Abstract
We report for the first time a proof-of-concept experiment employing Raman spectroscopy to detect intracerebral tumors in vivo by brain surface mapping. Raman spectroscopy is a non-destructive biophotonic method which probes molecular vibrations. It provides a specific fingerprint of the biochemical composition and structure of tissue without using any labels. Here, the Raman system was coupled to a fiber-optic probe. Metastatic brain tumors were induced by injection of murine melanoma cells into the carotid artery of mice, which led to subcortical and cortical tumor growth within 14 days. Before data acquisition, the cortex was exposed by creating a bony window covered by a calcium fluoride window. Spectral contributions were assigned to proteins, lipids, blood, water, bone, and melanin. Based on the spectral information, Raman images enabled the localization of cortical and subcortical tumor cell aggregates with accuracy of roughly 250 μm. This study demonstrates the prospects of Raman spectroscopy as an intravital tool to detect cerebral pathologies and opens the field for biophotonic imaging of the living brain. Future investigations aim to reduce the exposure time from minutes to seconds and improve the lateral resolution.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The principal obstacle during open surgery for intrinsic brain tumors and metastatic deposits of extracranial tumors is the inability to delineate infiltrating tumor from normal tissue. In addition, since the brain has a high functional density, the removal of any lesion is only possible at the very border of the lesion. Safety margins cannot be tolerated as for the treatment of extracerebral diseases. On the other hand, complete removal improves the recurrence-free interval and opens the possibility for adjunct therapeutic strategies. The identification and visualization of residual tumor is, therefore, of paramount importance to improve the outcome of patients with brain tumors.
In general, biophotonic imaging methods give information regarding intrinsic optical properties of tissue and the presence or absence of endogenous or exogenous chromophores. The benefits of using light in imaging living tissue include light providing high sensitivity to functional changes and revealing the dynamics of cells in the nervous system, either via the absorption of light, emission of light, and elastic or inelastic scattering. Further advantages are diffraction-limited spatial resolution in the micrometer, and even submicrometer range, non-destructive sampling, use of non-ionizing radiation, fast data collection, relatively inexpensive instrumentation, and reduced infrastructure requirements compared to other clinical imaging methods. Raman spectroscopy is a biophotonic method which probes molecular vibrations based on inelastic scattering of monochromatic radiation. It provides a specific fingerprint of the biochemical composition and structure of tissue without using any labels.
Among the promising applications of biophotonic imaging is the delineation of brain tumor margin in vivo. We initiated a research program to detect brain tumors by Raman spectroscopic imaging. Raman images were presented from brain tissue sections [1] and pristine brain tissue [2] and whole mouse brains ex vivo [3]. Here, we demonstrated the successful setup for in vivo Raman mapping to localize brain metastases.
Materials and methods
Murine brain tumor model
Six- to 8-week-old female NMRInu/nu nude mice were used in a hematogenous brain metastases model [4]. Mice were maintained under pathogen-free conditions according to international and institutional guidelines. Mice were anesthetized by intraperitoneal injection of ketamine and xylazine. Approximately 100,000 tumor cells suspended in 0.1 mL phosphate-buffered salt solution were injected in the right internal carotid artery of the mice, which produced hematogenous brain metastases [5]. Two cell lines were selected: the human melanoma cell line A375 (obtained from American Type Culture Collection, ATCC, Manassas, Virginia, USA) and murine melanoma cell line K1735 (kindly provided by I. Fidler, M.D. Anderson Cancer Center, Houston, Texas, USA). Metastases were allowed to grow for 3 weeks, until which the mice had symptoms of tumor growth. Five animals were prepared. For cortical in vivo imaging, the mice were anesthetized, and via a median skin incision, a bilateral craniotomy was performed which had a diameter of 6–8 mm. The dura was opened and removed, leaving the midline venous sagittal sinus intact. A transparent, 1-mm-thick window made of calcium fluoride was fixed to the skull using zinc phosphate cement (Harvard Dental International GmbH, Hoppegarten, Germany). The procedures were approved by the local and state ethics committees (AZ 24-9168.11-1-2002-11). Melanotic tumors were visible under normal light in two animals, therefore serving as a positive control for Raman spectroscopic findings. Tumors were not found at the cortical surface of the other three animals, and Raman images were not collected from these mice. Following image acquisition, the animals were killed, the brains were removed, embedded in OCT, quickly frozen in a dry ice, and stored at −80 °C for histological staining.
Raman spectroscopic imaging
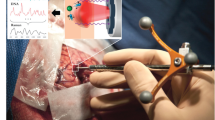
The experimental setup which is shown in Fig. 1 was similar to an earlier ex vivo study of mouse brains [3]. Instead of a filtered fiber-optic probe with a 50-μm excitation fiber and 100-μm collection fiber, a probe with a 100-μm excitation fiber and 200-μm collection fiber was used (both from Inphotonics, USA). A bandpass filter, a beam splitter, a lens, a mirror, and a longpass filter are integrated in the compact probe head. Both fibers had a focal distance of 5 mm. The excitation fiber was connected to a 785-nm diode laser (model BRM-785, BW TEC, USA). The collection fiber was connected to an f/1.8 spectrograph (Kaiser Optical Systems, USA) with a back-illuminated, deep-depletion, liquid nitrogen-cooled CCD detector (Roper Scientific, USA). The larger excitation fiber gave a larger focus of 120 μm as indicated by the manufacturer. Consequently, higher laser intensities of 200 mW could be used for excitation without inducing thermal damages. The larger collection fiber in combination with higher laser intensities enabled reducing the exposure time down to 4 s per spectrum. Raman images were recorded by serial acquisition of spectra at the step size of 100 μm using a motorized sample stage (Prior Scientific, UK). The spectral resolution in the interval 126–3,560 cm−1 was 4 cm−1. The Raman spectrometer was controlled using HoloGrams (Kaiser) software, which also performed automatic cosmic-ray correction, intensity calibration, and wavenumber calibration. The resulting Raman images were processed by in-house written programs under a Matlab platform (The Mathworks Inc., USA). First, a background was subtracted from each spectrum, then the spectra were normalized by simple multiplicative signal correction, and, finally, the spectra were segmented by k-means cluster analysis in the spectral range 900–1,700 cm−1. Details have been published previously [6].
Schematic setup of the Raman system used in this study (a). The fiber-optic probe head integrated a bandpass filter (BP), beam splitter (BS), mirror (M), longpass filter (LP), and a lens. The probe was coupled by an excitation fiber (EF) with the laser and by a collection fiber (CF) with the spectrometer. The mouse was placed on a motorized stage to collect Raman images in the mapping mode (b)
Results
The head of the animal was embedded in a mold of aluminum foil to reduce motion (Fig. 1). However, in future experiments, massive motion artifacts due to pulsations and breathing excursions have to be compensated by rigid fixation using ear bars. We used melanotic tumors to have a reproducible, easily verifiable control in the current proof-of-concept experiments. The presence of melanin is irrelevant to Raman spectroscopic imaging as any tumor has a specific signature. The pigment enabled selecting the areas of interest by visual inspection. The regions were supposed to cross the large midline venous blood vessel and the superior sinus and to cover macroscopically visible tumor as well as normal brain. Photos were recorded using a digital camera for documentation because the fiber probe was not equipped with a white light source and a camera. Two representative examples are shown in Figs. 2 and 3.
Deceased mouse with exposed cortex (a) after Raman spectroscopic images were acquired. The Raman image consisted of 37 × 33 spectra and probed an area of 3.6 × 3.2 mm. The sampling area involved the large tumor mass in the right hemisphere, the midline, and the left brain hemisphere. Raman images were segmented by cluster analysis (b) and displayed using false color coding: Normal brain tissue is depicted in blue, cyan, and yellow, the red midline structures include a central venous blood vessel termed sagittal sinus, and the tumor and tumor margin are shown in gray and black, respectively. In c, an overlay of the photomicrograph and the Raman image is shown, verifying the correct classification and the macroscopic diagnosis
Living mouse with cranial window (a). The box indicates the area which was studied by Raman imaging. Raman image segmented by cluster analysis (b): normal brain tissue (blue, cyan), midline (red), bone (brown), and tumor (black) in the left hemisphere at the border of the analyzed area. An H&E-stained coronal section through the brain is shown in c, the magnification in d
As depicted in Fig. 2, the Raman image consisted of 37 × 33 spectra and probed an area of 3.6 × 3.2 mm. The sampling area involved the large tumor mass in the right hemisphere, the midline, and the left brain hemisphere (Fig. 2a). The result of the cluster analysis was color coded (Fig. 2b). Normal brain tissue is represented by blue, cyan, and yellow. The tumor margin is colored black, the midline red, and the main tumor mass gray. An overlay of the Raman image and the photomicrograph is shown in Fig. 2c. It is evident that next to the large tumor mass in the right hemisphere, a small tumor mass is present in the left hemisphere. This small tumor was not visible in the photomicrograph because it was below the surface. The detection of the subsurface tumor demonstrates that Raman spectroscopy is able to penetrate deeper layers in the brain tissue. The exact penetration depth depends on factors such as excitation and collection geometry, the laser wavelength, laser intensity, and tissue composition. In the current backscattering geometry, the accessible depths in living tissue are typically only several hundreds of micrometer. Recently, an approach called spatially offset Raman spectroscopy was introduced, which enables applying the technique as a tomographic tool [7]. The feasibility of 3D tomographic imaging through a canine hind limp with thickness up to 45 mm was demonstrated using transmission Raman spectroscopy [8].
Although the width of the midline was 1 pixel in most parts of the data set, it is difficult to measure the exact spatial resolution on the basis of the cluster analysis results. Here, the lateral resolution was restricted by the data point spacing (100 μm) and laser focus diameter (120 μm). Taking these parameters into account together with distortions due to the strongly scattering tissue, the actual lateral resolution can be estimated as roughly 250 μm. Higher lateral resolution would require a smaller data point spacing and a smaller laser focus.
Figure 3a displays how a cranial window was placed on the head of a living mouse. The sampling area is indicated by a box. Data collection was terminated after 10 × 41 spectra because the mouse had increased motion artifacts that limited further investigations. Cluster analysis of the Raman image (Fig. 3b) resolved a tumor in the left brain hemisphere, the midline harboring a venous vessel termed superior sagittal sinus, bone, and normal brain tissue. The midline is depicted broader than the diameter of the sagittal sinus; in addition, the segmentation is somehow irregular toward the lower portion of the Raman image. This experiment points to the importance of motion artifacts of the mouse head during data acquisition. In forthcoming experiments, the animal will be physically fixed using ear bars and a moth clamp adaptor, which is the usual setup in our laboratory. The corresponding histological hematoxylin and eosin (H&E)-stained coronal section (Fig. 3c, magnification in Fig. 3d) reveals dense and sparse cortical tumor growth, rendering part of the brain surface black. These sections give a rough approximation of gross tumor growth, although they are not exactly orthogonal to the surface.
The Raman spectra in Fig. 4 were averaged without preprocessing or EMSC. For comparison and display in Fig. 4a, the Raman spectra representing the clusters of Fig. 2 have been background-corrected and normalized to the intensity of the band near 2,933 cm−1, which is assigned to valence vibrations of CH2 and CH3 moieties in proteins and lipids. The spectra of normal brain tissue slightly differ in the intensities of valence vibrations of OH bonds near 3,245 and 3,425 cm−1 that are mainly assigned to water. Therefore, the different classifications of normal brain might be due to various amounts of water overlying the curved murine brain. In order to provide a good contact between tissue and the calcium fluoride window, a drop of aqueous buffer was applied to the exposed brain. This procedure could affect the water content, and, consequently, the spectral range was not considered for cluster analysis and the region from 900 to 1,700 cm−1 was used instead because it contains only a weak Raman band of water near 1,640 cm−1. In general, bands in the lower wavenumber region from 500 to 1,800 cm−1 show lower intensities. Most prominent bands are assigned to the aromatic amino acid phenylalanine (1,004 cm−1), unsaturated fatty acids (1,267 and 1,661 cm−1), CH2 and CH3 deformation vibrations (1,301 and 1,445 cm−1), and the amide bands of protein backbones (1,267 and 1,661 cm−1) that overlap with spectral contributions of fatty acids. The Raman spectrum of the midline contains additional bands due to oxygenated hemoglobin near 750, 1,002, 1,212, 1,546, 1,605, and 1,619 cm−1. The intensities of these bands are enhanced due to a resonance effect known as resonance Raman spectroscopy. This is caused by the chromophore heme of the hemoglobin molecule which partly absorbs the excitation radiation, resulting in increased Raman cross-sections of heme-associated vibrations.
The Raman spectrum of the tumor displays additional spectral contributions due to the pigment melanin near 597, 976, 1,404, and 1,595 cm−1, which has also previously been found [3]. The Raman resonance enhancement amplifies the intensities, so the bands in the lower wavenumber region are even more intense than in the higher wavenumber region. The pigment is expressed by tumor cells and can be used as an inherent marker of these brain metastases. In the Raman spectrum of the tumor margin, spectral contributions of normal brain tissue and the pigment melanin overlap, indicating the infiltrative nature of tumor growth independent of its visibility. Furthermore, scattering from subsurface tumor portions might be responsible for weak spectral contributions of melanin in the spectrum of the cyan cluster of Fig. 4a. It cannot be excluded that the limited lateral resolution might give mixed clusters. That means the fiber probe excites and collects both tissues simultaneously at the transition between tumor and normal brain.
The Raman spectra representing the clusters in Fig. 3 are included in Fig. 4b. Here, the unprocessed spectra are displayed, which means without background correction and normalization. The Raman spectrum of the midline venous sagittal sinus is characterized by the lowest background and spectral contributions of hemoglobin. On an overall perspective, the background of normal brain tissue is obviously increased. As before, these spectra are dominated by spectral contributions of proteins, lipids, and water. Autofluorescence is not an issue for normal brain tissue and blood with 785-nm excitation. The background is increased in the spectra representing the brown and the black clusters. A band at 960 cm−1 in the Raman spectrum of the brown cluster is typical for hydroxyapatite of bone. The enhanced spectral contributions of the pigment melanin in tumor cells are accompanied by the highest background. Such a background is consistent with fluorescence emission, which is simultaneously excited by the laser. The overlap with fluorescence is even more problematic using excitation lasers in the wavelength range 400–700 nm. This is the main reason to select a near-infrared (NIR) laser with 785-nm emission as excitation source. In addition, the lower absorption of NIR radiation in tissues is advantageous to penetrate deeper sections of the tissue, as demonstrated for subsurface tumor detection in Fig. 2.
Discussion
Intense NIR lasers, innovative optical filters, and sensitive detectors were developed in the past and improved the sensitivity of Raman spectroscopy for biomedical studies (reviewed in [9]). The potential implications of in vivo Raman spectroscopy for online diagnostic purposes and guidance of therapies are promising. Due to technical limitations, most previous in vivo Raman studies simply recorded single spectra. In vivo applications have been reported to detect dysplasia in the rat palatum [10] and in the human gastrointestinal tract [11], precancers in the cervical epithelium [12], lesions of breast tissue [13], and polyps in colon [14] and to determine the molecular composition of artery wall [15]. Another in vivo study reported the detection of glioma brain tumors by Raman microscopy coupled to a microprobe [16]. The authors induced brain tumors in rats by direct injecting tumor cells in the brain. Tissue sections from this tumor model were previously studied by Raman spectral imaging [17]. In contrast, our proof-of-concept study obtained Raman images from the brains of living mice. Raman images enabled the identification of cortical and subcortical tumors, therefore visible and invisible tumors to the unshielded eye. The reported probe geometries [10–16] used excitation fibers with diameters of more than 200 μm, which gave a laser focus diameter of more than 200 μm depending on the numerical aperture. The lensed fiber-optic probe in this study had a smaller focus diameter near 100 μm, which enabled, in combination with a step size of 100 μm, a lateral resolution of roughly 250 μm. The resolution of these probes is not sufficient to identify single cells. From a clinical point of view, single cell resolution is beyond surgical capabilities and—if adopted as a standard—needs to be combined with an automated cell-based therapeutic strategy such as single cell laser ablations. Relevant surgical dexterity ranges from 100 to 1,000 μm if an operating microscope is used. Usually, tumor margins within the brain parenchyma can be estimated to a level of 1–3 mm, although this depends on purely visual inspection—better diagnostics would improve the surgical capabilities. The focused laser beam could induce carbonization and significant spectral changes if the local intensity is above the damage threshold. However, we concluded from the unchanged and non-distorted spectra that no visible thermal damage occurred. In addition, H&E staining did not reveal any burning in the measured area of living mice. Viable tissue is able to dissipate absorbed thermal energy to a greater extent than non-viable tissue. Prior to clinical application, toxic levels of laser irradiation have to be defined in more detail. Obviously, an exposure time of 4 s per spectrum and image acquisition in the serial mapping mode adding up to a total exposure of several minutes are impractical in intraoperative clinical applications. A technology that can be used in a clinical setting should either perform an automated scan or use navigated mapping of, e.g., a resection cavity. Therefore, the scope of further developments is to reduce exposure time and to increase lateral resolution.
The fiber-optic Raman probe from Inphotonics, which was used in this study, is too large for endoscopic applications. Miniaturized fiber-optic probes are required for use with endoscopes. In spite of more than 10 years of development by numerous research groups, such fiber-optic probes for biomedical application are not commercially available yet. A new mini-fiber-optic Raman probe suitable for insertion into the working channel of a bronchoscope from Emvision LCC (USA) was recently introduced and might fill the gap [18]. To avoid the technical complexity of Raman probes, unfiltered fused silica fibers have been introduced to probe the high wavenumber region from 2,400 to 3,800 cm−1 [19] and applied for Raman spectroscopic characterization of porcine brain tissue [20]. Even more useful for Raman imaging would be a probe made of fiber bundles. A similar probe was recently introduced for fluorescence imaging [21] which enables collecting images from brains of freely moving mice. In the current clinical environment, spectroscopic information could be introduced as part of multimodal imaging to guide brain tumor resection and visualize normal from infiltrated cancerous brain.
In general, a common set of classification algorithms need to be developed that can cover a broad range of pathologies. We have previously shown that the different grades of gliomas can be distinguished by optical spectroscopy, as well as various substructures within the normal human, porcine, and murine brain [3, 22, 23]. Another recent report demonstrated Raman spectroscopic classification of rat brain hippocampus suffering from Alzheimer’s disease [24]. Therefore, additional applications are the biochemical verification of target areas not only of tumors but also, e.g., stereotactic operations guided by a fiber-optic device for degenerative lesions, such as in Parkinson’s disease.
Alternative photonic methods are under investigation to offer intravital measurements. Nonlinear variants of Raman spectroscopy such as coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS) offer signal enhancement of several orders of magnitude. CARS was already applied in the context of brain tumors [25]. SRS is a new approach which was recently demonstrated for lipid vesicles in single cells [26]. In combination with laser scanning microscopes, images can be acquired at video time frame rates with diffraction-limited lateral resolution. Linear Raman spectroscopy is still a valuable experimental tool because in contrast to CARS and SRS, Raman spectra offer full spectral information [27]. Future progress in nonlinear Raman spectroscopy and fiber probe development will pave the way to enable promising prospects of Raman spectroscopy in diagnostic applications. The next step in our research program is to localize and monitor growth of brain metastases of various primary organs in living mice.
References
Krafft C, Sobottka SB, Schackert G, Salzer R (2006) Raman and infrared spectroscopic mapping of human primary intracranial tumors: a comparative study. J Raman Spectrosc 37:367–375
Krafft C, Sobottka SB, Schackert G, Salzer R (2005) Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors. Analyst 130:1070–1077
Krafft C, Kirsch M, Beleites C, Schackert G, Salzer R (2007) Methodology for fiber-optic Raman mapping and FTIR imaging of metastases in mouse brains. Anal Bioanal Chem 389:1133–1142
Schackert G, Fidler IJ (1988) Development of in vivo models for studies of brain metastasis. Int J Cancer 41:589–594
Kirsch M, Weigel P, Pinzer T, Carroll RS, Black PM, Schackert HK, Schackert G (2005) Therapy of hematogenous melanoma brain metastases with endostatin. Clin Cancer Res 11:1259–1267
Krafft C, Codrich D, Pelizzo G, Sergo V (2008) Raman and FTIR imaging of lung tissue: methodology for control samples. Vib Spectrosc 46:141–149
Matousek P, Morris MD, Everall N, Clark IP, Towrie M, Draper E, Goodship A, Parker AW (2005) Numerical simulations of subsurface probing in diffusely scattering media using spatially offset Raman spectroscopy. Appl Spectrosc 59:1485–1492
Schulmerich MV, Cole JH, Dooley KA, Morris MD, Kreider JM, Goldstein SA, Srinivasan S, Pogue BW (2008) Noninvasive Raman tomographic imaging of canine bone tissue. J Biomed Opt 13:020506
Krafft C, Dietzek B, Popp J (2009) Raman and CARS microspectroscopy of cells and tissues. Analyst 134:1046–1057
Bakker Schut TC, Witjes MJ, Sterenborg HJ, Speelman OC, Roodenburg JL, Marple ET, Bruining HA, Puppels GJ (2000) In vivo detection of dysplastic tissue by Raman spectroscopy. Anal Chem 72:6010–6018
Huang Z, Bergholt MS, Zheng W, Lin K, Ho KY, The M, Yeoh KG (2010) In vivo early diagnosis of gastric dysplasia using narrow-band image-guided Raman endoscopy. J Biomed Opt 15:037017
Utzinger U, Heintzelmann DL, Mahadevan-Jansen A, Malpica A, Follen M, Richards-Kortum R (2001) Near-infrared Raman spectroscopy for in vivo detection of cervical precancers. Appl Spectrosc 55:955–959
Motz JT, Gandhi SJ, Scepanovic OR, Haka AS, Kramer JR, Dasari RR, Feld MS (2005) Real-time Raman system for in vivo disease diagnosis. J Biomed Opt 10:031113
Molckovsky A, Song LM, Shim MG, Marcon NE, Wilson BC (2003) Diagnostic potential of near-infrared Raman spectroscopy in the colon: differentiating adenomatous from hyperplastic polyps. Gastrointest Endosc 57:396–402
Buschman HP, Marple ET, Wach ML, Bennett B, Schut TC, Bruining HA, Bruschke AV, van der Laarse A, Puppels GJ (2000) In vivo determination of the molecular composition of artery wall by intravascular Raman spectroscopy. Anal Chem 72:3771–3775
Beljebbar A, Dukic S, Amharref N, Manfait M (2010) Ex vivo and in vivo diagnosis of C6 glioblastoma development by Raman spectroscopy coupled to microprobe. Anal Bioanal Chem 398:477–487
Amharref N, Beljebbar A, Dukic S, Venteo L, Schneider L, Pluot M, Manfait M (2007) Discriminating healthy from tumor and necrosis tissue in rat brain tissue samples by Raman spectral imaging. Biochim Biophys Acta 1768:2605–2615
Magee ND, Villaumie JS, Marple ET, Ennis M, Elborn JS, McGarvey JJ (2009) Ex vivo diagnosis of lung cancer using a Raman miniprobe. J Phys Chem B 113:8137–8141
Santos LF, Wolthuis R, Koljenovic S, Almeida RM, Puppels GJ (2005) Fiber-optic probes for in vivo Raman spectroscopy in the high-wavenumber region. Anal Chem 77:6747–6752
Koljenovic S, Bakker Schut TC, Wolthuis R, Vincent AJPE, Hendriks-Hagevi G, Santos LF, Kros JM, Puppels GJ (2007) Raman spectroscopic characterization of porcine brain tissue using a single fiber-optic probe. Anal Chem 79:557–564
Flusberg BA, Nimmerjahn A, Cocker ED, Mukamel EA, Barretto RP, Ko TH, Burns LD, Jung JC, Schnitzer MJ (2008) High-speed, miniaturized fluorescence microscopy in freely moving mice. Nat Methods 5:935–938
Dreissig I, Machill S, Salzer R, Krafft C (2009) Quantification of brain lipids by FTIR spectroscopy and partial least squares regression. Spectrochim Acta A Mol Biomol Spectrosc 71:2069–2075
Kohler M, Machill S, Salzer R, Krafft C (2009) Characterization of lipid extracts from brain tissue and tumors using Raman spectroscopy and mass spectrometry. Anal Bioanal Chem 393:1513–1520
Chen P, Shen A, Zhao W, Baek SJ, Yuan H, Hu J (2009) Raman signature from brain hippocampus could aid Alzheimer's disease diagnosis. Appl Opt 48:4743–4748
Evans CL, Xu X, Kesari S, Xie XS, Wong ST, Young GS (2007) Chemically-selective imaging of brain structures with CARS microscopy. Opt Express 15:12076–12087
Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, Tsai JC, Kang JX, Xie XS (2008) Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science 322:1857–1861
Krafft C, Ramoji AA, Bielecki C, Vogler N, Meyer T, Akimov D, Rosch P, Schmitt M, Dietzek B, Petersen I, Stallmach A, Popp J (2009) A comparative Raman and CARS imaging study of colon tissue. J Biophotonics 2:303–312
Acknowledgments
We would like to thank Dr. Daniel Martin and Dipl. Biol Elke Leipnitz for their help. The research is funded by the German Research Foundation (DFG) within the project “Real time diagnosis of brain metastasis in animal models using Raman imaging” and by the MedDrive Start-up funds of the Medical Faculty, Dresden University of Technology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kirsch, M., Schackert, G., Salzer, R. et al. Raman spectroscopic imaging for in vivo detection of cerebral brain metastases. Anal Bioanal Chem 398, 1707–1713 (2010). https://doi.org/10.1007/s00216-010-4116-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-4116-7