Abstract
The heart is a complex multicellular organ comprising both cardiomyocytes (CM), which make up the majority of the cardiac volume, and non-myocytes (NM), which represent the majority of cardiac cells. CM drive the pumping action of the heart, triggered via rhythmic electrical activity. NM, on the other hand, have many essential functions including generating extracellular matrix, regulating CM activity, and aiding in repair following injury. NM include neurons and interstitial, immune, and endothelial cells. Understanding the role of specific cell types and their interactions with one another may be key to developing new therapies with minimal side effects to treat cardiac disease. However, assessing cell-type-specific behavior in situ using standard techniques is challenging. Optogenetics enables population-specific observation and control, facilitating studies into the role of specific cell types and subtypes. Optogenetic models targeting the most important cardiac cell types have been generated and used to investigate non-canonical roles of those cell populations, e.g., to better understand how cardiac pacing occurs and to assess potential translational possibilities of optogenetics. So far, cardiac optogenetic studies have primarily focused on validating models and tools in the healthy heart. The field is now in a position where animal models and tools should be utilized to improve our understanding of the complex heterocellular nature of the heart, how this changes in disease, and from there to enable the development of cell-specific therapies and improved treatments.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Cardiac optogenetics
- Interstitial cells
- Immune cells
- Endothelial cells
- Neurons
- Heterocellular interactions
1 Introduction
The vertebrate heart is the central muscular organ maintaining blood flow throughout an organism’s entire lifespan. Coordinated contraction–relaxation cycles are achieved by concerted electrical activation from the sinus node via the atria, the atrioventricular (AV) node, and the ventricular conduction system to the working myocardium of the ventricles. Cardiomyocytes (CM) are the predominant cell type driving the electromechanical activity of the heart. CM form a functional syncytium that enables depolarization-induced activation of cellular action potentials, leading to Ca2+-induced Ca2+ release, which in turn triggers CM contraction. CM are not only crucial for heart function but also occupy the majority of the heart volume. However, they are embedded in a complex and intricate network of other cells, non-myocytes (NM), including cardiac endothelial cells, fibroblasts, immune cells, and neurons. In fact, NM are significantly more numerous than CM in the healthy heart and the NM to CM ratio is further increased in cardiac disease. NM have diverse functions ranging from structural support and regulation of CM activity to driving repair following cardiac injury. Thus, in order to understand cardiac function in health and disease, we need to not only unravel the role of each individual cardiac cell type but also to understand how these cell types interact with one another.
Classically, cardiac electrical activity has been monitored with body surface electrodes, which report overall atrial and ventricular depolarization–repolarization cycles (the electrocardiogram—ECG). In basic research, ECG recordings have been complemented by dye-based optical mapping of membrane voltage, providing time-resolved, near-epicardial conduction maps of isolated Langendorff-perfused hearts. Modulation of electrical activity has been performed via implantable electrodes, e.g., electrical pacemakers for cardiac pacing and implantable cardioverter–defibrillators for electrical shock–based restoration of heart rhythm (defibrillation). Thus, classic cardiac monitoring and intervention tools report/modulate overall electrical activity that is dominated by CM, without the possibility to target cell-specific behavior of CM or NM.
Cell-specific targeting is possible, however, by combining advanced optical methods and state-of-the-art genetic engineering. The underlying optogenetic approaches enable manipulation and/or monitoring of the activity of specific cells, cell populations, or cell types. Cardiac optogenetics thus represents an emerging method in basic research for deciphering cell-specific functions and heterocellular interactions in intact myocardium, laying the foundation for future development of cell-specific therapies to treat cardiac diseases.
2 Introduction to Cardiac Optogenetics
Eight years after the discovery of directly light-gated ion channels in motile green algae (Nagel et al. 2002, 2003; Sineshchekov et al. 2002; Suzuki et al. 2003), and five years after ground-breaking experiments using channelrhodopsins in neurons and brain tissue (Li et al. 2005; Nagel et al. 2005; Boyden et al. 2005; Ishizuka et al. 2006), channelrhodopsin-2 (ChR2) and halorhodopsin from Natromonas pharaonis (NpHR) were first applied for contact-free alteration of cardiac electrophysiology by illumination (Arrenberg et al. 2010; Bruegmann et al. 2010). However, optogenetics includes both observation and control of cell function with light (Miesenböck 2009). Therefore, first cardiac optogenetic experiments were performed as early as in 2006, when Tallini et al. used the genetically encoded Ca2+ sensor GCaMP2 to measure CM Ca2+ transients in murine hearts in vivo (Tallini et al. 2006).
In the last decade, a handful of optogenetic actuators and reporters have been applied to the study of cardiac electrophysiology. Actuators used include ChR2 and the red-shifted channelrhodopsin chimera ReaChR, the chloride pump NpHR and the proton pump ArchT as well as the more recently found anion channelrhodopsins (ACR) (Arrenberg et al. 2010; Bruegmann et al. 2010; Govorunova et al. 2016; Nyns et al. 2017; Kopton et al. 2018; Funken et al. 2019). Established reporters are Ca2+ sensors of the GCaMP family and genetically encoded voltage sensors such as the voltage-sensitive fluorescent protein (VSFP), which have been used to image Ca2+ and voltage dynamics in cardiac cells and tissue (Tallini et al. 2006, 2007; Liao et al. 2015).
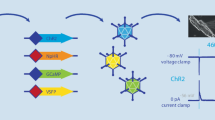
Proof-of-principle optogenetic studies were initially focused on overall cardiac function, showing the feasibility of optical pacing, atrial and ventricular defibrillation, as well as cardiac resynchronization (Bruegmann et al. 2010, 2016, 2018; Nussinovitch and Gepstein 2015; Nyns et al. 2017). Similarly, optical reporters have primarily been used to study CM function, in particular examining cardiac development (Chi et al. 2008, 2010; Hou et al. 2014; Weber et al. 2017) and, using subcellular targeting of sensors, Ca2+-induced Ca2+ release (Lu et al. 2013; Despa et al. 2014; Ljubojevic et al. 2014; Shang et al. 2014). Global activation of optogenetic sensors or actuators expressed in CM thus enables testing optogenetic approaches to study and/or steer overall cardiac electrophysiology with light. However, it provides limited insight into the role of CM subpopulations or other electrophysiologically relevant cell types. This book chapter focuses on optogenetic studies aiming to unravel basic research questions regarding cell-type-specific functions and interactions in the vertebrate heart in situ. We outline strategies to target different classes of cells and how these attempts have already widened our understanding of the heterocellular heart (Fig. 24.1).
3 Targeting CM Subpopulations
3.1 Global Expression and Localized Illumination
Arrenberg et al. combined global overexpression of either NpHR or ChR2 in CM with patterned illumination, generated with a digital micromirror device, to identify pacemaker cells of the developing zebrafish heart, and to optically induce cardiac rhythm disturbances such as AV block, tachycardia, and bradycardia (Arrenberg et al. 2010). Similarly, Crocini et al. developed an optical platform capable of simultaneously mapping and controlling the electrical activity of CM subsets within whole murine hearts at sub-millisecond temporal resolution (Crocini et al. 2016). Their optical stimulation setup consisted of a macroscope equipped with a laser scanning system based on acousto-optic deflectors. This platform enabled testing different geometric patterns for light activation of ChR2, with the aim of identifying optimal patterns of CM depolarization to terminate ventricular reentrant arrhythmias. In follow-up research, the system was further refined, allowing for closed-loop control of cardiac electrical activity, e.g., to restore normal conduction after AV block and to manipulate intraventricular wavefront propagation (Scardigli et al. 2018).
Complementary to patterned illumination for ChR activation, Weber et al. developed an imaging system for cell-accurate recording of Ca2+ dynamics of the entire developing zebrafish heart over extended time frames (here 36–52 h postfertilization) (Weber et al. 2017). To this end, they combined GCaMP5G expression in CM with nucleus-targeted mCherry labeling for identification of single cells. A custom-built light-sheet microscope enabled fast (400 Hz) high-resolution (0.5μm × 0.5μm × 1μm voxels) imaging of reporter fluorescence, and post-acquisition synchronization was used to visualize Ca2+ transients in CM across the embryonic myocardium.
3.2 Local Virus Delivery or Cell-Type-Specific Viral Serotypes
An alternative strategy for spatially defined, optogenetic activation was recently presented by Nyns et al. (2019). ReaChR-encoding adeno-associated viral (AAV) particles were locally applied to the right atrial (RA) epicardium of rat hearts (“gene painting”), resulting in efficient transmural transduction of RA myocytes, with minimal off-target expression in other cardiac compartments. Accordingly, RA illumination enabled optical pacing, as well as termination of atrial tachyarrhythmias, while illumination of non-RA areas did not affect heart rhythm.
Localized illumination or viral delivery allows for activation or recording from spatially restricted subsets of cells, but they do not necessarily enable targeting of specific cell classes within a region. With the advent of high-throughput single-cell sequencing approaches, the range of subtypes within each cell population is increasingly being recognized, and the functional relevance of each subpopulation needs to be elucidated.
The use of viral shuttles by itself may open up the possibility to target specific cell types, as certain viral serotypes preferentially transduce different cell populations (viral tropism), even upon systemic application. For example, the adeno-associated virus 2/9 has been shown to be cardiotropic, and CM specificity can be further increased by the use of a CM-specific promoter to drive transgene expression (Pacak et al. 2006; Bish et al. 2008; Prasad et al. 2011). In line, Vogt et al. showed that AAV2/9-mediated systemic delivery of Cop4 (coding for ChR2) ensured efficient and stable ChR2 expression in CM, allowing for blue-light triggered pacing of murine hearts (Vogt et al. 2015).
3.3 Genetic Targeting of Cardiomyocyte Subsets
Cell populations can be genetically targeted either by expressing transgenes under direct control of cell-specific promoters or using recombination systems such as the Cre-LoxP system. The latter approach was used by Zaglia et al. to drive ChR2 expression in CM (using α-myosin heavy chain-Cre) or in the specialized cells of the conduction system (using connexin-40-Cre) to probe the minimal cell number that needs to be simultaneously depolarized to induce ventricular extrasystoles (Zaglia et al. 2015). They found that focal ectopy requires depolarization of 1300–1800 working CM, supporting earlier computational predictions that inhibition of the Na+/K+ ATPase in approximately 1000 atrial CM would be sufficient to cause spontaneous ectopic beats (Winslow et al. 1993). In contrast, optogenetic depolarization of only 90–160 Purkinje fibers was sufficient to elicit focal ectopies in the murine ventricle. A Cre recombinase-based mouse model was also used by Wang et al. to elucidate the morphology and electromechanical function of phenylethanolamine N-methyltransferase (pnmt)-positive cells, a subset of CM able to convert noradrenaline to adrenaline, as well as their descendants (Wang et al. 2017).
As mentioned previously, NM constitute the majority of cardiac cells and play important roles in preserving and regulating cardiac structure and function—yet they were long regarded as “inert,” in as far as cardiac electrophysiology is concerned (acting as barriers, but not as active players). In recent years, a steadily rising number of studies have used optogenetic tools to target-specific NM populations, including interstitial cells, resident cardiac immune cells, cells forming the vasculature, and, last but not least, intracardiac neurons.
4 Interstitial Cells and Resident Cardiac Immune Cells
Interstitial fibroblasts not only generate the scaffold of the myocardium, but have been implicated in regulating cardiac function, both via biochemical and biophysical signaling (Gourdie et al. 2016). In vitro studies indicated direct electrotonic coupling between fibroblasts and CM as early as 50 years ago (Goshima and Tonomura 1969). However, only recently have optogenetic approaches provided direct evidence of such coupling in native myocardium. By selectively expressing VSFP in cardiac NM (using the Wilm’s tumor protein 1-Cre driver line) Quinn et al. observed action potential like depolarizations in NM of ventricular scar border zone tissue, indicating that these NM follow rhythmic de- and repolarization of electrotonically coupled CM in murine hearts after cryoinjury (Quinn et al. 2016). Similar findings were described for myofibroblasts, where combined myofibroblast-specific expression of the fluorescent reporter Zsgreen (driven by Periostin-Cre) and global loading with a red-shifted, voltage-sensitive dye, allowed for visualization of CM-myofibroblast coupling in the infarct border zone of mouse hearts (Rubart et al. 2017).
Resident macrophages constitute the most abundant immune cell type in the heart, with distinct functions in steady state and during myocardial remodeling following cardiac injury (Hulsmans et al. 2016). Notably, cardiac macrophages express connexin-43, indicating that they might be coupled to adjacent CM (Hulsmans et al. 2017). To assess functional macrophage-CM coupling, Hulsmans et al. expressed ChR2 specifically in resident cardiac macrophages (using the Cx3Cr1-Cre mouse line). Upon ChR2-mediated depolarization of macrophages, they observed improved AV node conduction at high-pacing frequencies, supporting the hypothesis that changes in the membrane potential of macrophages could potentially impact normal cardiac function even in healthy murine myocardium.
The above-described studies show how cell-specific optogenetic approaches can be used to probe the existence and functional relevance of heterocellular electrotonic coupling, both in healthy and remodeled mouse hearts.
5 Optogenetic Manipulation of the Cardiac Vasculature
The cardiac circulatory system provides oxygen and nutrients to the heart and facilitates the transport of immune and other circulatory cells to the four heart chambers. The principal cellular components of coronary vessels are endothelial and perivascular cells (vascular smooth muscle cells and pericytes), together providing the structure and regulating the development, stability, and contractile function of the vessels (Kapuria et al. 2018). Cardiac endothelial cells were optogenetically targeted by CreLoxP recombination of Cre-dependent ChR2 mice, bred with the cadherin (Cdh5)-Cre driver line (Zhang et al. 2015). Interestingly, blue-light mediated ChR2 activation in vascular endothelial cells led to vasoconstriction, thereby increasing the perfusion pressure of the Langendorff-perfused mouse heart. Vasoconstriction was also observed when activating ChR2 in cardiac vascular smooth muscle cells (driven by transgelin [Tagln]-Cre), with increasing constriction during sustained ChR2 activation, potentially indicating depolarization-induced changes in transmembrane ionic gradients (e.g., cellular Ca2+ overload) (Wu et al. 2015). Sustained ChR2 activation also elicited severe ventricular arrhythmias, suggesting insufficient myocardial blood perfusion.
6 Intrinsic Cardiac Nervous System
On top of centrally derived sympathetic and parasympathetic innervation of the heart, the heart has a complex intrinsic network of neurons, termed the intrinsic cardiac nervous system (ICNS). Traditionally, it was assumed that the ICNS acted as a relay system for the central nervous system, however, a number of subsequent studies confirmed that the ICNS plays a distinct and significant independent role in cardiac physiology in both health and disease (for a full review, see Wake and Brack 2016). However, probing the exact pathways and mechanisms by which the ICNS affects cardiac physiology using standard electrical stimulation techniques, presents with the difficulty of ensuring that only the specific neuron population of interest is excited. Optogenetics overcomes this issue by enabling cell-type-specific stimulation and recording, thus allowing the assessment of the role and interactions of different neuronal populations in the heart.
Studies using optogenetic tools to assess cardiac innervation initially focused on cardiac effects of optical activation of neurons in the central nervous system (Mastitskaya et al. 2012; Marina et al. 2013; Yu et al. 2017). Wengrowski et al. (2015) performed the first study looking directly at the role of the ICNS (Wengrowski et al. 2015). They expressed ChR2 in sympathetic neurons (SN; using the tyrosine hydroxylase-Cre driver line) and assessed how optical excitation of SN altered force production, heart rate, action potential duration, and arrhythmia susceptibility in Langendorff-perfused hearts. This study demonstrated that isolated hearts can be utilized to characterize cardiac responses to intracardiac neural activation through the use of optogenetics, simplifying experimental models, and opening up the path to a better understanding of cardiac innervation.
Prando et al. (2018) subsequently investigated the mechanisms underlying communication between SN and CM (Prando et al. 2018). Confocal microscopy demonstrated close contacts between SN and CM, and electron microscopy revealed sympathetic varicosities at an intermembrane distance of 70 nm from CM, and clustering of norepinephrine-containing vesicles in the SN close to the SN–CM interface. To test whether these points of contact act as spatially restricted domains for communication, an optogenetic sensor for cAMP (H187) was used in vitro, confirming that only the directly innervated cell was excited in response to SN stimulation. They further assessed the in vivo response to optogenetic activation of ChR2 in SN. By combining functional optogenetic experiments with same-site histological analysis, they identified a subset of SN that forms quasi-synaptic clefts with sinoatrial node CM. Presence of such quasi-synaptic junctions alters our understanding of cell–cell communication in the heart, a prerequisite for developing novel therapies, e.g., for treating atrial and ventricular fibrillation, both of which have been linked to abnormal nerve activity (Scherlag and Po 2006; Lu et al. 2009; He et al. 2013).
In 2019, the first reports targeting parasympathetic neurons (PN) were published. Moreno et al. expressed ChR2 in PN (using choline acetyltransferase-Cre mice) and demonstrated optically induced changes in heart rate and conduction (Moreno et al. 2019). Rajendran et al. took this a step further by examining both PN and SN innervation of the sinoatrial node (Rajendran et al. 2019). They demonstrated the ability to accurately trace cardiac innervation through the use of optical clearing, immunostaining, and genetic labeling, providing a high-resolution map of the murine ICNS. They further compared optical stimulation of PN to SN in terms of its effects on cardiac electrophysiology and confirmed the utility of optogenetics by comparing optical to electrical stimulation of the vagus nerve. Their data suggest that electrical stimulation affects both afferent and efferent fibers in the vagus nerve, whereas optical stimulation, due to genetic expression of probes in particular neuronal subsets, can be used to only stimulate efferent fibers.
The above studies exemplify how optogenetic approaches can greatly facilitate probing the roles of different cardiac cell types, including CM, fibroblasts, macrophages, endothelial cells, smooth muscle cells, and intracardiac neurons, and subpopulations thereof. In the following paragraphs, we will discuss several challenges of cardiac optogenetics at present, and propose how they might be overcome in the near future.
7 Current Challenges and Future Directions
A range of optogenetic sensors and actuators have been applied to record and modulate cell-type-specific cardiac activity. Despite genetically targetable optogenetic sensors first being used in the heart in 2006, and actuators in 2010, cardiac optogenetics research has primarily utilized CM-specific expression in healthy hearts. The ability to target-specific cell types, subtypes, and intracellular compartments has been shown, but primarily as a part of proof-of-principle studies. Thus far, the use of optogenetics to address fundamental scientific questions in the cardiac field is lagging behind neurosciences, with only a handful of published reports, largely focusing on the healthy heart (Quinn et al. 2016; Hulsmans et al. 2017; Prando et al. 2018; Rajendran et al. 2019). The advent of new models, and demonstration of their utility for assessing the specific roles of cardiac endothelial cells, smooth muscle cells, neurons, and interstitial cells in recent years, will hopefully encourage addressing more fundamental questions regarding cardiac biology in health and disease.
In particular, optogenetics could be used to modulate different NM populations in the diseased heart to help understand their role in cardiac homeostasis and repair and how these differ between localized lesions (e.g., those occurring due to myocardial infarction or ablation therapy) and global remodeling (e.g., as occurs in atrial fibrillation or cardiac hypertrophy). Potentially exciting applications of cardiac optogenetics—such as for defibrillation—have been explored in “healthy” hearts. These may not apply equally to a diseased heart, where cells die (e.g., CM), proliferate (e.g., interstitial cells), and/or invade from external sources (e.g., immune cells), changing tissue electrophysiological properties. Resulting spatial heterogeneities may act as organizing centers for arrhythmias, making them more difficult to be terminated. Furthermore, poor cell health may affect ion distributions, so that optogenetic activation of ion channels may have different effects in diseased, compared to healthy myocardium. In regard to optogenetic defibrilliation, it would be most interesting to explore which cell type(s) might be best suited to terminate arrhythmias with light, as targeting NM might be a safer option than directly targeting CM, and may allow for targeting lesion-specific activated cell populations. A further key issue with the application of optogenetics for treating cardiac disease is that no study has shown the ability to perform long-term cardiac optical stimulation or observation on vertebrates in vivo. Weber et al. recorded calcium transients in zebrafish embryos over 16 h (Weber et al. 2017), and no group has exceeded this period to our knowledge. A number of groups have demonstrated optical pacing or resynchronization in vivo with open-chest models (Bruegmann et al. 2010; Nussinovitch and Gepstein 2015; Vogt et al. 2015), and Nyns et al. recently demonstrated the ability to pace and defibrillate in a closed-chest model (Nyns et al. 2017). However, transitioning from using optogenetics in an anesthetized to a freely moving animal remains challenging, due to continuous motion (heartbeat, respiration) of the heart, and its location in the body.
The use of larger animals would potentially lower the technical threshold to implementing chronic studies by offering more space for implanting flexible light sources. Furthermore, larger mammals such as rabbits or pigs are considered more suitable models of the human heart compared to rodent species. This is especially true for cardiac electrophysiology, with large interspecies differences, e.g., in action potential shape and heart rate. However, taking cardiac optogenetics to non-rodent hearts poses additional challenges. One of the main hurdles is limited light penetration into tissue, as blue light is attenuated by 80% within 1–2 mm of the myocardial surface (Baxter et al. 2001; Zaglia et al. 2015). Tissue-penetrating light sources, which allow transmural illumination, are available, but their use would still be restricted by light penetration in 3D (Zgierski-Johnston et al. 2019). Alternatively, developments in the field of nanoparticles offer the possibility for exciting deeper tissue layers through either photo-upconversion (see review by All et al. 2019, where long-wavelength light is used to excite nanoparticles which emit lower wavelengths), or ultrasound-triggered light emission (Wu et al. 2019). However, the need to produce such nanoparticles limits their widespread adoption and their application and turnover pose nontrivial problems for research and development. A further approach would be the use of ultralight-sensitive optogenetic tools, such as the recently presented optogenetic Ca2+ modulator monSTIM1, which can be noninvasively activated by illuminating the animal with blue light of moderate intensity (Kim et al. 2020). Finally, newly developed genetically encoded infrared reporters (Monakhov et al. 2019) or red-shifted actuators (Oda et al. 2018) are set to enable the use of longer wavelength light for excitation, thereby opening up the possibility for transmural observation and steering in larger hearts. Red-shifted proteins offer the additional advantage of allowing for combined use of optogenetic tools, an area that has traditionally been challenging due to spectral overlap.
Utilizing newly generated optogenetic tools remains difficult. The majority of researchers using cardiac optogenetics—including ourselves—still utilize ChR2; despite the available repertoire of ChR variants with improved/tuned properties, including action spectra, photocycle kinetics, ion selectivity, and channel membrane targeting (Prigge et al. 2012; Schneider et al. 2015; Rost et al. 2017). This is largely due to the availability of floxed ChR2 mouse lines and ChR2-encoding viral vectors, while the development of new transgenic mouse lines is both cost- and time-consuming. We still encourage researchers to identify the optogenetic probe most suitable for each specific experimental design, as this may reduce unwanted side effects of optogenetic activation. For example, ChR2 has been shown to effectively silence CM activity during prolonged illumination, but at the cost of depolarizing CM to the reversal potential of ChR2 (near 0 mV), activating secondary voltage-gated ion channels. This may, in particular in diseased myocardium, aggravate the conditions (such as Ca2+-overload-induced arrhythmogenesis) that one might hope to control or treat. Other tools might be better suited to arrest cardiac excitation, in particular, hyperpolarizing light-driven pumps and light-gated Cl− and K+ channels (Arrenberg et al. 2010; Govorunova et al. 2016; Kopton et al. 2018; Bernal Sierra et al. 2018; Funken et al. 2019).
An alternative model system is zebrafish, as the effort involved in generating transgenic fish is much lower than for mammals. Furthermore, a number of recent publications suggest that fish may even be a more suitable cardiac model than mice (see editorial by Stoyek and Quinn 2018). A major advantage is the speed and relative simplicity of gene manipulation in zebrafish. Zebrafish are, of course, not suitable for all studies, due to their small size and different biology (e.g., two-chamber heart, potential to regenerate heart tissue), but they can serve as a good model for generating hypotheses and for testing novel optogenetic tools.
8 Summary
Experiments targeting a range of different cardiac cell types and subtypes have demonstrated the utility of optogenetics for teasing apart the role of specific cell populations in the heterocellular heart. The studies discussed above lay the groundwork for future research, focusing on understanding the interactions between different cell populations, and their role in development, homeostasis, disease, and therapy.
Abbreviations
- AAV:
-
Adeno-associated viral [particles]
- ACR:
-
Anion channelrhodopsins
- AV:
-
Atrioventricular
- ChR2:
-
Chlamyodomonas reinhardtii channelrhodopsin-2
- CM:
-
Cardiomyocytes
- ECG:
-
Electrocardiogram
- ICNS:
-
Intrinsic cardiac nervous system
- NM:
-
Non-myocytes
- NpHR:
-
Natromonas pharaonis halorhodopsin
- PN:
-
Parasympathetic neurons
- RA:
-
Right atrial
- SN:
-
Sympathetic neurons
- VSFP:
-
Voltage-sensitive fluorescent protein
References
All AH, Zeng X, Teh DBL et al (2019) Expanding the toolbox of upconversion nanoparticles for in vivo optogenetics and neuromodulation. Adv Mater 31:1–15. https://doi.org/10.1002/adma.201803474
Arrenberg AB, Stainier DYR, Baier H, Huisken J (2010) Optogenetic control of cardiac function. Science 330:971–974. https://doi.org/10.1126/science.1195929
Baxter WT, Mironov SF, Zaitsev AV et al (2001) Visualizing excitation waves inside cardiac muscle using transillumination. Biophys J 80:516–530. https://doi.org/10.1016/S0006-3495(01)76034-1
Bernal Sierra YA, Rost BR, Pofahl M et al (2018) Potassium channel-based optogenetic silencing. Nat Commun 9:4611. https://doi.org/10.1038/s41467-018-07038-8
Bish LT, Morine K, Sleeper MM et al (2008) Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum Gene Ther 19:1359–1368. https://doi.org/10.1089/hum.2008.123
Boyden ES, Zhang F, Bamberg E et al (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8:1263–1268. https://doi.org/10.1038/nn1525
Bruegmann T, Malan D, Hesse M et al (2010) Optogenetic control of heart muscle in vitro and in vivo. Nat Methods 7:897–900. https://doi.org/10.1038/nmeth.1512
Bruegmann T, Boyle PM, Vogt CC et al (2016) Optogenetic defibrillation terminates ventricular arrhythmia in mouse hearts and human simulations. J Clin Invest 126:3894–3904. https://doi.org/10.1172/JCI88950
Bruegmann T, Beiert T, Vogt CC et al (2018) Optogenetic termination of atrial fibrillation in mice. Cardiovasc Res 114:713–723. https://doi.org/10.1093/cvr/cvx250
Chi NC, Shaw RM, Jungblut B et al (2008) Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol 6:1006–1019. https://doi.org/10.1371/journal.pbio.0060109
Chi NC, Bussen M, Brand-Arzamendi K et al (2010) Cardiac conduction is required to preserve cardiac chamber morphology. Proc Natl Acad Sci U S A 107:14662–14667. https://doi.org/10.1073/pnas.0909432107
Crocini C, Ferrantini C, Coppini R et al (2016) Optogenetics design of mechanistically-based stimulation patterns for cardiac defibrillation. Sci Rep 6:1–7. https://doi.org/10.1038/srep35628
Despa S, Shui B, Bossuyt J et al (2014) Junctional cleft [Ca2+]i measurements using novel cleft-targeted Ca2+ sensors. Circ Res 115:339–347. https://doi.org/10.1161/CIRCRESAHA.115.303582
Funken M, Malan D, Sasse P, Bruegmann T (2019) Optogenetic hyperpolarization of cardiomyocytes terminates ventricular arrhythmia. Front Physiol 10:1–7. https://doi.org/10.3389/fphys.2019.00498
Goshima K, Tonomura Y (1969) Synchronized beating of embryonic mouse myocardial cells mediated by FL cells in monolayer culture. Exp Cell Res 56:387–392. https://doi.org/10.1016/0014-4827(69)90029-9
Gourdie RG, Dimmeler S, Kohl P (2016) Novel therapeutic strategies targeting fibroblasts and fibrosis in heart disease. Nat Rev Drug Discov 15(9):620–638. https://www.nature.com/articles/nrd.2016.89. Accessed 10 Jan 2020
Govorunova EG, Cunha SR, Sineshchekov OA, Spudich JL (2016) Anion channelrhodopsins for inhibitory cardiac optogenetics. Sci Rep 6:33530. https://doi.org/10.1038/srep33530
He B, Lu Z, He W et al (2013) The effects of atrial ganglionated plexi stimulation on ventricular electrophysiology in a normal canine heart. J Interv Card Electrophysiol 37:1–8. https://doi.org/10.1007/s10840-012-9774-2
Hou JH, Kralj JM, Douglass AD et al (2014) Simultaneous mapping of membrane voltage and calcium in zebrafish heart in vivo reveals chamber-specific developmental transitions in ionic currents. Front Physiol 5:344. https://doi.org/10.3389/fphys.2014.00344
Hulsmans M, Sam F, Nahrendorf M (2016) Monocyte and macrophage contributions to cardiac remodeling. J Mol Cell Cardiol 93:149–155. https://doi.org/10.1016/j.yjmcc.2015.11.015
Hulsmans M, Clauss S, Xiao L et al (2017) Macrophages facilitate electrical conduction in the heart. Cell 169:510–522.e20. https://doi.org/10.1016/j.cell.2017.03.050
Ishizuka T, Kakuda M, Araki R, Yawo H (2006) Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res 54:85–94. https://doi.org/10.1016/j.neures.2005.10.009
Kapuria S, Yoshida T, Lien C-L (2018) Coronary Vasculature in Cardiac Development and Regeneration. J Cardiovasc Dev Dis 5:59. https://doi.org/10.3390/jcdd5040059
Kim S, Kyung T, Chung J et al (2020) Non-invasive optical control of endogenous Ca2+ channels in awake mice. Nat Commun 11:210. https://doi.org/10.1038/s41467-019-14005-4
Kopton RA, Baillie JS, Rafferty SA et al (2018) Cardiac electrophysiological effects of light-activated chloride channels. Front Physiol 9:1806. https://doi.org/10.3389/FPHYS.2018.01806
Li X, Gutierrez DV, Hanson MG et al (2005) Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci 102:17816–17821. https://doi.org/10.1073/pnas.0509030102
Liao MLC, De Boer TP, Mutoh H et al (2015) Sensing cardiac electrical activity with a cardiac myocyte-targeted optogenetic voltage indicator. Circ Res 117:401–412. https://doi.org/10.1161/CIRCRESAHA.117.306143
Ljubojevic S, Radulovic S, Leitinger G et al (2014) Early remodeling of perinuclear Ca2+ stores and nucleoplasmic Ca2+ signaling during the development of hypertrophy and heart failure. Circulation 130:244–255. https://doi.org/10.1161/CIRCULATIONAHA.114.008927
Lu Z, Scherlag BJ, Lin J et al (2009) Autonomic mechanism for initiation of rapid firing from atria and pulmonary veins: evidence by ablation of ganglionated plexi. Cardiovasc Res 84:245–252. https://doi.org/10.1093/cvr/cvp194
Lu X, Ginsburg KS, Kettlewell S et al (2013) Measuring local gradients of intramitochondrial [Ca2+] in cardiac myocytes during sarcoplasmic reticulum Ca2+ release. Circ Res 112:424–431. https://doi.org/10.1161/CIRCRESAHA.111.300501
Marina N, Tang F, Figueiredo M et al (2013) Purinergic signalling in the rostral ventro-lateral medulla controls sympathetic drive and contributes to the progression of heart failure following myocardial infarction in rats. Basic Res Cardiol 108:1–10. https://doi.org/10.1007/s00395-012-0317-x
Mastitskaya S, Marina N, Gourine A et al (2012) Cardioprotection evoked by remote ischaemic preconditioning is critically dependent on the activity of vagal pre-ganglionic neurones. Cardiovasc Res 95:487–494. https://doi.org/10.1093/cvr/cvs212
Miesenböck G (2009) The optogenetic catechism. Science 326:395–399
Monakhov M, Matlashov M, Colavita M, et al (2019) Bright near-infrared genetically encoded voltage indicator for all-optical electrophysiology. bioRxiv 536359. https://doi.org/10.1101/536359
Moreno A, Endicott K, Skancke M et al (2019) Sudden heart rate reduction upon optogenetic release of acetylcholine from cardiac parasympathetic neurons in perfused hearts. Front Physiol 10:16. https://doi.org/10.3389/fphys.2019.00016
Nagel G, Ollig D, Fuhrmann M et al (2002) Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296:2395–2398. https://doi.org/10.1126/science.1072068
Nagel G, Szellas T, Huhn W et al (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A 100:13940–13945. https://doi.org/10.1073/pnas.1936192100
Nagel G, Brauner M, Liewald JF et al (2005) Light activation of Channelrhodopsin-2 in excitable cells of caenorhabditis elegans triggers rapid behavioral responses. Curr Biol 15:2279–2284. https://doi.org/10.1016/j.cub.2005.11.032
Nussinovitch U, Gepstein L (2015) Optogenetics for in vivo cardiac pacing and resynchronization therapies. Nat Biotechnol 33:750–754. https://doi.org/10.1038/nbt.3268
Nyns ECA, Kip A, Bart CI et al (2017) Optogenetic termination of ventricular arrhythmias in the whole heart: towards biological cardiac rhythm management. Eur Heart J 38:2132–2136. https://doi.org/10.1093/eurheartj/ehw574
Nyns ECA, Poelma RH, Volkers L et al (2019) An automated hybrid bioelectronic system for autogenous restoration of sinus rhythm in atrial fibrillation. Sci Transl Med 11:1–12. https://doi.org/10.1126/scitranslmed.aau6447
Oda K, Vierock J, Oishi S et al (2018) Crystal structure of the red light-activated channelrhodopsin Chrimson. Nat Commun 9:1–11. https://doi.org/10.1038/s41467-018-06421-9
Pacak CA, Mah CS, Thattaliyath BD et al (2006) Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ Res 99:e3–e9. https://doi.org/10.1161/01.RES.0000237661.18885.f6
Prando V, Da Broi F, Franzoso M et al (2018) Dynamics of neuroeffector coupling at cardiac sympathetic synapses. J Physiol 596:2055–2075. https://doi.org/10.1113/JP275693
Prasad K-MR, Xu Y, Yang Z et al (2011) Robust cardiomyocyte-specific gene expression following systemic injection of AAV: in vivo gene delivery follows a Poisson distribution. Gene Ther 18:43–52. https://doi.org/10.1038/gt.2010.105
Prigge M, Schneider F, Tsunoda SP et al (2012) Color-tuned channelrhodopsins for multiwavelength optogenetics. J Biol Chem 287:31804–31812. https://doi.org/10.1074/jbc.M112.391185
Quinn TA, Camelliti P, Rog-Zielinska EA et al (2016) Electrotonic coupling of excitable and nonexcitable cells in the heart revealed by optogenetics. Proc Natl Acad Sci 113:14852–14857. https://doi.org/10.1073/pnas.1611184114
Rajendran PS, Challis RC, Fowlkes CC et al (2019) Identification of peripheral neural circuits that regulate heart rate using optogenetic and viral vector strategies. Nat Commun 10:1–13. https://doi.org/10.1038/s41467-019-09770-1
Rost BR, Schneider-Warme F, Schmitz D, Hegemann P (2017) Optogenetic tools for subcellular applications in neuroscience. Neuron 96:572–603. https://doi.org/10.1016/j.neuron.2017.09.047
Rubart M, Tao W, Lu X-L et al (2017) Electrical coupling between ventricular myocytes and myofibroblasts in the infarcted mouse heart. Cardiovasc Res 107:1011–1020. https://doi.org/10.1093/cvr/cvx163
Scardigli M, Müllenbroich C, Margoni E et al (2018) Real-time optical manipulation of cardiac conduction in intact hearts. J Physiol 596:3841–3858. https://doi.org/10.1113/JP276283
Scherlag BJ, Po S (2006) The intrinsic cardiac nervous system and atrial fibrillation. Curr Opin Cardiol 21:51–54. https://doi.org/10.1097/01.hco.0000198980.40390.e4
Schneider F, Grimm C, Hegemann P (2015) Biophysics of channelrhodopsin. Annu Rev Biophys 44:167–186. https://doi.org/10.1146/annurev-biophys-060414-034014
Shang W, Lu F, Sun T et al (2014) Imaging Ca2+ nanosparks in heart with a new targeted biosensor. Circ Res 114:412–420. https://doi.org/10.1161/CIRCRESAHA.114.302938
Sineshchekov OA, Jung K-H, Spudich JL (2002) Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A 99:8689–8694. https://doi.org/10.1073/pnas.122243399
Stoyek MR, Quinn TA (2018) One fish, two fish, red fish, blue fish*: zebrafish as a model for cardiac research. Prog Biophys Mol Biol 138:1–2. https://doi.org/10.1016/j.pbiomolbio.2018.11.003
Suzuki T, Yamasaki K, Fujita S et al (2003) Archaeal-type rhodopsins in chlamydomonas: model structure and intracellular localization. Biochem Biophys Res Commun 301:711–717. https://doi.org/10.1016/S0006-291X(02)03079-6
Tallini YN, Ohkura M, Choi B-R et al (2006) Imaging cellular signals in the heart in vivo: cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc Natl Acad Sci U S A 103:4753–4758. https://doi.org/10.1073/pnas.0509378103
Tallini YN, Brekke JF, Shui B et al (2007) Propagated endothelial Ca2+ waves and arteriolar dilation in vivo: measurements in Cx40BAC-GCaMP2 transgenic mice. Circ Res 101:1300–1309. https://doi.org/10.1161/CIRCRESAHA.107.149484
Vogt CC, Bruegmann T, Malan D et al (2015) Systemic gene transfer enables optogenetic pacing of mouse hearts. Cardiovasc Res 106:338–343. https://doi.org/10.1093/cvr/cvv004
Wake E, Brack K (2016) Characterization of the intrinsic cardiac nervous system. Auton Neurosci Basic Clin 199:3–16. https://doi.org/10.1016/j.autneu.2016.08.006
Wang Y, Lin WK, Crawford W et al (2017) Optogenetic control of heart rhythm by selective stimulation of cardiomyocytes derived from pnmt + cells in murine heart. Sci Rep 7:40687. https://doi.org/10.1038/srep40687
Weber M, Scherf N, Meyer AM et al (2017) Cell-accurate optical mapping across the entire developing heart. elife 6:1–23. https://doi.org/10.7554/eLife.28307
Wengrowski AM, Wang X, Tapa S et al (2015) Optogenetic release of norepinephrine from cardiac sympathetic neurons alters mechanical and electrical function. Cardiovasc Res 105:143–150. https://doi.org/10.1093/cvr/cvu258
Winslow RL, Varghese A, Noble D et al (1993) Generation and propagation of ectopic beats induced by spatially localized Na-K pump inhibition in atrial network models. Proc R Soc B Biol Sci 253:55–61. https://doi.org/10.1098/rspb.1993.0126
Wu Y, Li SS, Jin X et al (2015) Optogenetic approach for functional assays of the cardiovascular system by light activation of the vascular smooth muscle. Vasc Pharmacol 71:192–200. https://doi.org/10.1016/j.vph.2015.03.006
Wu X, Zhu X, Chong P et al (2019) Sono-optogenetics facilitated by a circulation delivered rechargeable light source for minimally invasive optogenetics. Proc Natl Acad Sci U S A 116:26332–26342. https://doi.org/10.1073/pnas.1914387116
Yu L, Zhou L, Cao G et al (2017) Optogenetic modulation of cardiac sympathetic nerve activity to prevent ventricular arrhythmias. J Am Coll Cardiol 70:2778–2790. https://doi.org/10.1016/j.jacc.2017.09.1107
Zaglia T, Pianca N, Borile G et al (2015) Optogenetic determination of the myocardial requirements for extrasystoles by cell type-specific targeting of Channel Rhodopsin-2. Proc Natl Acad Sci 112:E4495–E4504. https://doi.org/10.1073/pnas.1509380112
Zgierski-Johnston CM, Ayub S, Fernández MC et al (2019) Cardiac pacing using transmural multi-LED probes in channelrhodopsin-expressing mouse hearts. Prog Biophys Mol Biol 154:51–61. https://doi.org/10.1016/j.pbiomolbio.2019.11.004
Zhang S, Cui N, Wu Y et al (2015) Optogenetic intervention to the vascular endothelium. Vasc Pharmacol 74:122–129. https://doi.org/10.1016/j.vph.2015.05.009
Acknowledgments
We thank all members of the Institute of Experimental Cardiovascular Medicine for critical discussion of the manuscript. This research was funded by the German Research Foundation DFG (SPP1926: FS1486/1-2, ZG58/1-1, and an Emmy-Noether-Fellowship: FS1486/2-1). Both authors are members of the DFG-funded Collaborative Research Centre 1425.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Zgierski-Johnston, C.M., Schneider-Warme, F. (2021). Observing and Manipulating Cell-Specific Cardiac Function with Light. In: Yawo, H., Kandori, H., Koizumi, A., Kageyama, R. (eds) Optogenetics. Advances in Experimental Medicine and Biology, vol 1293. Springer, Singapore. https://doi.org/10.1007/978-981-15-8763-4_24
Download citation
DOI: https://doi.org/10.1007/978-981-15-8763-4_24
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-8762-7
Online ISBN: 978-981-15-8763-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)