basal cell carcinoma and hair follicles in skin tissue sections by Raman micro-spectroscopy. J Mol Struct 993(1–3):57–61
Lee YJ et al (2013) Label-free and quantitative evaluation of cytotoxicity based on surface nanostructure and biophysical property of cells utilizing AFM. Micron 49:54–59
Lee S et al (2018) Local-dependency of morphological and optical properties between breast cancer cell lines. Spectrochim Acta A Mol Biomol Spectrosc 205:132–138
Li QS et al (2008) AFM indentation study of breast cancer cells. Biochem Biophys Res Commun 374(4):609–613
Li Y et al (2010) Research on the Raman spectral character and diagnostic value of squamous cell carcinoma of oral mucosa. J Raman Spectrosc 41(2):142–147
Liu TY et al (2011) Functionalized arrays of Raman-enhancing nanoparticles for capture and culture-free analysis of bacteria in human blood. Nat Commun 2
Lyng FM et al (2007) Vibrational spectroscopy for cervical cancer pathology, from biochemical analysis to diagnostic tool. Exp Mol Pathol 82(2):121–129
Maiti NC et al (2004) Raman spectroscopic characterization of secondary structure in natively unfolded proteins: alpha-synuclein. J Am Chem Soc 126(8):2399–2408
Marro M et al (2014) Dynamic molecular monitoring of retina inflammation by in vivo Raman spectroscopy coupled with multivariate analysis. J Biophotonics 7(9):724–734
McQueenie R et al (2012) Detection of inflammation in vivo by surface-enhanced Raman scattering provides higher sensitivity than conventional fluorescence imaging. Anal Chem 84(14):5968–5975
Neugebauer U et al (2010) Towards detection and identification of circulating tumour cells using Raman spectroscopy. Analyst 135(12):3178–3182
Oshima Y et al (2010) Discrimination analysis of human lung cancer cells associated with histological type and malignancy using Raman spectroscopy. J Biomed Opt 15(1)
Rehman S et al (2007) Raman spectroscopic analysis of breast cancer tissues: identifying differences between normal, invasive ductal carcinoma and ductal carcinoma in situ of the breast tissue. J Raman Spectrosc 38(10):1345–1351
Rygula A et al (2013) Raman spectroscopy of proteins: a review. J Raman Spectrosc 44(8):1061–1076
Saha A et al (2011) Raman spectroscopy: a real-time tool for identifying microcalcifications during stereotactic breast core needle biopsies. Biomed Opt Express 2(10):2792–2803
Sato H et al (2009) Raman study of brain functions in live mice and rats: a pilot study. Vib Spectrosc 50(1):125–130
Shapiro A et al (2011) Raman molecular imaging: a novel spectroscopic technique for diagnosis of bladder cancer in urine specimens. Eur Urol 59(1):106–112
Singh B et al (2012) Application of vibrational microspectroscopy to biology and medicine. Curr Sci 102(2):232–244
Smith R, Wright KL, Ashton L (2016) Raman spectroscopy: an evolving technique for live cell studies. Analyst 141(12):3590–3600
Synytsya A et al (2014) Raman spectroscopy at different excitation wavelengths (1064, 785 and 532 nm) as a tool for diagnosis of colon cancer. J Raman Spectrosc 45(10):903–911
Talari ACS et al (2015) Raman spectroscopic analysis differentiates between breast cancer cell lines. J Raman Spectrosc 46(5):421–427
Zhou HB et al (2014) SERS detection of bacteria in water by in situ coating with Ag nanoparticles. Anal Chem 86(3):1525–1533
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Lee, S., Kim, J.K. (2021). Label-Free Raman Spectroscopic Techniques with Morphological and Optical Characterization for Cancer Cell Analysis. In: Kim, J.K., Kim, J.K., Pack, CG. (eds) Advanced Imaging and Bio Techniques for Convergence Science. Advances in Experimental Medicine and Biology, vol 1310. Springer, Singapore. https://doi.org/10.1007/978-981-33-6064-8_14
Download citation
DOI: https://doi.org/10.1007/978-981-33-6064-8_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-33-6063-1
Online ISBN: 978-981-33-6064-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)