Abstract
Enzymes are fascinating the researchers because of their enormous power of catalysis and eco-friendly nature. In biotechnological processes, diversity of microbes is studied, and different metabolic reactions entitle a potential repository that direct valuable production of desirable products. Since community demands are getting more intensified, there is a continuous need to evolve the enzymes. There has been an immense development in techniques and computational tools that has developed the industries to meet the growing demands. The techniques such as protein engineering help in development of quality products by mutating the amino acids to make more stable and efficient product. Further, the techniques like enzyme immobilization give the opportunity to reuse the used enzyme with the same efficiency, thus a cost-effective measure for the industrial enzyme. Nanotechnology and CLEA formation are also incorporated in enzyme engineering to increase enzyme efficiency and their characteristics.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
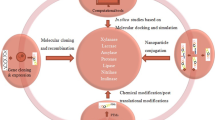
The microbial enzymes are natural biocatalysts which are produced by different forms of microorganisms such as bacteria, fungi, algae, yeast, and actinomycetes to accelerate the rate biochemical reactions taking place in vivo system (Baweja et al. 2016; Kumar et al. 2014; Singh et al. 2016). These enzymes have been involved in all the processes such as DNA replication, transcription, translation, protein synthesis, metabolic reactions, and cycles which are essential for life, and their unique ability to carry out substrate-specific transformation has made them suitable for industrial processes. Enzymes are omnipresent ranging from living organisms to industries which catalyze the biochemical reactions to maintain the living system or synthesis of a product or degradation of a substance (Shrivastava et al. 2012, 2013; Kumar et al. 2014, 2016). The enzymes have replaced the chemicals from the industries, which are more eco-friendly and help to step up toward green environment. The industries are now totally enzyme dependent, and there is continuous demand for a number of microbial enzymes with novel characteristics and stability under industrial extreme conditions (Kumar and Shukla 2015). There is a continuous progress in microbiology, biotechnology, and bioinformatics to provide microbial enzymes with novel characteristics for the development of better industrial processes (Singh and Shukla 2012, 2015). The techniques of recombinant DNA technology, protein engineering, immobilization, nanotechnology, and metabolic engineering have progressed enough to support the industrial enzyme load. The industry demands operation stable enzymes with novel biochemical properties which should be economic and environment friendly. The continuous efforts are being done to meet all objectives of industrial demand of enzymes by utilizing a combination of various modern techniques of biotechnology, bioinformatics, microbiology, and nanotechnology. The enzyme engineering is proving as one of the finest techniques to improve the enzyme properties like operational stability, physical stability of enzyme, substrate specificity, and enhanced activity. There are several enzyme engineering techniques such as protein engineering, metabolic engineering, immobilization, and nanotechnology, and in silico methods; in this chapter, all these aspects will be described briefly. It will be helpful to understand the revolution in the microbial enzyme by changing at certain level or complete redesigning of the enzymes.
2 Improvement of Enzymes by Tailoring Their Protein Sequences
The industries continuously demand enzymes with high stabilities and substrate specificities. Since the native enzyme lacks the efficient system that can cope with hostile industrial conditions, there is a need to improve the enzyme that could withstand the harsh industrial conditions. With progress in recombinant technology and enzyme engineering, it has become possible to obtain customized enzymes. The improved novel enzymes that can fit into industry can be achieved by genetic manipulated microorganisms such as recombinant insulin production by using Escherichia coli as host. The recombinant DNA technology makes possible to 100-fold increase in production of enzymes than the native expression, making them available at low cost and in large quantities. Thus, various food-processing enzymes and laundry enzymes can be tailored as per demand of the industrial process.
The protein engineering allows modification in protein itself to improve the properties for suitability to industrial process. In protein engineering, mutation is the key to improve the enzyme properties and to explore protein function. It is a method to alter a protein sequence to obtain a desired effect, such as change in the substrate specificity with increased stability toward extremes of pH and the temperature and in organic solvents. The protein engineering is divided into two types: (1) site-directed mutagenesis or rational design and (2) random mutagenesis.
2.1 Improvement of Enzyme Properties by Site-Directed Mutagenesis
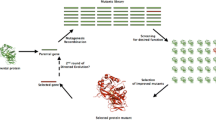
The protein engineering is proving as stupendous technique to modify the enzyme to achieve customized biocatalyst. The major drawback with wild-type enzyme is that they cannot bear harsh experimental conditions. To overcome the snag of native enzyme, researchers are continuously adopting various methods to obtain refined industrial enzymes. Protein engineering is one such technique that works at the level of nucleotide to evolve the functional aspect of the protein. A comparison of native and engineered enzyme production has been described in Fig. 13.1. Site-directed mutagenesis, as the name suggests, is site-specific technique to improve enzyme. Thus, it is the technique for the proteins with full knowledge of structure and mechanism of action. The mutation type may vary as per requisite like point mutation, insertion, deletion, and substitution. Depending upon the number of mutation in a gene, it can be single site-directed mutagenesis and multiple site-directed mutageneses. The major application of site-directed mutagenesis is to introduce novel properties like enhanced specificity, stability, activity, solubility, expression, etc. to the biocatalyst. There are number of reports on improvement of industrial enzyme. A study was conducted to improve α-galactosidase features; the protein was mutated at specific site that improved the enzyme properties and also added the structural and functional information (Xu et al. 2014). In a similar study, the thermostability of the immobilized protease was improved by introducing Cys residues on surface of a cysteine-free mutant of a thermolysin-like protease from B. stearothermophilus and thus facilitated the site-directed immobilization of protease via single thiol group onto thiol Sepharose (Eijsink et al. 1995). It was reported by Rahimi et al. 2016 that mutation at nearby active site region is more promising in improving the protein function. A study was conducted to improve the keratinase enzyme to enhance its application at industrial level. Although the native enzyme itself had immense activity and pH stability, a truncation of PPC domain improved the tolerance to alkalinity, salt, chaotropic agents, and detergents (Fang et al. 2012). A study deduced that substitution of conserved residue Asn by arginine of γ-glutamyltranspeptidase (BlGGT) by site-directed mutagenesis resulted in reduction in the catalytic activity (Lin et al. 2016). Wang and coworkers also elucidated the role of conserved amino acid residues by generating mutants by site-directed mutagenesis (Wang et al. 2015). The thermophilic archeal protein ST0452 was studied to comprehend the molecular machinery; after analyses, the researchers identified certain amino residues important for the glucosamine-1-phosphate and galactosamine-1-phosphate activities, viz., His308 is necessary for both GalN-1-P and GlcN-1-P AcTase activities, whereas Asn331 and Tyr311 are important only for the GalN-1-P AcTase activity (Zhang et al. 2015).
2.2 Improvement of Enzyme Properties by Random Mutagenesis
Random mutagenesis mimics the nature’s process of variant generation following the unbiased approach. Since it involves mutation in randomized manner, thus it is a method of choice for those proteins whose structure has not been deduced (Baweja et al. 2016). Thus, it is quite an easy technique to employ but has cumbersome screening process since a number of variants produced are often large. There are various physical and chemical methods to create random mutation, such as chemical agents like ethyl methane sulfonate (EMS), methylnitronitrosoguanidine (MNNG), and ethylnitrosourea (ENU), using error-prone PCR by using less FideliTaq polymerase instead of using pfu polymerase; using base analogs, altering the concentration of nucleotides; using heavy water during PCR, mutator strain, and many more; or using genetic recombination techniques like those based on gene recombination which are DNA shuffling, random chimeragenesis on transient templates (RACHITT), staggered extension process (StEP), recombined extension on truncated templates (RETT), and iterative truncation for the creation of hybrid enzymes (ITCHY) (Sen et al. 2007; Rasila et al. 2009; Baweja et al. 2015). The error-prone PCR is the most common technique in random mutagenesis with high success rates. The primary motto behind random mutagenesis is to characterize the open reading frame (ORF) and to modify the gene to obtain the desired product (Ramli et al. 2011).
There are various computational tools available that guide the library diversity and design, viz., ConSurf-HSSP GLUE, PEDEL, DRIVeR, and SCHEMA (Labrou 2010). There are various bioinformatics techniques available that reduce the cumbersome process of screening out libraries. Techniques like modeling and docking of enzymes prescreen the variants by giving the docking score that evaluates the enzyme-substrate relationship and effectiveness of their binding. The modeling and docking studies have been done in various enzymes like inulinase and xylanase (Karthik and Shukla 2012; Singh et al. 2016). The molecular dynamics simulation helps evaluate the stability of particular protein in particular milieu and thus filters out the variants during library screening (Singh et al. 2016).
3 Changing Pathways: Synthetic Metabolic Engineering
Industrial biotechnology promises to revolutionize conventional chemical manufacturing in the years ahead, largely owing to the excellent progress to reengineer cellular metabolism. It was evidenced that production of food stuffs and biofuels was enhanced after the era of metabolic engineering (Yadav et al. 2012). Metabolic engineering involves modification of metabolic pathways to screen the effect on the production of desired metabolite. To owe successful metabolic engineering, the first step is to crack metabolic pathway involved in the production of particular metabolite and to preclude the rate-limiting step in the reaction. The alteration in the rate-limiting step can be done by either overexpression of heterologous gene contributing the rate limitation or inhibiting the pathway in the network that halts the formation of desired product as shown in Fig. 13.2. To renew the production of metabolite, various experimental and computational tools are used to add beneficial traits to the system (Stafford and Gregory 2001). Metabolic flux plays an important role in valuation of the cellular phenotype; thus flux determination methods are inevitable components of metabolic engineering. Nowadays isotopic tracers are used to evaluate the balance of intracellular and extracellular metabolites. The 13C and 14C compounds were used to track the changes in and out of the cell (Klapa et al. 1999; Schmidt et al. 1997; Stafford et al. 2001). The introduction of multigene pathways into a host for heterologous production often faces flux imbalance because the host usually does not possess complex regulatory machinery to maintain such pathways. However efforts have been done to resolve such problems by combining metabolic engineering tools with combinatorial genetics (Ajikumar et al. 2010; Lee et al. 2011). Although metabolic flux helps to deduce the pathway feature, it is not enough to decipher the system. Various high-throughput probes are used for complete revelation of metabolic networks. Among various molecular biological tools, efficient transformation system, viz., plasmids, hosts, and efficient promoters, is crucial for efficient product development and its modification using gene-editing approaches (Gupta and Shukla 2016; Keasling 1999). Gupta and Shukla (2015) described E.coli as suitable host during transformation.
4 Immobilization of Microbial Enzyme
Microbial enzymes catalyze a number of biochemical reactions efficiently and selectively; that’s why they possess the ability to synthesize or to convert one compound to another. Immobilized forms of microbial enzymes have several industrial applications that are clear-cut as they provide a recycling method for the production of various compounds through biocatalyzed reactions. An immobilized microbial enzyme is the stable form of enzyme which is bound to an inert, insoluble material such as silica, chitosan, calcium alginate, copper alginate, agarose, polyacrylamide, etc. which provides increased stability in changing conditions of pH or temperature during industrial processes. It allows enzymes to stay held on supported material throughout the reaction following which they are separated for recycling and reuse. Along with synthesis of desired compounds, immobilized microbial enzymes also have the ability to decompose harmful compounds making them suitable for additional field of industrial application in bioremediation and purification. Besides these applications, the enzyme immobilization techniques are basis to synthesize a number of biotechnological products that have various applications in biosensors and bioaffinity chromatography and diagnostics (Guibault et al. 1991). A therapeutic application of immobilized enzymes is in extracorporeal shunts (Chang 1991). In the history of three or four decades, immobilization techniques have been developed swiftly, but still there is a need for further development. Immobilization technique is systematically studied with the probability of modification and improvement of enzyme stability and characteristics for economic purposes. There are a number of microbial enzymes such as xylanase, phytase, laccase, inulinase, cellulase, and amylase which have been immobilized on various materials. Among this series, microbial xylanase, which catalyzes the hydrolysis of xylan, is considered one of the most significant hydrolases. It has numerous applications, but most extensively it is utilized in paper and pulp industry as a biobleaching and biodeinking agent. Kapoor and Kuhad in 2007 used a number of matrices to immobilize the xylanase enzyme using various methods from Bacillus pumilus such as entrapment using gelatin, physical adsorption on chitin, ionic binding with Q-sepharose, and covalent binding with HP-20 beads with maximum xylanase immobilization efficiency. Similarly, Nagar et al. (2012) used the immobilized xylanase enzymes to improve the digestibility of poultry feed. Aluminum oxide pellets charged with glutaraldehyde were used for the immobilization which results in increase of enzyme temperature optima from 50 to 60 °C and V max from 3333.33 to 5000 IU/mL. Immobilized xylanase was biochemically active up to ten consecutive cycles with 60% of its initial activity. In the same series, xylanase enzyme has also been covalently immobilized on the beads of glutaraldehyde-alginate exteriorly which retains their efficiency more than 91% with an increase in kinetic parameters V max (7092–8000 IU/ml) and K m (0.9–1.49%) and an increase in pH optima 5–5.5 and temperature optima from 40 to 45 ° (Pal and Khanum 2011). The enzyme has been reused five times while retaining >85% of its starting activity. Recently, matrix entrapment method was carried out by Bibi et al. (2015) to immobilize microbial endo-β-1,4-xylanase produced by Geobacillus stearothermophilus KIBGE-IB29 within agar-agar gel beads.
Among the industrial enzymes, protease has taken a pivotal position in detergent industry and leather industry. The alkaline protease from Bacillus mycoides was immobilized on different carriers using various immobilization methods including physical adsorption, covalent binding, entrapment, and ionic binding. An alkaline protease preparation was physically adsorbed on chitosan, entrapped in 2% cross-linked polyacrylamide, covalently bonded on chitin and ionically bonded on Amberlite IR-120 that were observed with highest activities by Abdel-Naby et al. (1998). In previous year, chitosan-immobilized protease from Bacillus licheniformis was applied in therapeutic use by Elchinger et al. (2015). They synthesized protease gains anti-biofilm activities after immobilization and was explored against biofilms formed by Listeria monocytogenes, Pseudomonas aeruginosa, Staphylococcus aureus, etc. Similarly, immobilization techniques also employed for laccase enzyme to make them suitable for various bioremediation application and wastewater treatment. The laccase beads were synthesized by immobilizing laccase enzyme on copper-alginate beads, and additionally Fe2O3 was incorporated in the bead through magnetic force. These lac beads have been used for bioremediation of triclosan and Remazol Brilliant Blue R and subsequently for wastewater treatment (Thanh le et al. 2016). Laccase from Trametes versicolor was also covalently immobilized on the composite polymer particles of poly(2-chloroethyl acrylate), p(CEA), which were grafted on zeolite particles via surface-initiated atom transfer radical polymerization (SI-ATRP). The immobilized laccase on the zeolite-g-p (CEA) particles was applied in biodegradation of dye Reactive Red 120. Besides these enzymes, immobilization was carried out with several other microbial enzymes which have been summarized in Table 13.1 with their application and support material being used.
4.1 Microbial Enzyme Immobilization Using Nanotechnology
Nanoparticles exhibit some attractive properties like elevated surface reactivity, high catalytic efficiency, tough adsorption ability, and great surface-to-volume ratio which make them attractive agent for immobilization. Adsorption of microbial enzymes on nanoparticles leads to enhanced performance of microbial enzymes in terms of its catalytic activity (Lynch and Dawson 2008). The application of enzyme immobilized nanoparticles was started during the 1980s (Pereira et al. 2002; Soriano et al. 2005). A number of microbial enzymes such as xylanase, protease, amylases, and phytase have been immobilized on various nanoparticles such as Fe3O4-coated chitosan, 1,3,5-triazine-functionalized Fe3O4@SiO2 nanoparticles, gold nanoparticles, carbon nanoparticle, etc. In order to characterize the structure, size, and magnetic properties of the immobilized xylanase, Fourier transform infrared spectra (FTIR), thermo-gravimetric analysis (TGA), transmission electron microscopy (TEM), vibrating sample magnetometer (VSM), and X-ray photoelectron spectroscopy (XPS) were used for analysis. The enzyme activity, thermostability, storage stability, pH stability, and reusability of the nanoparticles of microbial enzymes have exhibited significant superiority to the free microbial enzymes. The xylanase MNPs showed quite impressive stability after nine reaction cycles with about 65% of its initial activity (Soozanipour et al. 2015). Experimental results by Soozanipour et al. (2015) suggested that the 1,3,5-triazine-functionalized Fe3O4@SiO2 nanoparticles could be the novel convenient magnetic carrier for xylanase immobilization. Similarly, recently, Shahrestani et al. (2016) synthesized 1,3,5-triazine-functionalized silica encapsulated magnetic nanoparticles to immobilize xylanase enzyme to apply in clarification of bear and juices with impressive stability even after ten reaction cycle. In the same sequence, xylanase from Aspergillus niger that has been immobilized covalently on the surface Fe3O4-coated chitosan magnetic nanoparticles showed a high binding capacity (Liu et al. 2015). Xylanase MNPs can be used in a number of industrial applications under broader pH and temperature ranges, having long-term storage capability and permitting magnetically recycling of the enzyme for purification or reuse of the product. Similarly, cellulase enzyme has also been physically adsorbed through ionic bond on superparamagnetic nanoparticles with binding efficiency of 95% and used for long-term storage (Khoshnevisan et al. 2011). Amylase enzymes from Streptomyces sp. MBRC-82 also have been immobilized on gold nanoparticles which have various medicinal applications by Manivasagan et al. (2015).
4.2 Immobilization by Forming CLEAs
CLEAs are insoluble enzyme aggregates which are formed by cross-linking of protein precipitates using cross-linking reagents such as glutaraldehyde. CLEAs exhibited high stability and high activity in aqueous medium as well as in nonaqueous medium. These enzyme aggregates have also showed a high stability at high temperature (Sheldon 2007). CLEAs may have a combination of several enzyme activities; such CLEAs are called multipurpose CLEAs or combi-CLEAs. Extent of cross-linking often influences their activity morphology, stability, and enantioselectivity. Nadar et al. (2016) evaluated the effects of various cross-linkers and precipitating agent on amylase activity recovery of macromolecular cross-linked enzyme aggregates (M-CLEAs) of α-amylase. Precipitates of amylase enzyme cross-linked by dextran showed 91% activity, ammonium sulfate used as precipitating agent, but glutaraldehyde CLEAs (G-CLEAs) exhibited only 42% activity. Recently, Mahmod et al. (2015) manufactured multipurpose cross-linked enzyme aggregate (multi-CLEA) with lipase and protease activity.
5 Homologue Augmentation and Substitution
The rate-limiting step is the major issue in enhanced production of desired metabolite which can be altered by importing the homologous enzyme from different hosts, the process known as homologous augmentation. Furthermore, turnover of the heterologous pathway can also be increased by using homologues of nonnative enzymes, known as homologue substitution. Both of these techniques were employed to deduce the carotenoid production in E. coli (Yoon et al. 2009). Yadav et al. proposed chimeric pathways involving each enzyme from different host to construct MVA pathway for carotenoid production.
6 Conclusion and Future Perspectives
In this chapter a number of enzyme engineering techniques have been briefed to improve the enzymatic characteristics together with its recycling methods. Microbial enzymes with enhanced physiological properties have greater commercial application in the industries. Protein engineering technology along with other molecular techniques and nanotechnology approaches has occupied the major position in the proteomic studies. Here it is shown that microbial enzymes, isolated from a number of sources such as fungi, bacteria, actinobacteria, yeast, and metagenomic sample, were subjected to protein engineering to modify their characteristics such as activity, temperature, and pH stability to make them economic for industrial applications. A number of modern genomic techniques such as genome-walking PCR, TAIL-PCR, error-prone PCR, StEP recombination, and metagenomic approaches have been proved as successful tools to create structural modifications at protein translational level. A few reports on site-directed mutagenesis and directed evolution have been explained as successful techniques to enhance thermostability and pH stability of microbial enzyme. The main focus of the chapter is laid upon modern enzyme engineering techniques to obtain a microbial protein with greater operational stability together with reusability. A combinatorial approach of metagenomic, proteomics, genomics, nanotechnology, and bioinformatics is required to obtain our goal of enzyme engineering.
References
Abdel-Naby M, Ismail AMS, Ahmed SA, Abdel Fattah AF (1998) Production and Immobilization of alkaline protease from Bacillus Mycoides. Bioresour Technol 64(3):205–210
Ajikumar PK, Xiao W-H, Keith EJ, Tyo YW, Fritz S, Effendi L, Oliver M, Too HP, Blaine P, Gregory S (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74
Baweja M, Singh PK, Shukla P (2015) Enzyme technology, functional proteomics and systems biology towards unravelling molecular basis for functionality and interactions in biotechnological processes. In: Shukla P (ed) Frontier discoveries and innovations in interdisciplinary microbiology. Springer/Verlag, Berlin/Heidelberg, pp 207–212
Baweja M, Nain L, Kawarabayasi Y, Shukla P (2016) Current technological improvements in enzymes toward their biotechnological applications. Front Microbiol 7:965. doi:10.3389/fmicb.2016.00965
Bibi Z, Shahid F, Ul Qader SA, Aman A (2015) Agar–agar entrapment increases the stability of endo-β-1,4-xylanase for repeated biodegradation of xylan. Int J Biol Macromol 57:121–127
Celikbicak O, Bayramoglu G, Yılmaz M, Ersoy G, Bicak N, Salih B, Arica MY (2014) Immobilization of laccase on hairy polymer grafted zeolite particles: Degradation of a model dye and product analysis with MALDI–ToF-MS. Microporous Mesoporous Mater 199:57–65
Chang MS (1991) Therapeutic applications of immobilized proteins and cells. Bioprocess Technol 14:305–318
Eijsink VGH, Veltman OR, Aukema W, Vriend G, Venema G (1995) Structural determinants of the stability of thermolysin-like proteinases. Nat Struct Biol 2:374–379
Elchinger PH, Delattre C, Faure S, Roy O, Badel S, Bernardi T, Taillefumier C, Michaud P (2015) Immobilization of proteases on chitosan for the development of films with anti-biofilm properties. Int J Biol Macromol 72:1063–1068
Fang ZM, Li TL, Chang F, Zhou P, Fang W, Hong YZ, Zhang XC, Peng H, Xiao YZ (2012) A new marine bacterial laccase with chloride-enhancing, alkaline-dependent activity and dye decolorization ability. Bioresour Technol 111:36–41
Guibault GG, Kauffmann JM, Patriarche GJ (1991) Immobilized enzyme electrodes as biosensors. Bioprocess Technol 14:209–262
Gupta SK, Shukla P (2015) Advanced technologies for improved expression of recombinant proteins in bacteria: perspectives and applications. Crit Rev Biotechnol 18:1–10
Gupta SK, Shukla P (2016) Microbial platform technology for recombinant antibody fragment production. A review. Crit Rev Microbiol 43:1–12. doi:10.3109/1040841X.2016.1150959
Kapoor M, Kuhad RC (2007) Immobilization of xylanase from Bacillus pumilus strain MK001 and its application in production of xylo-oligosaccharides. Appl Biochem Biotechnol 142(2):125–138
Karthik MVK, Shukla P (2012) Computational strategies towards improved protein function prophecy of xylanases from Thermomyces lanuginosus. Springer, New York. doi:10.1007/978-1-4614-4723-8
Keasling JD (1999) Gene expression tools for the metabolic engineering of bacteria. Trends Biotechnol 17:452–460
Khoshnevisan K, Bordbar AK, Zare D, Davoodi D, Noruzi M, Barkhi M, Tabatabaei M (2011) Immobilization of cellulase enzyme on superparamagnetic nanoparticles and determination of its activity and stability. Int J Biol Macromol 171(2):669–673
Klapa MI, Park SM, Sinskey AJ (1999) Stephanopoulos G: metabolite and isotopomer balancing in the analysis of metabolic cycles: I. Theory. Biotechnol Bioeng 62:375–391
Kumar V, Shukla P (2015) Functional aspects of xylanases toward industrial applications. In: Shukla P (ed) Frontier discoveries and innovations in interdisciplinary microbiology. Springer/Verlag, Berlin/Heidelberg, pp 157–165
Kumar V, Pandey P, Gupta S, Shukla P (2014) A reviving preliminary evoke on few xylanase producing fungal isolates from different ecological niche. Int J Curr Microbiol App Sci 3(4):501–506
Kumar V, Marín-Navarro J, Shukla P (2016) Thermostable microbial xylanases for pulp and paper industries: trends, applications and further perspectives. World J Microbiol Biotechnol 32(2):34. doi:10.1007/s11274-015-2005-0
Labrou NE (2010) Random Mutagenesis Methods for In Vitro Directed Enzyme Evolution. Curr Protein Pept Sci 11:91–100
Lee JY, Yang KS, Jang SA, Sung BH, Kim SC (2011) Engineering butanol-tolerance in Escherichia coli with artificial transcription factor libraries. Biotechnol Bioeng 108:742–749
Lin MG, Chi MC, Chen YY, Wang TF, Lo HF, Lin LL (2016) Site-directed mutagenesis of a conserved Asn450 residue of Bacillus licheniformis γ-glutamyl transpeptidase. Int J Biol Macromol 91:416–425
Liu MQ, Huo WK, Xu X, Jin DF (2015) An immobilized bifunctional xylanase on carbon-coated chitosan nanoparticles with a potential application in xylan-rich biomass bioconversion. J Mol Catal B Enzym 120:119–126
Lynch I, Dawson KA (2008) Protein-nanoparticle interactions. NanoToday 3:40–47
Mahmod SS, Yusof F, Jami MS, Khanahmadi S, Shah H (2015) Development of an immobilized biocatalyst with lipase and protease activities as a multipurpose cross-linked enzyme aggregate (multi-CLEA). Process Biochem 50(12):2144–2157
Manivasagan P, Venkatesan J, Kang KH, Sivakumar K, Park SJ, Kim SK (2015) Production of α-amylase for the biosynthesis of gold nanoparticles using Streptomyces sp. MBRC-82. Int J Biol Macromol 72:71–78
Nadar SS, Muley AB, Ladole MR, Joshi PU (2016) Macromolecular cross-linked enzyme aggregates (M-CLEAs) of α-amylase. Int J Biol Macromol 84:69–78
Nagar S, Mittal A, Kumar D, Gupta VK (2012) Immobilization of xylanase on glutaraldehyde activated aluminum oxide pellets for increasing digestibility of poultry feed. Process Biochem 47(9):1402–1410
Pal A, Khanum F (2011) Covalent immobilization of xylanase on glutaraldehyde activated alginate beads using response surface methodology: characterization of immobilized enzyme. Process Biochem 46(6):1315–1322
Pereira JF, de Queiroz MV, Gomes EA, Muro-Abad JI, de Ara’ujo EF (2002) Molecular characterization and evaluation of pectinase and cellulose production of Penicillium spp. Biotechnol Lett 24(10):831–838
Rahimi M, van der Meer JY, Geertsema EM, Poddar H, Baas BJ, Poelarends GJ (2016) Corrigendum: mutations closer to the active site improve the promiscuous aldolase activity of 4-oxalocrotonate tautomerase more effectively than distant mutations. Chem Bio Chem 17(13):1290. doi:10.1002/cbic.201600321
Ramli AM, Muhammad NM, Rabu A, Munir A, Murad A, Diba FAB, Illias RM (2011) Molecular cloning, expression, and biochemical characterisation of a cold-adapted novel recombinant chitinase from Glaciozyma antarctica PI12. Microb Cell Factories 10:94. doi:10.1186/1475-2859-10-94
Rasila TS, Pajunen MI, Savilahti H (2009) Critical evaluation of random mutagenesis by error-prone polymerase chain reaction protocols, Escherichia coli mutator strain, and hydroxylamine treatment. Anal Biochem 388:71–80
Schmidt K, Carlsen M, Nielsen J, Villadsen J (1997) Modeling isotopomer distributions in biochemical networks using isotopomer mapping matrices. Biotechnol Bioeng 55:831–840
Sen S, Dasu VV, Mandal B (2007) Developments in directed evolution for improving enzyme functions. Appl Biochem Biotechnol 143:212–223
Shahrestani H, Taheri-Kafrani A, Soozanipour A, Tavakoli O (2016) Enzymatic clarification of fruit juices using xylanase immobilized on 1,3,5-triazine-functionalized silica-encapsulated magnetic nanoparticles. Biochem Eng J 109:51–58
Sheldon RA (2007) Cross-linked enzyme aggregates (CLEAs): stable and recyclable biocatalysts. Biochem Soc Trans 35(6):1583–1587
Shrivastava S, Lata S, Shukla P (2012) An insight on recent advances on immobilization methods for Industrial enzymes and its relevance to xylanases. Dyn Biochem Process Biotechnol Mol Biol 6(1):57–61
Shrivastava S, Shukla P, Deepalakshmi PD, Mukhopadhyay K (2013) Characterization, cloning and functional expression of novel xylanase from Thermomyces lanuginosus SS-8 isolated from self-heating plant wreckage material. World J Microbiol Biotechnol 12:2407–2415
Silva MF, Rigo D, Mossi V, Treichel H (2013) Evaluation of enzymatic activity of commercial inulinase from Aspergillus niger immobilized in polyurethane foam. Food Bioprod Process 91(1):54–59
Singh PK, Shukla P (2012) Molecular modeling and docking of microbial inulinases towards perceptive enzyme-substrate interactions. Indian J Microbiol 52:373–380
Singh P, Shukla P (2015) Systems biology as an approach for deciphering microbial interactions. Brief Funct Genomics 14:166–168
Singh PK, Joseph J, Goyal S, Grover A, Shukla P (2016) Functional analysis of the binding model of microbial inulinases using docking and molecular dynamics simulation. J Mol Model 22(4):1–7
Soleimani M, Khani A, Najafzadeh K (2012) α-Amylase immobilization on the silica nanoparticles for cleaning performance towards starch soils in laundry detergents. J Mol Catal B Enzym 74(1–2):1–5
Soozanipour A, Taheri-Kafrani A, Isfahani AL (2015) Covalent attachment of xylanase on functionalized magnetic nanoparticles and determination of its activity and stability. Chem Eng J 270:235–243
Soriano M, Diaz P, Pastor FI (2005) Pectinolytic systems of two aerobic sporogenous bacterial strains with high activity on pectin. Curr Microbiol 50(2):114–118
Stafford DE, Gregory S (2001) Metabolic engineering as an integrating platform for strain development. Curr Opin Microbiol 4:336–340
Stafford DE, Yanagimachi KS, Stephanopoulos G (2001) Metabolic engineering of indene bioconversion in Rhodococcus sp. Adv Biochem Eng Biotechnol 73:85–101
Strakšys A, Kochane T, Budriene S (2016) Catalytic properties of maltogenic α-amylase from Bacillus stearothermophilus immobilized onto poly (urethane urea) microparticles. Food Chem 211:294–299
Thanh le T, Murugesan K, Lee CS, Vu CH, Chang YS, Jeon JR (2016) Degradation of synthetic pollutants in real wastewater using laccase encapsulated in core-shell magnetic copper alginate beads. Bioresour Technol 216:203–210
Wang S, Fu X, Liu Y, Liu X, Wang L, Fang J, Wang PG (2015) Probing the roles of conserved residues in uridyltransferase domain of Escherichia coli K12 GlmU by site-directed mutagenesis. Carbohydr Res 413(2):70–74
Xu H, Qin Y, Huang Z, Liu Z (2014) Characterization and site-directed mutagenesis of an α-galactosidase from the deep-sea bacterium Bacillus megaterium. Enzym Microb Technol 56:46–52
Yadav VG, Mey MD, Lim CG, Ajikumar PK, Stephanopoulos G (2012) The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng 14(3):233–241
Yewale T, Singhal RS, Vaidya A (2013) Immobilization of inulinase from Aspergillus niger NCIM 945 on chitosan and its application in continuous inulin hydrolysis. Biocatal Agric Biotechnol 2:96–101
Yoon SH, Lee SH, Das A, Ryu HK, Jang HJ, Kim JY, Oh DK, Keasling JD, Kim SW (2009) Combinatorial expression of bacterial whole mevalonate pathway for the production of beta-carotene in E. coli. J Biotechnol 140:218–226
Zhang Z, Shimizu Y, Kawarabayasi Y (2015) Characterization of the amino acid residues mediating the unique amino-sugar-1-phosphate acetyltransferase activity of the archaeal ST0452 protein. Extremophiles 19:417–427
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kumar, V., Baweja, M., Liu, H., Shukla, P. (2017). Microbial Enzyme Engineering: Applications and Perspectives. In: Shukla, P. (eds) Recent advances in Applied Microbiology . Springer, Singapore. https://doi.org/10.1007/978-981-10-5275-0_13
Download citation
DOI: https://doi.org/10.1007/978-981-10-5275-0_13
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-5274-3
Online ISBN: 978-981-10-5275-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)