Abstract
Many types of human cancers overexpress MDM2 protein. A common characteristic among these cancers is an associated increase in mdm2 splice variants. Provided here is a comprehensive list, based on a literature review, of over 70 mdm2 variants. These variants are grouped according to in-frame versus out-of-frame status and their potential (or ability) to be translated into isoform proteins. We describe the putative functions for these mdm2 splice variant mRNAs, as well as the mechanistic drivers associated with increased mdm2 transcription and splicing. The paradoxical signal transduction functions of the most commonly studied variants mdm2-a,-b and -c are addressed for their outcomes in the presence and absence of wild-type p53. These outcomes vary from tumor promotion to growth arrest. Finally, we present issues in the detection of endogenous MDM2 protein and how many of the antibodies commonly used to detect MDM2 do not present a full picture of the cellular representation of the isoform proteins. This review provides a focusing lens for individuals interested in learning about the complexities of mdm2 mRNAs and their protein isoforms as well as the roles MDM2 isoforms may play in cancer progression.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
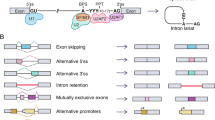
Many human cancers over-express MDM2 and the mdm2 gene locus produces a diverse array of mdm2 splice variants [1, 2]. Alternative splicing is predominantly co-transcriptional [3] with approximately 6.3 alternatively spliced transcripts occurring per human gene [4]. The coordination of splicing with transcription highlights the importance of alternative splicing in signal transduction. Unlike the average human gene locus, the mdm2 gene gives rise to more than the average 6.3 alternative spliced transcripts. Increased transcription and splicing of mdm2 is associated with increased tumorigenesis [1]. However, it is unclear exactly what biological functions the alternative spliced mdm2 transcripts contribute to tumorigenesis. We reviewed the literature describing mdm2 splice variants associated with oncogenesis and found that at least 72 have been described. It has not been determined how many of these splice variants express protein in cancer cells.
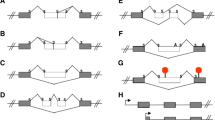
The association of high MDM2 expression with tumorigenic potential was identified in 1991 in the Donna George laboratory [5]. In 1996, the Lunec group detected increased expression of mdm2 splice variants in multiple tumor types [6]. The alternative spliced transcripts were named mdm2-a, mdm2-b, mdm2-c, mdm2-d and mdm2-e in order to contrast them with the full-length version called mdm2-fl [6]. Since 1996 the list of mdm2 alternative spliced variants related to oncogenesis has vastly increased (see Table 14.1). We will focus on the three most commonly found, and best studied, exon skipped transcripts that can be translated into protein [23].
The most commonly detected isoforms are mdm2-a, mdm2-b and mdm2-c. These three mdm2 splice variants are found in human leukemia, soft tissue sarcoma, Hodgkin’s lymphoma, glioblastoma, rhabdomyosarcoma, liposarcoma, and many different carcinomas including ovarian, breast, bladder, lung and oral squamous cell carcinoma (see Table 14.2). It is highly likely that the protein products MDM2-A, MDM2-B and MDM2-C contribute to the diversity of the human cancer proteome [23]. In addition to mdm2 spliced variants potentiating cancer proteome diversity, alternative spliced mdm2 products have also been associated with a small nuclear RNA processed form called hdm365 [7]. This suggests that some human mdm2 transcripts may possess RNA-based functions.
Transcripts of Human mdm2 in Cancer
Two different promoters, P1 and P2, control transcription of the mdm2 gene, giving rise to two different mRNA messages that encode MDM2-FL [28]. The mdm2 gene has 12 exons and the P1 promoter drives transcription from upstream of exon 1 and coordinates the splicing out of exon 2. The P1 promoter is responsible for basal mdm2 transcription and is controlled in part by NF-κB binding sites [29]. The P2 promoter drives transcription from upstream of exon 2 and is controlled by numerous transcription factors. Directly adjacent to the P2 dependent promoter are two binding sites for the transcription factor p53 and in response to stress p53 activates mdm2 transcription [28, 30]. Other transcription factor binding sites adjacent to the P2 promoter include the Ets/Ap-1, E-box, RXR and Smad binding sites and GC boxes (reviewed in [31]). The Ras signaling pathway can stimulate mdm2 transcription via the Ap-1/Ets sites [32]. TGF-β signaling stimulates transcription via Smad 2/3 transcription factors binding to the Smad binding sites and MYCN in neuroblastoma cells binds to the E-box near the P2 promoter [33, 34]. The GC boxes are the regions that bind the Sp1 transcription factor. A single nucleotide polymorphism in the GC box region, at position 309, that changes a T to G increases the affinity for Sp1 binding to drive mdm2 transcription [35]. Patients who are homozygous G/G SNP 309 in the mdm2 gene have increased susceptibility to multiple cancers [35, 36]. In addition, the tissue-specific RXRγ transcription factor and binding region in retinoblastoma cells can activate P2 dependent mdm2 expression [37]. High levels of estrogen also activate mdm2 by activating P2 promoter transcription [38, 39]. While promoter usage has not been shown to be a factor in the alternative splicing of mdm2 transcripts, the robust signaling of oncogenes present in cancers often drives P2 mdm2 oncogene mediated transcription [40, 41]. We hypothesize that this increased transcription from the P2 promoter changes the mdm2 splice variant isoform ratio.
In 2002, Bartel, Taubert and Harris summarized the existence of over 40 different human tumor associated mdm2 splice variants [2]. At that time the list of distinctive mdm2 mRNAs was as follows: mdm2-fl, mdm2-a, mdm2-b, mdm2-c, mdm2-d, mdm2-e, mdm2-a1, mdm2-kb2, mdm2-kb3, mdm2-jn1, mdm2-ds2, mdm2-ds3, mdm2-is1, mdm2-gk1, mdm2-pm2, mdm2-eu2, mdm2-bl, mdm2-n, mdm2-fb25, mdm2-fb26, mdm2-fb28, mdm2-fb29, mdm2-fb30, mdm2-fb55, mdm2-281 bp, mdm2-219 bp, mdm2-254 bp, mdm2-f, mdm2-g, mdm2-h, mdm2-ln229a, mdm2-ln229b, mdm2-ln18, mdm2-g116, mdm2-g150, mdm2-var2, mdm2-var1, mdm2-delF, mdm2-delE, and mdm2-fb60 (see Table 14.1). These transcripts are found in human cancers, but until recently, no corresponding endogenous protein products had been detected. Many of the mdm2 splice variant transcripts produced by exon skipping lack the coding region for the p53 interacting domain [2]. The most common variants associated with many different types of human cancers (and missing the p53 interacting domain) are: mdm2-a (lacking exons 4–9), mdm2-b (lacking exons 4–11), and mdm2-c (lacking exons 5–9) (see Table 14.2). All three of these have the potential to endogenously encode oncogenic proteins (see Table 14.3) (reviewed in [23]).
Alternatively spliced mdm2 transcripts in human cancer continue to be detected. The list has increased in number beyond the previously identified 40 (see Table 14.1). The most recent additions to the list of mdm2 splice variants come from a study of oral squamous cell carcinoma (OSCC). This study shows that mdm2 splice variants associate with increased likelihood to form OSCC [8] as mdm2 splice variants are detected in 89 % of oral squamous cell carcinoma [40]. Four splice variants in OSCC are the previously identified mdm2-b, mdm2-c, mdm2-pm2 and mdm2-eu2 (with mdm2-b found most often). Interestingly, 26 mdm2 OSCC variants are novel isoforms. Those found to be in-frame range in size from 252 bp to 1,095 bp, and were named: MYO-1, MYO-4, MYO-8, MYO-9, MYO-11, MYO-12, MYO-18, MYO-20, MYO-23, MYO-24, MYO-25 and MYO-32. What is consistent for the in-frame oral cancer variants is that they retain the MDM2 ring-finger binding domain. A significant number of OSCC mdm2 transcripts are out-of-frame. These range in size from 262 bp to 1,385 bp and were named: MYO-2, MYO-3, MYO-5, MYO-6, MYO-7, MYO-10, MYO-13, MYO-14, MYO-17, MYO-19, MYO-21, MYO-27, MYO-28, MYO-29, MYO-30, MYO-31 and MYO-33.
At least 72 mdm2 alternative spliced transcripts have been identified in human cancers. This number of 72 includes those 40 compiled in 2002 [2], the OSCC transcripts [8], two novel transcripts that we documented that are driven from the P2 promoter (P2mdm2-10 and P2mdm2-C1 [16]) and the RNA-based functional form, hdm365 [7]. It is likely that the tally of 72 variable mdm2 transcripts is an underestimate because they continue to be identified. Moreover, while many mdm2 alternatively spliced transcripts have been detected, the identification of endogenous MDM2 splice variant polypeptides is still lacking. This is partially due to a lack of specific antibodies to detect them. However, the fact that many of the alternative and aberrantly spliced mdm2 messages are not competent to encode protein suggests that some mdm2 splice variants might function as regulatory RNA molecules.
The ENCODE project consortium guidelines for functional elements of the genome demonstrates that only a small percentage of the genome (2.9 %) covers areas of protein-coding exons. Furthermore, 62 % of the genome represents RNA molecules with only 5.5 % accounted for in protein-annotated regions [4]. Therefore, the majority of the functional RNA molecules encoded by the human genome represent non-coding regions and for mdm2 transcripts may indicate a major RNA-based function.
Out-of-Frame Versus In-Frame mdm2 Transcripts
There are over 70 known splice variant transcripts and they represent alternatively and aberrantly spliced mRNAs (see Table 14.1). Alternatively spliced mdm2 transcripts are those that result due to exon-exon splicing and give rise, more often than not, to in-frame transcripts with the potential to produce protein [2]. Aberrantly spliced transcripts represent those that result due to the use of cryptic internal splice sites within the mdm2 exon or intron sequences [2]. Aberrant mdm2 splicing produces transcripts that are mainly out-of-frame and these do not have the potential to generate protein.
Of the known mdm2 transcripts, approximately 46 % do not encode protein and all but one of these is spliced out-of-frame to the full-length mdm2 transcript (Table 14.1, group A). Group A represents this subset of numerous mdm2 transcripts generated in human cells. One major product in this group is the hdm365 transcript (in Table 14.1) that potentially has an RNA-based function [7]. This transcript is initiated from the P2 promoter of mdm2 and retains exons 2, 3, 4 and 5 [7]. The hdm365 transcript resides in the nucleus and is located at sites of mdm2 transcription [7]. This localization suggests a role for this mdm2 transcript in splicing or regulation of the mdm2 mRNA message.
The mdm2 transcripts that are assumed, but not proven, to encode protein make up approximately 41.7 % of the identified mdm2 transcripts (Table 14.1, group B). Group B represents both alternatively and aberrantly spliced mdm2 transcripts. It is not clear if these transcripts form protein in the cell, as the tools to properly identify each potential MDM2 protein isoform need to be developed.
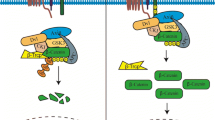
The final category of mdm2 transcripts accounts for 12.5 % of the known mdm2 transcripts. They have been confirmed by in vitro translation assays to encode MDM2 protein isoforms (Table 14.1, group C). Interestingly, only MDM2-FL, MDM2-A, MDM2-B, MDM2-C, MDM2-D and MDM2-E have been shown to have a biological function in vitro or in vivo [6, 18, 24, 26, 42–44]. Furthermore, none of these MDM2 protein isoforms except for the full-length MDM2 (MDM2-FL) have been detected as expressed endogenously in cancer cells. Although there are high levels of mdm2 transcripts found in cancers, the level of transcripts do not correlate with high MDM2 protein levels [12, 17]. The reason for this may be due to the absence of proper antibody epitope recognition since antibodies detect some, but not all, MDM2 isoforms within the background of MDM2-FL.
Full-length MDM2, translated from exons 3–12, possesses both oncogenic and tumor suppressive properties [31]. Translation of the MDM2 protein begins in exon 3 and P2-derived transcripts are more efficiently translated than P1-derived transcripts [30]. Some oncogenic properties of MDM2 come from the ability of the protein to interact with the tumor suppressor p53 and target it for proteasome-mediated degradation [47]. However, MDM2 also interacts with the p53 mRNA and increases the translation of p53 protein [48]. This apparent paradox for MDM2 function is increased in complexity by the fact that some mdm2 splice variants have the capacity to encode polypeptides that lack portions of the p53 interacting domain [2, 31]. Therefore, some of this paradoxical behavior may be explained by determining the functions of specific MDM2 splice variant isoforms.
Mechanisms That Drive Alternative Splicing of mdm2 Transcripts in Cancer
It is common to find a loss of splicing fidelity in cancer cells [49]. The mechanisms responsible for changes in splicing in cancer continue to emerge. Evidence attributes some of these changes to variations in cis-regulatory elements, sequences within the RNA which effect splice-site usage and recognition [50]. Many Serine/Arginine rich (SR) and heterogeneous ribonucleoprotein (hnRNP) splicing factor proteins are up-regulated in cancers and these trans- acting splicing factors can increase splicing events [49, 51, 52]. The oncogene c-MYC drives upregulation of specific splicing factors including polypyrimidine-tract binding protein (PTB) and hnRNP A1 and A2 (reviewed in [51–53]). With oncogenes driving alternative splicing, it is not surprising that alternatively spliced transcripts of mdm2 are found in many different cancers (see Tables 14.1 and 14.2). The mdm2 splice variants mdm2-a , mdm2-b and mdm2-c result from exon skipping. This exon skipping occurs because some mdm2 introns have a defective polypyrimidine tract, a cis-regulatory element important for splicing factor binding and 3′ splice site recognition [21]. The splice variants mdm2-d and mdm2–e on the other hand result from an aberrant splicing mechanism that does not use the normal exon-intron boundaries [21]. Interestingly, some known aberrantly spliced mdm2 transcripts have a common splicing pattern due to sequences of high homology in the mdm2 transcript that serve as cryptic splice donor and acceptor sites for splicing factor binding [2].
Alternative splicing of mdm2 transcripts and transcription from the P2 promoter are also driven by genotoxic stress conditions such as cisplatin or ultraviolet radiation [22, 25]. Some splice variants produced under genotoxic stress conditions, like mdm2-b, are seen at high frequency in cancers [40]. A conserved cis-regulatory element in intron 11 of the mdm2 gene promotes this stress-induced regulation of mdm2 splicing [40]. Stress-induced splicing, in particular that seen with cisplatin treatment, induces co-transcriptional mdm2 exon skipping through disruption of the EWS-YB1 interaction [41]. EWS is a protein that interacts with the RBP7 subunit of RNA pol II and YB1 interacts with the spliceosome [54, 55]. The stress-induced cotranscriptional exon skipping of mdm2 produces mdm2 variants missing the p53 interaction domains. Therefore, exon skipping may help to promote a more robust p53 response by inhibiting the production of MDM2 that interacts with p53 [41].
The Biological Functions of Ectopically Expressed MDM2-A, MDM2-B, and MDM2-C
The biological outcomes of ectopically expressed MDM2-A, MDM2-B, and MDM2-C range from growth activation to growth inhibition under different circumstances (see Table 14.3). The variable outcomes are associated with the presence or absence of wild-type p53 protein. For example, if wild-type p53 is expressed then MDM2-A transgenic homozygous mouse pups die of unknown causes shortly after birth [43]. The only mice that survive with MDM2-A are hemizygous [43]. However, in a p53-null background homozygous mice survive and the expression of MDM2-A alters the tumor spectrum of transgenic p53-null mice toward increased T-cell lymphomagenesis [44]. Additionally, p53 heterozygous mice crossed with MDM2-A expressing transgenic mice develop aggressive mammary tumors [44]. Furthermore, the expression of MDM2-A in p53-null mouse embryo fibroblasts (MEFs) promotes cell transformation [44]. The exogenous expression of MDM2-A in wild-type MEFs inhibits cell growth. This inhibition of cell growth correlates with an increase in p53 transcriptional activity and high p21 protein levels [43]. Similarly, in the immortalized primary BJ fibroblast cell line ectopic expression of MDM2-A up-regulates p21 along with Cyclin D1 and Cyclin E [26]. Exogenously expressed MDM2-A interacts with endogenous MDM2-FL and activates wild-type p53 activity thus explaining some of the differences seen in a p53-null background [43].
Similar to MDM2-A, exogenous expression of MDM2-B also has differential outcomes in the presence or absence of wild-type p53. The exogenous expression of MDM2-B in transgenic mice is not compatible with normal development [42]. Only when MDM2-B is expressed under a promoter with limited tissue expression are mice able to survive. The transfection of the mdm2-b into NIH/3T3 cells increases cell proliferation and transformation capabilities [42]. Interestingly, the expression of MDM2-B in NIH/3T3 cells interferes with the induction of apoptosis without affecting p53 stability or activity and is linked to an increase of p65 RelA protein levels [42]. Surviving MDM2-B transgenic mice with tissue specific expression have increased tumorigenesis that correlates with this increase in p65 protein levels [42]. MDM2-B expression also increases cell proliferation in p53-null, ARF-null and Rb-null MEFs, therefore indicating a p53-independent mechanism of action [42]. However, other studies show exogenously expressed MDM2-B interacts with MDM2-FL protein localizing MDM2-FL to the cytoplasm in numerous cell lines to allow wild-type p53 protein to be activated [24, 46]. Ectopic expression of MDM2-B also up-regulates p21 expression in immortalized BJ fibroblasts correlating with an inhibition of cell proliferation [26].
Our laboratory works on the MDM2-C splice variant. Unpublished studies from our laboratory were presented at the 2011 MDM2 Workshop in New York City and were recently published [45]. We have designed a specific antibody toward MDM2-C to detect the endogenous MDM2-C protein isoform and we have explored the biological functions of MDM2-C. Exogenous expression of MDM2-C in the presence or absence of p53 in H1299 lung carcinoma cells showed increased colony formation as compared to MDM2-FL or vector control [45]. Therefore, like MDM2-A and MDM2-B, MDM2-C shows a p53-independent transformation function. Furthermore, the transfection of mdm2-c in the presence or absence of p53 into H1299 cells increased colony formation, indicated by transforming ability. The co-transfection of mdm2-c and p53 into H1299 cells did not significantly decrease p53 transcriptional activity or change p53 protein levels and MDM2-C was also able to interact with MDM2-FL [45]. Therefore, unlike MDM2-A and MDM2-B, MDM2-C does not increase the activity of wild-type p53. An in vivo mouse model has yet to be carried out for MDM2-C. Until this is done, we will not know the full biological functions of MDM2-C.
Detection of Endogenous MDM2
There are a number of MDM2 specific antibodies that detect the endogenous MDM2 protein in cancer cells and cancer tissues (reviewed in [23]). These MDM2 antibodies recognize epitopes of multiple MDM2 domains including the amino terminus, the central region, and carboxyl terminus of the protein. However, the MDM2 antibodies utilized to determine MDM2 protein levels in cancers are often to the central region. Therefore, they are not appropriate to detect the majority of MDM2 splice variant isoforms. This is especially true since the main antibody used in immunohistochemistry of cancer tissues for MDM2 protein expression is IF2 (Ab-1). The epitope of recognition for the IF2 antibody lies within amino acids 26–169, which represents the p53-binding domain of the MDM2 protein and spans exons 4 and 5 [19, 56]. Therefore, using the IF2 antibody (or any other antibody to a region deleted by a splicing event) will not show a true representation of the levels of MDM2 protein present in the cancer tissue.
Work to examine the expression of endogenous MDM2 splice variant protein isoforms is being carried out in our laboratory. We generated a rabbit polyclonal antibody to the MDM2-C isoform. The MDM2-C rabbit polyclonal antibody specifically detects MDM2-C, and not MDM2-FL, expressed by an in vitro translation system [45]. This MDM2-C specific antibody also detects endogenously expressed MDM2-C in cancer cell lines and cancer tissues. To our knowledge, we are the first group to generate an antibody specific for an MDM2 spliced variant protein isoform. The mdm2-c transcript is the third most common mdm2 transcript found in cancer cells and tissues [1]. Therefore, detection of the MDM2-C protein isoform may provide a new cancer biomarker. MDM2 endogenous expression undoubtedly results from a mixture of mdm2 transcripts such as mdm2-a, mdm2-b, and mdm2-c. The proteins expressed from these transcripts are all potential cancer biomarkers. It is important that these biomarkers be detected with the proper MDM2 antibodies that are specific for various isoforms. The use of antibodies to MDM2 in the clinic have led to the conclusion that cancers with high levels of spliced variant transcripts have less MDM2 protein [57]. In actuality, not detecting MDM2 protein in breast cancers with mdm2 splice variant transcripts is a false negative [57]. Future research needs to make use of MDM2 splice-variant specific antibodies, or antibodies to either the extreme amino or carboxyl terminus of MDM2, in order to evaluate the true nature of MDM2 protein expression in cancer.
Summary
The diverse array of mdm2 splice variants in human cancers suggests they have functional significance and can serve as cancer biomarkers. To date, MDM2 protein biomarker studies have been carried out with antibodies that give false negative results for the accumulation of MDM2 isoforms lacking central regions of the polypeptide. Future MDM2 biomarker studies must be carried out with consideration given to detecting multiple isoforms. In order to detect multiple MDM2 isoforms, antibody reagents must recognize either the amino or carboxyl terminus of MDM2 because as shown in Table 14.1 most mdm2 splice variants retain these regions. Alternatively, future MDM2 biomarker studies could make use of mixtures of antibodies with specificity to the MDM2 amino and carboxyl termini as well as the specific amino acid splice junction residues for focused splice variants. Recommendations for future MDM2 biomarker studies should combine new methods for the detection of mdm2 splice variant RNA messages along with the detection of multiple MDM2 isoform proteins. This is because the MDM2 polypeptides and RNA sequences may cooperate in the transformation process. The oncogenic MDM2 pathway is a central node in cancer progression that may make use of many isoforms of the MDM2 protein and mdm2 RNA and future research should center on this exciting oncogenic hub.
References
Jeyaraj S, O’Brien DM, Chandler DS (2009) MDM2 and MDM4 splicing: an integral part of the cancer spliceome. Front Biosci 14:2647–2656, doi:3402 [pii]
Bartel F, Taubert H, Harris LC (2002) Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2(1):9–15
Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R (2012) Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 22(9):1616–1625. doi:10.1101/gr.134445.111
Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Kheradpour P, Lassman T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Good PJ, Feingold EA, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Giddings MC, Gingeras TR, Guigo R, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Eaton ML, Dobin A, Lassmann T, Tanzer A, Lagarde J, Lin W, Xue C, Williams BA, Zaleski C, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Hayashizaki Y, Reymond A, Antonarakis SE, Hannon GJ, Ruan Y, Carninci P, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Grasfeder LL, Giresi PG, Battenhouse A, Sheffield NC, Showers KA, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Iyer VR, Sandhu KS, Zheng M, Wang P, Gertz J, Vielmetter J, Partridge EC, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, Muratet MA, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Newberry JS, Levy SE, Absher DM, Wong WH, Blow MJ, Visel A, Pennachio LA, Elnitski L, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Davidson C, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Hunt T, Jungreis I, Kay M, Khurana E, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tapanari E, Tress ML, van Baren MJ, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Valencia A, Raymond A, Addleman N, Alexander RP, Auerbach RK, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O’Geen H, Ouyang Z, Patacsil D, Raha D, Ramirez L, Reed B, Shi M, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Struhl K, Weissman SM, Tenebaum SA, Penalva LO, Karmakar S, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Victorsen A, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Jain G, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Johnson AK, Johnson EM, Kutyavin TM, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sanchez ME, Sandstrom RS, Shafer AO, Stergachis AB, Thomas S, Vernot B, Vierstra J, Vong S, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Stamatoyannopoulos JA, Beal K, Brazma A, Flicek P, Johnson N, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Miller W, Bickel PJ, Banfai B, Boley NP, Huang H, Li JJ, Noble WS, Bilmes JA, Buske OJ, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Lochovsky L (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489(7414):57–74. doi:10.1038/nature11247
Fakharzadeh SS, Trusko SP, George DL (1991) Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. Embo J 10(6):1565–1569
Sigalas I, Calvert AH, Anderson JJ, Neal DE, Lunec J (1996) Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med 2(8):912–917
Bartl S, Ban J, Weninger H, Jug G, Kovar H (2003) A small nuclear RNA, hdm365, is the major processing product of the human mdm2 gene. Nucleic Acids Res 31(4):1136–1147
Sam KK, Gan CP, Yee PS, Chong CE, Lim KP, Karen-Ng LP, Chang WS, Nathan S, Rahman ZA, Ismail SM, Cheong SC (2012) Novel MDM2 splice variants identified from oral squamous cell carcinoma. Oral Oncol 48(11):1128–1135. doi:10.1016/j.oraloncology.2012.05.016
Yu Z, Zhang B, Cui B, Wang Y, Han P, Wang X (2012) Identification of spliced variants of the proto-oncogene HDM2 in colorectal cancer. Cancer 118(4):1110–1118. doi:10.1002/cncr.26330
Bartel F, Taylor AC, Taubert H, Harris LC (2001) Novel mdm2 splice variants identified in pediatric rhabdomyosarcoma tumors and cell lines. Oncol Res 12(11–12):451–457
Schlott T, Nagel H, Laskawi R, Eiffert H, Droese M (2001) Genetic analysis of the human oncoprotein MDM2 in benign and malignant tumors of the salivary gland. Pathobiology 69(2):67–76
Lukas J, Gao DQ, Keshmeshian M, Wen WH, Tsao-Wei D, Rosenberg S, Press MF (2001) Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res 61(7):3212–3219
Hori M, Shimazaki J, Inagawa S, Itabashi M (2000) Alternatively spliced MDM2 transcripts in human breast cancer in relation to tumor necrosis and lymph node involvement. Pathol Int 50(10):786–792
Tamborini E, Della Torre G, Lavarino C, Azzarelli A, Carpinelli P, Pierotti MA, Pilotti S (2001) Analysis of the molecular species generated by MDM2 gene amplification in liposarcomas. Int J Cancer 92(6):790–796. doi:10.1002/ijc.1271
Kraus A, Neff F, Behn M, Schuermann M, Muenkel K, Schlegel J (1999) Expression of alternatively spliced mdm2 transcripts correlates with stabilized wild-type p53 protein in human glioblastoma cells. Int J Cancer 80(6):930–934
Arva NC, Talbott KE, Okoro DR, Brekman A, Qiu WG, Bargonetti J (2008) Disruption of the p53-Mdm2 complex by Nutlin-3 reveals different cancer cell phenotypes. Ethn Dis 18(2 Suppl 2):S2-1–8
Bartel F, Meye A, Wurl P, Kappler M, Bache M, Lautenschlager C, Grunbaum U, Schmidt H, Taubert H (2001) Amplification of the MDM2 gene, but not expression of splice variants of MDM2 MRNA, is associated with prognosis in soft tissue sarcoma. Int J Cancer 95(3):168–175
Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G (2001) An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene 20(30):4041–4049. doi:10.1038/sj.onc.1204533
Anderson JJ, Challen C, Atkins H, Suaeyun R, Crosier S, Lunec J (2007) MDM2 RNA binding is blocked by novel monoclonal antibody h-MDM2-F4-14. Int J Oncol 31(3):545–555
Matsumoto R, Tada M, Nozaki M, Zhang CL, Sawamura Y, Abe H (1998) Short alternative splice transcripts of the mdm2 oncogene correlate to malignancy in human astrocytic neoplasms. Cancer Res 58(4):609–613
Liang H, Atkins H, Abdel-Fattah R, Jones SN, Lunec J (2004) Genomic organisation of the human MDM2 oncogene and relationship to its alternatively spliced mRNAs. Gene 338(2):217–223
Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G (2006) Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res 66(19):9502–9508. doi:10.1158/0008-5472.CAN-05-4271
Okoro DR, Rosso M, Bargonetti J (2012) Splicing up mdm2 for cancer proteome diversity. Genes Cancer 3(3–4):311–319. doi:10.1177/1947601912455323
Dias C, Liu Y, Yau A, Westrick L, Evans S (2006) Regulation of hdm2 by stress-induced hdm2alt1 in tumor and nontumorigenic cell lines correlating with p53 stability. Cancer Res 66(19):9467–9473
Dias CS, Liu Y, Yau A, Westrick L, Evans SC (2006) Regulation of hdm2 by stress-induced hdm2alt1 in tumor and nontumorigenic cell lines correlating with p53 stability. Cancer Res 66(19):9467–9473. doi:10.1158/0008-5472.CAN-05-3013
Sanchez-Aguilera A, Garcia JF, Sanchez-Beato M, Piris MA (2006) Hodgkin’s lymphoma cells express alternatively spliced forms of HDM2 with multiple effects on cell cycle control. Oncogene 25(18):2565–2574. doi:10.1038/sj.onc.1209282
Weng MW, Lai JC, Hsu CP, Yu KY, Chen CY, Lin TS, Lai WW, Lee H, Ko JL (2005) Alternative splicing of MDM2 mRNA in lung carcinomas and lung cell lines. Environ Mol Mutagen 46(1):1–11. doi:10.1002/em.20118
Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M (1995) A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res 23(14):2584–2592
Busuttil V, Droin N, McCormick L, Bernassola F, Candi E, Melino G, Green DR (2010) NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc Natl Acad Sci U S A 107(42):18061–18066. doi:10.1073/pnas.1006163107
Barak Y, Gottlieb E, Juven-Gershon T, Oren M (1994) Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev 8:1739–1749
Manfredi J (2010) The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 24(15):1580–1589, 24/15/1580 [pii]. doi:10.1101/gad.1941710
Ries S, Biederer C, Woods D, Shifman O, Shirasawa S, Sasazuki T, McMahon M, Oren M, McCormick F (2000) Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell 103(2):321–330
Slack A, Lozano G, Shohet JM (2005) MDM2 as MYCN transcriptional target: implications for neuroblastoma pathogenesis. Cancer letters 228(1–2):21–27, S0304-3835(05)00350-2 [pii]. doi:10.1016/j.canlet.2005.01.050
Araki S, Eitel JA, Batuello CN, Bijangi-Vishehsaraei K, Xie XJ, Danielpour D, Pollok KE, Boothman DA, Mayo LD (2010) TGF-beta1-induced expression of human Mdm2 correlates with late-stage metastatic breast cancer. J Clin Invest 120(1):290–302. doi:10.1172/JCI39194
Bond G, Hu W, Bond E, Robins H, Lutzker S, Arva N, Bargonetti J, Bartel F, Taubert H, Wuerl P, Onel K, Yip L, Hwang S, Strong L, Lozano G, Levine A (2004) A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119(5):591–602
Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, Toppmeyer D, Offit K, Levine AJ (2006) MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res 66(10):5104–5110
Xu X, Fang Y, Lee T, Forrest D, Gregory-Evans C, Almeida D, Liu A, Jhanwar S, Abramson D, Cobrinik D (2009) Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell 137(6):1018–1031, S0092-8674(09)00400-0 [pii]. doi:10.1016/j.cell.2009.03.051
Brekman A, Singh KE, Polotskaia A, Kundu N, Bargonetti J (2011) A p53-independent role of Mdm2 in estrogen-mediated activation of breast cancer cell proliferation. Breast Cancer Res: BCR 13(1):R3. doi:10.1186/bcr2804
Phelps M, Darley M, Primrose JN, Blaydes JP (2003) p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor alpha-positive breast cancer cells. Cancer Res 63(10):2616–2623
Singh RK, Tapia-Santos A, Bebee TW, Chandler DS (2009) Conserved sequences in the final intron of MDM2 are essential for the regulation of alternative splicing of MDM2 in response to stress. Exp Cell Res 315(19):3419–3432. doi:10.1016/j.yexcr.2009.07.017
Dutertre M, Sanchez G, De Cian MC, Barbier J, Dardenne E, Gratadou L, Dujardin G, Le Jossic-Corcos C, Corcos L, Auboeuf D (2010) Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol 17(11):1358–1366. doi:10.1038/nsmb.1912
Steinman HA, Burstein E, Lengner C, Gosselin J, Pihan G, Duckett CS, Jones SN (2004) An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J Biol Chem 279(6):4877–4886
Volk EL, Schuster K, Nemeth KM, Fan L, Harris LC (2009) MDM2-A, a common Mdm2 splice variant, causes perinatal lethality, reduced longevity and enhanced senescence. Dis Model Mech 2(1–2):47–55. doi:10.1242/dmm.000992
Volk EL, Fan L, Schuster K, Rehg JE, Harris LC (2009) The MDM2-a splice variant of MDM2 alters transformation in vitro and the tumor spectrum in both Arf- and p53-null models of tumorigenesis. Mol Cancer Res 7(6):863–869. doi:10.1158/1541-7786.MCR-08-0418
Okoro D, Arva N, Gao C, Polotskaia A, Puente C, Rosso M, Bargonetti J (2013) Endogenous human MDM2-C is highly expressed in human cancers and functions as a p53-independent growth activator. PLoS One 8(10):e77643
Evans S, Viswanathan M, Grier J, Narayana M, El-Naggar A, Lozano G (2001) An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene 20(30):4041–4049
Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387:296–299
Naski N, Gajjar M, Bourougaa K, Malbert-Colas L, Fahraeus R, Candeias MM (2009) The p53 mRNA-Mdm2 interaction. Cell Cycle 8(1):31–34
Ghigna C, Valacca C, Biamonti G (2008) Alternative splicing and tumor progression. Curr Genomics 9(8):556–570. doi:10.2174/138920208786847971
Kalnina Z, Zayakin P, Silina K, Line A (2005) Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer 42(4):342–357. doi:10.1002/gcc.20156
David CJ, Manley JL (2010) Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 24(21):2343–2364. doi:10.1101/gad.1973010
Grosso AR, Martins S, Carmo-Fonseca M (2008) The emerging role of splicing factors in cancer. EMBO Rep 9(11):1087–1093. doi:10.1038/embor.2008.189
David CJ, Chen M, Assanah M, Canoll P, Manley JL (2010) HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463(7279):364–368. doi:10.1038/nature08697
Chansky HA, Hu M, Hickstein DD, Yang L (2001) Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res 61(9):3586–3590
Petermann R, Mossier BM, Aryee DN, Khazak V, Golemis EA, Kovar H (1998) Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene 17(5):603–610. doi:10.1038/sj.onc.1201964
Leach FS, Tokino T, Meltzer P, Burrell M, Oliner JD, Smith S, Hill DE, Sidransky D, Kinzler KW, Vogelstein B (1993) p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res 53(10 Suppl):2231–2234
Hori M, Shimazaki J, Inagawa S, Itabashi M (2002) Overexpression of MDM2 oncoprotein correlates with possession of estrogen receptor alpha and lack of MDM2 mRNA splice variants in human breast cancer. Breast Cancer Res Treat 71(1):77–83
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Rosso, M., Okoro, D.E., Bargonetti, J. (2014). Splice Variants of MDM2 in Oncogenesis. In: Deb, S., Deb, S. (eds) Mutant p53 and MDM2 in Cancer. Subcellular Biochemistry, vol 85. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9211-0_14
Download citation
DOI: https://doi.org/10.1007/978-94-017-9211-0_14
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-9210-3
Online ISBN: 978-94-017-9211-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)