Abstract
MicroRNAs (miRNAs) negatively regulate gene expression level of mRNA post-transcriptionally. Deep sequencing and large-scale screening methods have yielded about 1,500 miRNA sequences in human. Each miRNA contains a seed sequence that is required, but not sufficient, for the correct matching with its targets. Recent technological advances make it possible to capture the miRNAs with their cognate mRNAs at the RISC complex. These experiments have revealed thousands of validated mRNA-miRNA pairing events. In the context of human stem cells, 90% of the identified transcripts appear to be paired with at least two different miRNAs.
In this chapter, we present a comprehensive outline for a combinatorial regulation mode by miRNAs. Initially, we summarize the computational and experimental evidence that support a combined effect of multiple miRNAs. Then, we describe miRror2.0, a platform specifically convened to consider the likelihood of miRNAs cooperativity in view of the targets, tissues and cell lines. We show that results from miRror2.0 can be further refined by an iterative procedure, calls Psi-miRror that gauges the robustness of the regulation. We illustrate the combinatorial regulation projected onto graphs of human pathways and show that these pathways are amenable to disruption by a small set of miRNAs. Finally, we propose that miRNA combinatorial regulation is an attractive regulatory strategy not only at the level of single target, but also at the level of pathways and cellular homeostasis. The joint operation of miRNAs is a powerful means to overcome the low specificity inherent in each individual miRNA.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
16.1 General Overview
MicroRNAs (miRNAs) are short non-coding RNAs (ncRNAs) that negatively regulate gene expression post-transcriptionally [1]. Recent miRNA detection techniques confirmed the presence of hundreds of miRNAs in healthy and diseased tissues [2, 3]. An estimate across animal genomes suggests that almost 1% of the genes in human and C. elegans consists of miRNAs. These estimates are derived from a combination of computational and experimental methods [4, 5].
In human and other metazoa, miRNA plays a role as an additional layer of post-transcriptional regulation [6]. Mechanistically, miRNAs exert their function via base-pair complementarity at the RNA-induced silencing complex (RISC) [7]. The binding of miRNA to mRNA leads to gene silencing. Silencing of a gene by miRNA leads to a change in the mRNA stability, enhanced degradation and to some degree also translational arrest [1, 8, 9]. It was originally proposed that the impact of miRNAs in animals is primarily at the translational level [10–12]. However, the current view argues that most of the miRNA effects are attributable to the post-transcriptional enhancement of mRNA degradation (through blocking of cap binding proteins, deadenylation and more).
A coherent picture of miRNA regulation is still highly fragmented, mainly due to gaps in the understanding of miRNA modes of action in-vivo [13]. Nevertheless, ample evidence indicates that deregulation of miRNAs leads to pathogenesis (i.e., obesity, cancer, neurodegenerative diseases). In fact, for viral infection and cancer, a coordinated change in the relative expression levels of miRNA-sets were shown to be a strong indicator of the pathological state [7, 14, 15]. Less is known on the role of miRNAs under normal physiology conditions, chronic metabolic stress and ageing [16, 17].
miRNAs are best known for the regulation of stem cell differentiation [18], immunological cell function [19], organogenesis [20], cell identity [21], apoptosis [22] and more. The study of miRNAs in the context of cancer biology shows that a disruption in miRNA biogenesis leads to tumorigenesis [23] and to a drastic change in the relative expression of a large number of mRNAs [24]. Furthermore, several miRNAs directly regulate cell cycle genes and thus induce oncogenic activity [3, 25, 26]. In other instances, the activity of miRNA resembles tumor suppression [27]. Interestingly, many of the miRNAs in human are located at fragile sites [28] in agreement with the prevalence of miRNAs in cancer progression.
16.1.1 Outline
This chapter focuses on the notion of combinatorial miRNA action. We consider this hypothesis by appealing to experimental data as well as computational evidence. We provide evidence that the concept of ‘miRNAs working together’ is valid for the different levels of cell regulation from an individual target to a set of targets and ultimately, at biological pathways. We present miRror2.0 as a computational, statistical platform that incorporates the concept of cooperativity when analyzing experimental results. We present the notion that within cells, the disruption of a network is best achieved by a coordinated action of a series of miRNAs. We conclude by suggesting some general trends and architectural design principles for cellular regulation by miRNAs.
16.1.1.1 Classification and Nomenclatures
In this section, we focus on the complete set of human miRNAs. There are over 1,500 miRNAs in humans and about 750 in mouse [29]. We introduce some classifications and accepted notations for the inventory of miRNAs:
-
(i)
Chromosomal organization: Each miRNA is annotated as ‘isolate’ or ‘cluster.’ Specifically, a miRNA that is located in the vicinity of another one (within 10 kb) belongs to a cluster. miRNA that belong to a cluster are encoded in a polycistronic transcript. Over 40% of human miRNAs are organized in such genomic clusters. The remaining miRNAs are considered ‘isolates’.
-
(ii)
Degree of seed sequence overlap: Two miRNAs that overlap in their seed sequences (6–8 nucleotides at the 5′ region of the mature miRNA) belong to the same family. The term ‘single’ refers to the appearance of the seed only once. The definition of a family (according to miRBase [29]) is across species. For our discussion, we follow a species centric view of a family. Illustratively, consider a seed sequence of a miRNA that appears once in human, once in mouse but not in the fly. This sequence is viewed as ‘single’ for human or mouse, but as a ‘family’ from a cross-taxa perspective.
-
(iii)
Degree of pre-miR overlap: The hairpin sequence of the pre-miR (ranges from 70 to 120 nucleotides) includes the mature miRNA sequence. miRNAs that share sequence identity beyond the seed region may be identical in the entire miRNA sequence (22–24 nucleotides), or even at the entire primary transcript (called pri-miR). Such classification is not limited to sequence identity and also considers secondary structure resemblance [29].
-
(iv)
IsomiR classes: Along the miRNA biogenesis and maturation process [30], two strands of the stem-loop structure are produced [31]. The accumulated short reads from deep sequencing experiments indicated the presence of IsomiRs. The IsomiRs refer to miRNAs variants that derive from the chromosomal location but are modified, mainly at the 3′ and 5′ tails to produce rich variants of mature miRNAs [32]. In such experiments, often the two complementary strands are identified, albeit at drastically different expression levels. The strands are called ‘guide/mature’ and ‘passenger/star’ strands. Traditionally, the non-preferred strand sequences were indicated as miR* (star, antisense) [33].
-
(v)
Genomic identity: Most miRNAs are located in intergenic regions, similar to any coding gene. Some miRNAs overlap genes and their sequence is at the same position as other coding or non-coding gene. A substantial number of miRNAs are located in introns of host genes (mirtrons, [34]). The different genomic location is indicative of variation in the maturation and regulation process [35, 36].
-
(vi)
Evolutionary evidence: miRNAs can be classified according to their degree of evolutionary conservation. While many miRNAs are human specific, some orthologs are found only in chimpanzees [37]. Other miRNAs span the entire animal tree (from human to hydra) [38]. The presence of miRNAs that are evolutionary conserved is in accord with their role in basic physiology and cell fate functions.
Figure 16.1 illustrates two forms of miRNA classifications (a chromosomal location and a family assignment) and their relation.
Classifications of the human miRNAs. The human miRNAs are classified according to chromosomal organization and family assignments. Focusing on the 1,527 human miRNAs (miRBase Ver. 18, November 2011) indicates 132 chromosomal clusters (covering 417 miRNAs). The rest are isolated miRNAs (separated by >10 kb). Genes that share the same seed in human are included in the same family. There are 139 families, covering 540 miRNAs. 310 of the miRNAs belong to families that intersect with clusters. The rest (230 miRNAs) are found among the ‘isolates’
The nomenclature of miRNAs is still evolving. The growth in the number of identified miRNAs is attributed to several deep sequencing datasets [33]. Results from this technology have led to the expansion in the number of known miRNAs, mainly by identifying the complementary strand (denoted -5p and -3p to identify the directionality of the sequence as the 5′-arm and the 3′-arm of the stem, respectively) (Table 16.1). Table 16.1 illustrates the guidelines for navigating among the different miRNA names. Unfortunately, different names may be used for the same sequence by different database. For example, microRNA.org [39], PicTar [40] and miRDB [41] refer to the same sequence as hsa-miR-19b-2-5p, hsa-miR-19b and hsa-miR-19b-2*, respectively. Agreement on canonical names is necessary for the task of comparing the performance of individual miRNA-target predictors. Inconsistency in miRNA naming is an unfortunate reality. A reduced consistency between different miRNA-target predictors is partially a result of the inconsistent use of miRNA naming by different miRNA-target DBs.
16.2 Pairing Between a miRNA and Its Target
The most studied recognition signal for miRNA-target pairing is restricted to 6–8 nucleotides (the seed and the immediate vicinity). With 1,500 miRNAs in human and about 10,000 candidate target genes (excluding their alternative splicing variants), the network of interactions is quite complex. Critically, the determinants of binding specificity are poorly understood. Contrary to the known activity of transcription factors, individual miRNAs often attenuate the expression of their direct targets very modestly (e.g., 25% decrease in expression level) [42]. Consequently, a definitive identification of miRNA mappings to their genuine targets is unlikely to succeed comprehensively with current technology [43]. The fraction of false positives across all prediction methods is high, and the number of false negatives is unknown [44, 45]. As a rough estimate, each miRNA is assumed to attenuate tens to hundreds of targets. More importantly, distinguishing between a direct and an indirect miRNA–target interaction remains a crucial challenge in the field.
16.2.1 Computational miRNA-Target Predictions
Currently, miRBase (version 18) is the most exhaustive collection of miRNAs with over 18,000 mature miRNA sequences in over 100 organisms [33]. The 1,500 miRNAs from human and 750 from mouse are estimated to target about half of the genes in human and rodents [46].
There are many databases, algorithms and resources that provide predictions of miRNAs and their direct targets. We will not explicitly discuss the differences among these major resources. We introduce the main resources that were applied while developing the concept of ‘combinatorial regulation’ and the associated software platform. However, it is important to note that several of these tools are meta-servers that combine results from a variety of individual resources [47].
Currently, there are over a dozen miRNA-target resources [48, 49]. While all resources use the knowledge of seed sequence complementarity, some algorithms add a weight to account for imperfect hybridization, context dependent features (e.g., accessibility of binding sites), species conservation, thermodynamic stability of the miRNA-mRNA duplex and any combination of the above. The stable miRNA-target prediction databases include: (i) TargetScan database [50]; (ii) microCosm which is based on the miRanda algorithm [51]; (iii) PicTar (with two settings, according to the degree of evolutionary conversation) [40]; (iv) DIANA–MicroT [52]; (v) PITA (with dual settings for stringency and coverage) [53]; (vi) MirZ [54]; (vii) microRNA.org that allows analysis of multiple miRNAs acting on the same gene-target using the miRanda algorithm [39]; (viii) miRDB resource [41]; (ix) TargetRank (either conserved or all miRNAs) [55]; (x) miRNAMap2 [56]; (xi) RNA22 (cbcsrv.watson.ibm.com/rna22.html), and (xii) the meta-predictor MAMI (http://mami.med.harvard.edu/). A total of 15 sets of predictions are available for human miRNAs. While this is far from an exhaustive list, the most stable and up-to-date DBs for miRNA-target prediction are listed.
Additional descriptive features associated with miRNAs include the distribution of miRNA binding sites, positioning of the binding sites on the mRNA sequences, transcript length, and energy of the secondary structures of the transcript [57]. These features are often excluded in the prediction algorithms, due to their sparse characterization in existing in-vivo studies and the difficulty to generalize these features across animals. Nevertheless, recent miRNA-prediction tools are based on the use of the more thoroughly characterized features of miRNAs in conjunction with algorithms that use statistical models such as a Bayesian models or machine learning (ML). In such schemes, hundreds of examples (negative and positive) are used to suggest the best separation between the true and false predictions. The consistency with validated results is highest among predictors that applied ML technologies [58]. Several of the resources (MirZ, microRNA.org) and miRBase [33] provide miRNA expression profiles for a large number of tissues and cell lines.
Rigorous assessment studies showed that the consistency among major miRNA-target prediction tools is rather poor, reflecting a huge fraction of false positives associated with each of them [59]. The ability of the most established predictors (PITA, DIANA-microT4.0, Miranda, Microcosm, TargetScan5.0, TargetScanS, Pictar and MirZ-ElMMo and RNA22) to explain the profile of the down-regulated genes in cells following overexpressing of individual miRNA has been reported. For example, the precision in recovering the results from hsa-miR-1 overexpression in HeLa cells [42] was 23–50% with a sensitivity level of 6–20%. The union or intersection of any five of these predictors had negligible effect on the overall success and, in fact, a slight reduction in the performance was noted.
16.2.2 Experimental Data for miRNAs in Cellular Systems
In recent years, experimental protocols and tailored chemical probes were developed for the study of miRNA regulation within a cellular context. Their development was necessary since the biochemical approaches initially used to identify miRNAs were biased towards the most abundant miRNAs. Other recently developed methods (e.g., those which provide detailed analysis of the factors determining the level for the regulation of a specific miRNA towards a specific target using, e.g., 3′-UTR segment-luciferase reporter systems will not be discussed but are reviewed in [60]).
The identification of miRNAs in a broad spectrum of metazoan calls for applying comparative genomics technologies. The evolutionary signal from sequence conservation and structural consideration was brought into consideration. Hence, the collection of reported miRNAs has expanded and was doubled between 2009 and 2011. We will only mention methods that provide a global view on the cellular regulation by miRNAs [61].
Functional confirmation of the miRNAs and their target genes is mostly based on in-vitro studies in which a specific miRNA is introduced to cells. In a mirror view, a candidate miRNA gene is knocked down (e.g., using anti-miR). A few hours after the cell manipulation (usually 12–72 h), global gene expression profiling is performed using transcriptomic DNA microarray. The differential expression levels of genes relative to mock-transfected cells are then recorded. The signal that is sought is a negative correlation between the overexpression of the miRNA of interest and the targeted genes. The results of such experiments are collected in the major gene expression archives including GEO [62] and ArrayExpress [63]. Over 30 large-scale experiments of this type were carried out (some with few controls and a minimal reproducibility in the experimental design). Major concern in such experiments is that a shift in the balance between the authentic binding sites and off-targets cannot be avoided [64].
In a more physiological paradigm, cells are exposed to some predetermined condition (hypoxia, glucose starvation, heat shock, drug) and a change in the expression profile of the miRNAs is monitored. The result from these cases is a list of a few hundred candidate miRNAs. Applying the commercially available miRNA DNA-array platforms, large-scale analyses are presented. In recent years, an experimental design has emerged which uses size fractionated RNA from treated cells as input for deep sequencing, thereby collecting a complete set of ‘small ncRNAs’, among them the miRNAs [65]. Several large-scale studies of human tissues in health and disease were reported using this approach. For example, a catalogue of the miRNAs was compiled by RNA-Seq from cancerous and normal cervical tissues [66], melanoma [67], human stem cells [68] and more. These experiments detected coordinated change in the expression of groups of miRNAs [5]. We expect to see more experiments that follow such experimental setting.
In human embryonic stem cells (hESC), the regulation of miRNAs was monitored by immunoprecipitation (IP) of the Argonaute (AGO) proteins [69]. Variations of this approach were reported using tagged AGO proteins. In such settings, the short miRNAs were eluted from the complexes and characterized by sequencing [70]. A similar approach was followed with other RISC proteins [71].
The methods described above focus on a miRNA paired with its cognate mRNA target. The potential effect of miRNAs at the proteome level is typically studied using Mass spectrometry (MS) based methods [11, 72]. Briefly, cells are metabolically labeled (e.g., stable isotope labeling by amino acids, SILAC) to allow a direct comparison of protein expression. While the results from such methodologies are non-conclusive, the expression level of hundreds of proteins appears to be affected [72]. It is likely that miRNAs do modify, whether directly or as a secondary effect, the expression levels of many proteins. This is probably achieved through attenuation of ribosomal initiation, elongation or translational rate.
16.2.3 Validation of miRNAs-Targets
Computational predictions and the experimental results are both compiled in the TarBase database [73]. TarBase is a manually compiled repository for achieving the validated miRNA-target pairs. It relies mainly on in-vitro miRNA overexpression experiments. Additionally, literature-based records for miRNA-targets are collected in miRecords [74]. An assessment using the TarBase benchmark [73] with ∼50 manually validated instances and the results of a quantitative Mass Spectrometry (MS) experiment confirmed the limited agreement between the target prediction algorithms [58]. A gain in prediction success was achieved by combining several prediction resources (using TarBase as a benchmark). However, the consistency in prediction of the different algorithms is rather poor [75, 76].
Many experimentalists tend to use their favorite miRNA-target predicting algorithm and resources (often with insufficient justification). The difference in the prediction results and hence, in the biological interpretation is a result of a substantial fraction of false positives present by all existing methods. To provide a functional relevance to miRNA regulation, several tools analyze the match of miRNA-targets in view of the cell processes, GO (gene ontology), diseases [77] and pathways [78, 79].
16.3 Missing Pieces in Understanding miRNA Regulation
Some aspects of miRNA regulation in cells remain open. For example, we lack an understanding of the kinetics and rate limiting steps throughout the maturation of the pri-miRNA into their active miRNA form [80, 81]. The mechanistic details of miRNA-target recognition remain elusive. For example, the functional relevance of the complementary strand in the pre-miRNA duplex, coined passenger miRNA, is a matter of debate. Initially, it was thought that this strand is simply destined for degradation. However, in some cases, the expression of the passenger miRNAs is associated with tissue specificity and developmental stages. RNA editing of miRNAs provides an additional level of diversity. IsomiRs are miRNAs sequences that are slightly different from those encoded by the DNA. It is postulate that isomiRs are active components of miRNA-based regulation [31]. An even broader questions concerns miRNAs in the context of the mRNAs and related miRNAs. The following aspects in miRNA biology remain for further investigations:
-
(i)
The dynamic aspects of the regulation. Kinetic parameters are missing for degradation, turnover and extrusion of miRNAs [82].
-
(ii)
The AGO occupancy in the cells. The link between induction of mRNA degradation and the AGO occupancy remains to be determined [83]. Specifically, the overlap between the miRNAs that are associated with a specific AGO (e.g., AGO-1 and AGO-2) is minimal [84].
-
(iii)
The quantitative nature of miRNA regulation. Specifically, the balance of miRNAs and mRNAs can be considered from the perspectives of titration [85], accessibility of binding sites and competition [86]. miRNAs in cells are probably sequestered by a “sponge” effect [87]. In such scenarios, a competition on binding sites may dominate the balance of miRNAs, AGO binding sites and eventually the cellular response [88].
-
(iv)
The localization of miRNA regulation. Most of the proteins necessary for miRNA gene silencing are localized to P-bodies. However, the knowledge regarding sub-cellular partition of RISC, mRNA and miRNA within the cells is very limited [89].
16.4 Working Together
Several of these open questions can be approached by a quantitative consideration of mRNAs and miRNAs guided by the concept of competing endogenous RNA (ceRNA) [90]. Accordingly, miRNA-binding sites are assumed to regulate the availability of miRNAs. A prediction from the ceRNA hypothesis claims that the induction of genes having specific miRNA binding sites will indirectly lead to a reduction in the potency of such miRNAs. This hypothesis is supported by experimental data [86]. Based on the ceRNA concept, viruses, pseudogenes and even duplicated genes should be considered as elements that may titrate out the miRNAs, leading to a relief of basal repression.
We will henceforth focus our discussion to the notion of combinatorial activity of miRNAs and the view of miRNAs ‘working together’ as part of a broader cellular design principle.
16.4.1 Evidence for miRNAs Working Together
The concept of combinatorial regulation by miRNAs was validated experimentally. Manipulating a target gene by adding multiple distinct miRNA binding sites on the same transcript augmented the regulation levels [8, 91, 92]. In a cellular context, a parallel overexpression of 2–3 miRNAs resulted in a synergistic effect on the transcriptional level of some candidate genes [93]. For example, in pancreatic cells, for the known target of miR-375, a combined addition of miR-124 and let-7b led to synergy in target inhibition [40]. Similarly, the expression of miR-16, miR-34a and miR-106b altered the cell cycle while no effect on the cell cycle is monitored by each of these miRNAs, separately [85]. The regulation of the tumor suppressor Fus1 in cancer cells depends on the presence of at least three miRNAs (miR-93, miR-98, miR-197) working together [94]. Importantly, introducing several miRNAs not only affected specific candidate targets but also had a measurable effect on specific pathways. For example, a complete block in cell cycle was achieved by a combination of three miRNAs, while the impact of each of these miRNAs alone was less pronounced [95]. A synergetic effect on cell death and the oncogenic properties of multiple miRNAs acting on the same target was recently established [96]. Despite the growing number of instances reported, the generality of the combinatorial phenomenon is yet to be fully established [97].
Some genes are known to have many (predicted) miRNA binding sites. Many of these genes are cell cycle regulators. Recently, several systematic analyses were performed in order to validate the cooperative action of miRNAs on candidate genes [61]. Cells were manipulated to express a luciferase reporter gene for the 3′-UTR of p21 which is a cyclin-dependent kinase inhibitor 1 (also called p21/WAF1). From over 250 different miRNAs that were tested, about 30 miRNAs showed a direct attenuation of the reporter gene [98]. Similar experiments for the CCND1 gene (G1/S-specific cyclin-D1) revealed similar results with seven miRNAs cooperatively attenuating the expression of CCND1, as established by using a 3′-UTR luciferase reporter construct [99].
16.4.2 Tools for the Detection of Combinatorial Regulation by miRNAs
The goal of most described miRNA-target predicting tools (Sect. 16.2.1) is to predict one-to-one relations, namely to determine a miRNA that matches a transcript (at a single or multiple sites). However, as discussed in Sect. 16.4.1, miRNAs most likely act as an ensemble that directly and specifically alters the expression of multiple gene-targets. Conversely, a collection of genes that are targeted coordinately in some experimental settings can be used as input to uncover the set of miRNAs that is most likely responsible for their measured level of gene expression. Along this principle, the MiRonTop [100], Diana-mirExTra [101] and GeneSet2miRNA [102] were developed.
We expanded this notion in the miRror Suite platform. miRror Suite transforms noisy miRNA predictions into a rational unified analysis. The miRror Suite is centered on the miRror2.0 algorithm [103]. The implementation is based on projecting most existing prediction tools into a unified statistical platform. Thus miRror2.0 can predict a coherent list of miRNAs that best explain the observed, complex signature of hundreds of down-regulated genes from experimental data. While our discussion on miRror2.0 focuses on miRNAs from human, the system supports the analysis of other animals (mouse, rat, fly, worm and zebrafish). There are a number of optional parameters that allow the miRror2.0 tool to restrict the analysis to more specific requests. For example, the prediction can operate on preselected tissues or cell-lines (from about 100 options). We demonstrate the generality of the miRror application and its inherent flexibility.
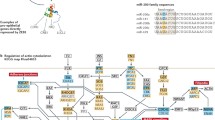
In practice, the miRror platform is used to connect a gene list to the minimal set of preferred miRNAs or a miRNA collection to a set of genes (Fig. 16.2a). We refer to these analyses as Gene2miR and miR2Gene, respectively. The core of the statistical basis underlying miRror is the miRtegrate algorithm (Fig. 16.2a). In a nutshell, for the Gene2miR mode, miRtegrate calculates the probability of matches between the experimental gene set and all miRNAs. This is done by comparing the gene set to the complete gene list that is reported by each of the miRNA-target prediction DBs. The probability of the miRNA’s interaction with the input gene set as opposed to the rest of the genes in each DB is calculated. Calculating a P-value for the set of input (miRNA or Genes) is performed according to the hypergeometric distribution [103]. The reported result is any set with ranked probabilities and scores that meet the statistical threshold (e.g., P-value = 0.01, corrected for multiple tests).
The workflow of the miRror2.0 platform. (a) miRror2.0 in the miR2Gene mode. There are two main modes of operations: the miR2Gene and the Gene2miR. The input for these modes is a set of miRNAs or genes of any size. Following a selection of an organism (human, mouse, rat, worm, fly and zebrafish) and the operational mode (miR2Gene, Gene2miR), some optional choices are available: (i) the tissue of interest or the preferred cell-lines. The information is processed from the atlas of gene expressions [63]. (ii) Selection of all genes or only highly expressed subset (above a predetermined value, typically it reduces the list of genes by ∼30%); (iii) the top scoring miRNA binding sites according to each DB. For each DB scoring method, a fraction that accounts for the top (i.e., 10, 25, 50% and all predictions) can be selected for the analysis. (iv) Select any combination of the DBs (15 in the case of human). (v) To initiate the miRror2.0 search, several free parameters that determine the stringency of the procedure are selected. These parameters include the choice of P-value threshold, the minimal number of supporting DBs and the minimal number of input ‘hits’. By changing these parameters, a relaxed or a strict search protocol is activated. (b) PSI-miRror in a Gene2Gene operational mode with two iterations (from Set-1 to Set-3). The Venn diagram shows the overlap of the input gene list (Set-1) and the output gene list (Set-3). The Venn allows focusing on genes that were removed from the input set or those that were added to the output set
Figure 16.2a illustrates the key principles of the miRror2.0 platform. In reality, the number of genes or miRNAs that result from any large-scale transcriptomic experiments is in the 10s–100s for miRNAs and the 100s–1,000s for genes. The platform is based on a large number of parameters allowing control over the statistical threshold of miRtegrate (which effectively translates to operation stringency). For example, in a case where the set of miRNAs or genes is derived from a specific tissues or cell line, the algorithm recalculates the likelihood of the input set in view of the candidate genes known to be represented in the selected tissues as obtained from Bio-GPS (http://biogps.gnf.org). In addition, the analysis can be restricted to any combinations of miRNA-target prediction DBs (for human, any combination of the 15 supported DBs). The platform may activate the miRror2.0 algorithm by applying only the top scoring predictions from each of the DBs used (e.g., the top 25% of predictions). The flexibility in the number of DBs and the choice of P-value threshold for the miRtegrate algorithm allows full control over the specificity and extent of the resulting analysis.
The combinatorial view implemented in miRror2.0 is further refined by the PSI-miRror operation. Schematically, PSI-miRror is an iterative protocol that aims to refine the input sets (Genes or miRNAs) by increasing the coherence of the input miRNA set to the set of genes and iteratively refining the list of the genes by re-applying the miRror2.0 cycle.
Figure 16.2b illustrates the operation of PSI-miRror in Gene2Gene mode. PSI-miRror can be activated in four modes: Gene2Gene and miR2miR but also miR2Gene and Gene2miR. Figure 16.2b shows that an input Gene Set (Set-1) results in a list of miRNAs that is then fed to an additional iteration that results in an intermediate gene set (Set-2). By analogy to PSI-BLAST [104], the procedure halts when no additional refinement is achieved. In most instances, genes are added or removed from the original list. The application of PSI-miRror is attractive for testing hypotheses. Genes that were added along the iterations of PSI-miRror and are reported in the final set (Set-3, Fig. 16.2b) are candidates for further investigation and experimental validation. The intuition is that genes or miRNAs that are not coherent with the experimental results (e.g., due to the indirect effect of miRNAs) will be removed by the PSI-miRror operation, while coherent genes or miRNAs that were missed, will be added. Often, the intersection of the initial set and the final set is the most coherent set that can be further analyzed (Fig. 16.2b, Venn diagram).
16.4.3 Testing the Predictive Power of miRror2.0
As opposed to the other predicting tools, miRror2.0 and its advanced application of PSI-miRror consider the ensemble rather than individual entities (miRNAs or genes) in the regulatory scheme. A crucial component is the associated scoring system. The performance and the predictions from miRror2.0 are ranked according to the miRror Internal Score (henceforth miRIS). miRIS aims to maximize the different constraints that are implemented by miRror2.0. Specifically, in the Gene2miR mode, we seek a maximal agreement among the selected DBs and high sensitivity in respect to the input. miRIS is composed of a balanced contribution of these two components. Sensitivity is defined as the number of hits from the entire input list. For example, consider an overexpression experiment of a specific miRNA in cells and assume that 400 genes were down-regulated (as measured by a DNA microarray). miRror application at the Gene2miR mode is applied after setting the desired level of stringency (determined by a P-value threshold) and selecting the number of predictors for the analysis. For this illustration, we assume that 12 predicting DBs are selected. miRIS is associated with any of the predicted miRNAs. For example, for a miRNA on the list, only 6 DBs support the prediction and only 200 out of the 400 genes in the input set are marked as relevant genes (which we refer to as ‘hits’), the calculated miRIS for this miRNA is therefore 0.5. A miRIS of 0.75 is calculated once all the 400 genes are reported (i.e., maximal sensitivity) or if only 200 hits are reported, but with a full agreement of all 12 DBs. As shown by this example, miRIS combines DB consistency and sensitivity into a single score.
The validity of the concept of miRNAs ‘working together’ by miRror platform was tested. To this end, we took advantage of the growing number of experiments in which (i) miRNAs were introduced into cell cultures; (ii) the entire transcriptome is compared to control (often cells introduced with a scrambled sequence or a mock transfection). We analyzed data from such experiments from a variety of cell lines. The analyses combined data from DNA microarray platforms (Affymetrix, Agilent) and quantitative mass spectrometry experiments. Specifically, we focused on experiments that report on miRNAs that were overexpressed in cells. Under such controlled conditions, we tested whether miRror can successfully identify the actual overexpressed miRNA using solely the gene expression profiles.
A growing number of such experiments are reported for which the whole transcriptome is compared to that of controlled cells. We assess the ability to recover the evidence of the transfected miRNA from the global transcriptomic profiles of the down-regulated genes (at a threshold >1.2-fold). Importantly, for some experiments hundreds of genes were used as input without reduction in the performance. For example, while only 270 genes were down regulated in an experiment of hsa-miR-145 transfection (DLD-1 cells, GSE18625), for hsa-miR-335 (LM2-Lung cells, GSE9586) this number was almost 10,000. Success was determined according to miRIS. Namely, reporting the position of the correct (the overexpressed) miRNA in the ranked list of all miRNA predictions.
Figure 16.3 shows the results from miRror for overexpression experiments of hsa-miR-124 (GSE6207), hsa-miR-155 and hsa-miR-1 (GSE2002). Ten percent of the down-regulated genes in these experiments (1,700–3,300 genes) were used as input for miRror2.0. Zooming on the top ten predictions (Fig. 16.3, bottom) shows that the actual miRNA is recovered as the top prediction (from 200 best predicted miRNAs). Moreover, the ten top miRNA predictions show a sharp drop in score. In some cases an additional miRNA reaches very significant miRIS (e.g., has-miR-1). Interestingly, the extent of down-regulated genes (i.e., fold change) is not a significant indicator for a successful recovery of the relevant miRNA by miRror2.0. We consider a success when the miRror prediction reports the correct (experimentally over-expressed) miRNA among the top five results, ranked according to miRIS.
Ranked analysis on miRror2.0. Data were collected from the GEO [62] and extracted from the SOFT files. The Affymetrix platform datasets include: GSE6207 (HepG2 cells, hsa-miR-124-24hr) and GSE22002 (HeLa cells, hsa-miR-1, hsa-miR-155). The results are shown in view of the miRror2.0 results, ranked according to miRIS for the top 200 predictions (top) and the top 10 predictions (bottom). The correct miRNA from the over-expression experiments are marked blue
We applied miRror to about 30 large-scale miRNA over-expression experiments. miRror successfully identified the relevant miRNA in 70% of the experiments. The success of the individual DBs ranges from 20 to 60%. Remarkably, miRror was fairly stable regarding the number of genes that were loaded, from 1 to 50% of the down-regulated genes (at a moderate fold change of ≥1.2).
In order to assess the high success in recovering the hidden miRNA from a noisy signal of hundreds of unfiltered genes (at a subtle repression level of >1.2), we repeated the tests with random sets of genes (genes must be reported in at least one DB) or the up-regulated genes (at the same expression ratio of ≥1.2). We show that selecting the objective miRNA failed by repeating the miRror protocol on randomized sets (multiple randomization of identical group size).
We attribute the source of stability in miRNA identification to (i) the predetermined statistical threshold that is applied for a dozen of miRNA-target predictors; (ii) the obligatory demand for a minimal consistency ≥2 DBs and (iii) a requirement of a minimal agreement on number of hits from the input gene list for each proposed miRNA.
16.4.4 Measurements of Direct Binding by miRNAs
The method of HITS-CLIP [45] was developed as a way to directly monitor protein–RNA interactions in living cells [105]. Briefly, the method is based on trapping by cross-linking RNA–protein complexes of interacting molecules that are within a minimal molecular distance. The protocol allows the collection of trapped molecules which are then subjected to trimming of the RNA hanging tails. The result of this protocol is a collection of minimally sized fragments that are suitable for deep sequencing. The use of the HITS-CLIP on AGO-based complexes provides genome wide miRNA–mRNA interaction maps. The first HITS-CLIP experiment was done on mouse brain under stringent conditions [45].
The AGO based HITS-CLIP results address some of the questions on the modus operandi of miRNAs in living cells. Specifically, the experiment (ideally) separates between a direct and an indirect effect of miRNAs. More importantly, the analysis only allows detection of RNA segments (mRNA or miRNA) that are within a short molecular distance and a narrow range of 50–60 nucleotides of the mRNA molecule. Considering the relatively high specificity of the AGO-mRNA (relying on the correlation among independent biological samples), the number of miRNAs that were trapped and identified per transcript is an approximation of in-vivo regulation and AGO-occupancy.
At present, the results from the CLIP-Seq [106] and PAR-CLIP [76] methodologies are limited to only few cellular settings. Nevertheless, some trends for the combinatorial activity of the miRNAs can already be demonstrated. Actually using HITS-CLIP [107], only 10% of the genes were regulated by a single miRNA. Results from the recent CLIP based methods (HITS-CLIP, PAR-CLIP and CLIP-Seq) [58] show that each of the genes that were identified to be subjected to miRNA regulation is in fact targeted by multiple miRNAs. These experiments allow, for the first time, the construction of a miRNA-mRNA interaction map, which supports the notion of combinatorial, cooperative action by miRNAs on targeted transcripts [45].
A collection of large-scale CLIP experiments is compiled in the StarBase database [108]. StarBase includes thousands of experimentally confirmed miRNA-target interactions and complementary data from other AGO proteins. In addition, it provides a genome browser for the reads that were collected during the deep sequencing phase. Figure 16.4 illustrates the complexity in CLIP-experimental interpretation. The deep sequencing reads are illustrated as ‘piles’ (Fig. 16.4a). Using a consensus of prediction algorithms and in some cases, direct sequencing of miRNAs, the match of the miRNA and the gene is reported. In Fig. 16.4a three different miRNAs regulate the presented gene (indicated as miR-a, miR-b and miR-c). However, the miRNA-target prediction algorithms (Fig. 16.4b, marked as Predictors A-D), predict nine miRNA binding sites (1–9). Recall that many of the binding sites were not validated experimentally (miR-e and miR-f). Additional information that became evident from the CLIP-based experiments concerns the intensity of the reads for each miRNA (indicated schematically by the + sign, Fig. 16.4b). The simplistic illustration (Fig. 16.4a) emphasizes the challenge in formulating a miRNA combinatorial view: (i) Some binding sites are more potent than others. (ii) The consistency between the different predictors is limited. (iii) Multiple binding sites for the same miRNA differ in the intensity of the reads (miR-a, purple). (iv) The miRNA-target prediction algorithms often support overlapping binding sites. In reality, the overlap with other binding sites may be excluded due to accessibility argumentation. (v) There is no direct evidence for cooperative binding on one molecule, instead, the scheme (Fig. 16.4a) is most likely a reflection of miRNAs bound to the population of mRNAs.
Interpretation of CLIP-based experiments. Data analyzed from StarBase [108]. (a) The deep sequencing reads are illustrated and the ‘piles’ along the 3′-UTR of the gene. (b) Each of the binding sites (1–9) is associated with predicting DB (marked A–D). Note that some binding sites are more potent than other (marked by + symbol). The consistency between the predicting DBs is only partial (see binding site 3,4). There are multiple binding sites (at different extent) for the same miRNA (miR-a, purple). The predicting DBs indicate miRNAs that overlap on the sequence of their binding sites (overlap, OL). The OL sites are often excluded due to accessibility argumentation
16.4.5 Looking Through the miRror – Predictions Versus Experimental Reality
The coherence between the miRNA-targets that are based on gene expression (Fig. 16.3) and those obtained from the CLIP-based experiments is surprisingly low [106]. The gene expression data and the CLIP data are collected from live cells. Still, over-expression of specific miRNA may be prone to non-physiologically high miRNA concentrations. On the other hand, a bias in the CLIP data may reflect the inability to capture transient interactions of AGO and mRNAs. Moreover, the identity of the apparently trapped miRNAs in the CLIP assays is largely based on the set of computational predicting tools that suffer from high false positive rates. It is suggested that the readouts of mRNA suppression (measured by gene expression profiles) and the initial pairing (measured by CLIP technologies) are complementary but non-overlapping assessments of the regulation by miRNAs. An analysis of the features that govern miRNA–target match is critical to improving the prediction power of the methods (both computational and experimental).
Figure 16.5 shows the statistical analysis of the data collected from StarBase [108]. The cumulative representation (combining several experiments) with a gene centric view on all the data shows that 50% of the 6,200 targeted genes are associated with up to eight miRNAs (Fig. 16.5a) and 90% of the genes are targeted by up to 35 miRNAs. A few genes are even targeted by more than 100 miRNAs. Interestingly, when all the potential sites that are predicted by all 12 human miRNA-target DBs (the union of all), the analysis shows that the experimental CLIP data captures only a relative small fraction of the predicted set (Fig. 16.5b).
Statistical analysis from CLIP-based experiments. (a) A cumulative view of the number of miRNAs regulating each gene, from CLIP data. The steep climb demonstrates how prevalent regulation by multiple miRNAs appears to be. (b) A comparison of CLIP data and predicted miRNA-targets. The combination of all 15 human predicting DBs is shown. The miRNA-targets that are covered by the CLIP data are shown separately in light blue. The distributions from the CLIP data and the union of the DB predictions are very different. It emphasizes the gap between the computational view on the targeting potential of genes by miRNAs and the observed gene targeting from the most up-to-date CLIP experiments
Figure 16.6 summarizes the potential of miRror2.0 to be used as an assessment tool for combinatorial regulation. Several tests are carried out to examine the coherence of the experimental data and miRror results. The results of the scheme illustrated in Fig. 16.6a are discussed (Fig. 16.3). The scheme in Fig. 16.6b is applied to the CLIP data from StarBase [108]. Specifically, the ensemble of miRNAs that is associated with each targeted gene was collected for each gene from the thousands of genes that are targeted by at least two miRNAs (Fig. 16.6b). For 98% of the genes miRror successfully identified the relevant gene. For 81%, the correct prediction was among the top 10% of predictions (typically from a ranked list of >1,000 predictions). This finding supports the predictive power of miRror2.0 for genes that are targeted by a high number of miRNAs. Furthermore, when considering genes that were targeted by >20 miRNAs, the performance of miRror reached 98% and a complementary view was noted for genes that were targeted by <10 miRNAs (the performance dropped slightly to 90% success).
Assessment of the combinatorial nature of miRNAs. (a) Using Gene2miR mode to assess the recovery of over-expression of miRNAs from the repressed genes from large-scale transcriptomic profiling platforms. (b) Using miR2Gene mode to assess the recovery of a gene from the collection of the available CLIP-based experimental data. (c) Assessment using the Jaccard Index (JI) of pairs of miRNAs. Data were from CLIP-based experiments and from the prediction according to selected predicting DBs. (d) Using miRror2.0 at a Gene2miR mode for KEGG human pathway graphs and testing the ability of small sets of miRNA to disrupt the connectivity of the pathway. For details see text
The tools and methodologies developed for the validation of miRror2.0 and the large CLIP dataset allowed us to test the minimal mode of combinatorial regulation – namely the concept of ‘miRNA pairs’. Figure 16.6c shows the protocol applied in formulating the concept of miRNA-pairs. Pairs are the simplest form of multiple miRNA co-regulation and thus are a natural starting point for a computational assessment. We wish to see how well each of the major prediction DBs matches the biological experimental data. The number of genes that are reported from the multiple CLIP data (compiled by StarBase [108]) is >6,200. A vast majority of them are regulated by multiple miRNAs (Fig. 16.5). There are two general modes that comply with regulation by pairs (Fig. 16.6c): (i) A pair of miRNAs that expands the set of targets, thus allowing a better coverage of the relevant genes. (ii) Each miRNA in the pair tightly overlaps the targets of the other miRNA. In this case, the pair of miRNAs acts in ‘backup’ mode, with a high degree of redundancy. The two extreme scenarios are formulated using the Jaccard Index (JI). Intuitively, JI is a simple measure for comparing the similarity (intersection) and diversity (union) of the sample sets. A low JI value is indicative of the expansion mode while high JI indicates the backup mode.
From this naïve view on all pairs of miRNAs that were reported in CLIP data, several observations are worth noting: (i) The data from the CLIP experiments are dominated by a very low JI. Although only less than 20% of the pairs have no shared targets, the JI is extremely low. When compared the same data for a sample of miRNA-target predicting DBs, each DB centers at a different JI (the arrows indicates the average JI value, Fig. 16.6c).
The analyses that are schematized in Fig. 16.6 further emphasize that a naive approach considering a single miRNA DB in order to extrapolate pairwise relations is insufficient. Using the miRror platform is a step toward such an extrapolation. Specifically, we observed that the targets covered by many miRNAs from the CLIP data provided us with higher prediction rates and scores relative to targets that were characterized by being targets of a relatively small number of miRNAs.
16.5 Working Together at the Pathway Level
The interpretation of gene sets that resulted from coordinated miRNAs (Fig. 16.6 miR2Gene), or from any other miRNA-target prediction protocol should be analyzed within a cellular context. A number of tools were developed that cover aspects of protein and functional interaction (STRING [109]), regulatory pathways (Reactome [110]) and functional annotations (PANDORA [111] and DAVID [112]).
The pathway representation best describes the biological processes in cells. The human regulatory pathways are compiled by the KEGG resource [113]. KEGG pathways are a collection of manually drawn pathway maps. These maps represent knowledge on the molecular interaction and reaction networks for domains including human diseases, organismal systems, cellular processes, and environmental information processing. The collection covers ∼100 pathway maps for human.
The notion of ‘miRNAs working together’ is tested in view of metabolic and regulatory pathways. Specifically, regulation of miRNAs was suggested at the level of pathway or biological process [114]. We assess the possibility that cooperative action by a small, selected group of miRNAs can alter the expression of genes that belong to the same pathway, without sacrificing specificity. An extension of the ‘working together’ concept argues that a disruption of the pathway’s topology by miRNAs has the potential to alter the outcome of the targeted pathway. It is known that various diseases and developmental stages are characterized by a coordinated alteration of a number of miRNAs.
With this idea, one can prioritize each pathway according to its susceptibility to regulation by a small group of miRNAs (for example pairs or triplets). Reliable resources for human pathways are the PID (NCI human pathways) [115] and KEGG [116]. The definition of pathways is somewhat vague as some resources describe modules rather than full pathways. For example, the Reactome database covers 1,100 pathways (cellular modules) [117] and the Human Pathway Database (HPD), that unifies the major resources, includes over 1,000 pathways [118]. Many of the pathways were previously analyzed in view of their modular nature, redundancy and robustness against perturbations [119, 120]. While recent studies have acknowledged the usefulness of miRNAs on regulation pathways [121, 122], adding the connectivity of genes in the pathway is a key determinant that was largely ignored. The intuition for the pathway disruption approach is that a quantitative change in a set of miRNAs is expected to alter the pathway outcome [90, 123].
A test case (Fig. 16.6d) using the 100 regulatory, disease oriented and metabolic pathways from KEGG that cover about 4,500 human genes revealed an intriguing principle of miRNA regulation. Analysis of the pathway via the concept of individual miRNA prediction databases results in hundreds of potential miRNAs. Therefore, matching a small number of miRNAs to a pathway is virtually impossible with the current prediction DBs. However, the potential of a small group of coordinated miRNAs to alter the integrity of human pathway can be challenged using the miRror2.0 combinatorial tool. The motivation of our approach is in assessing the potential of a set of miRNAs to disrupt a graph that represents a cellular pathway.
For the scheme in Fig. 16.6d, we start from a pathway and end up with a selected list of miRNA sets (i.e., pairs and triplets that work together) which preferentially disrupt the integrity of the pathway. The following steps are taken: (i) All high quality human pathways are converted to undirected graphs in which the nodes are the genes (or complexes) and the edges are the regulatory interactions. (ii) Each pathway is converted to a gene set that is subjected to miRror2.0 to determine the ranked list of possible miRNAs. Refinement by PSI-miRror further limits the list of possible miRNAs to the most relevant set. (iii) Designing a disconnecting score (DIS) that captures the degree of network disruption (for example, the partition into connected components, edge elimination and the like). (iv) Applying an exhaustive search for all pairs and triplets from all candidate miRNAs combinations. Finally, providing the sets that maximally impact graph connectivity (resulting in a high DIS score). As the miRNA combinatorial space is vast, for a pathway that reported by miRror2.0 to have 40 miRNAs, about 10,000 possible miR-Triplets need to be ranked (by DIS score) in order to identify the best sets. For a pathway with 50 potential miRNA candidates, the search space for miRNA triplets is 19,600. Of course, the topology of the pathway graph is a key determinant in this scheme. Note that the number of genes in the human KEGG pathways ranges from 10 to 250 and the number of initial candidates for miRNA disruption (according to miRror output) range from 2 to 60.
Several conclusions are derived from this approach. Most notably, 85% of all KEGG based pathways are amenable to disruption by a small miRNA set of pairs or triplets (the same results apply to PID pathways [115]). Analyzing all pathways revealed that typically, only 4–5 miRNAs are associated with the most potent set of miRNA pairs and triplets for each pathway. Several biological interpretations from the miRNA cooperative pathway disruption scheme can be drawn. Genes such as MAPK1, EGFR, AKT3, SRC that are prevalent in tens of regulatory pathways, are almost always included in the set of disrupted genes. Most likely, these proteins serve as connectors in the pathway graphs. Thus, targeting these signaling genes will lead to a substantial disruption of many pathways. Most surprisingly, the selected miRNA sets (pairs and triplets) with a maximal capacity to disrupt pathways show a minimal overlap. Therefore, it seems that a critical factor in selecting the most influential miRNA combinations is the graph topology rather than the identity of the individual nodes in the pathway graph.
16.6 Concluding Remarks
The concept of miRNAs working together is not new. In this chapter, we present experimental evidence while emphasizing tradeoffs in adopting a combinatorial mode of regulation for living cells under changing conditions.
We present a tool that incorporates the concept of ‘working together’ and describe some tests in view of the current experimental knowledge. The miRror Suite is a platform that empowers experimental biologists in gaining insights from a broad range of experimental protocols. It is based on a many-to-one and many-to-many approach. Namely, a group of miRNAs as an input leads to a minimized set of genes that best explain the observed gene expression profile. Similarly, this applies for a set of genes as input. The many-to-many optimization is performed by the PSI-miRror approach that provides a refined set of molecules by iterative application of the PSI-miRror algorithm. The miRror Suite provides an integrative, statistically based platform and exposes miRIS: a combined scoring system for a successful prediction of miRNA combinatorial regulations. miRror performance is discussed in [124].
Multiple layers of regulations in the cell are coordinated in governing cellular phenotypes. Most notable are: epigenetic chromatin marks, the transcription machinery of gene expression, the translation process, the degradation of transcripts and proteins, the metabolic balance and more. miRNAs constitute an additional layer of regulation that was carefully studied in stem cells, viral infection, cancer progression and other pathologies. It is likely that regulation of miRNAs is a key strategy of the cell as it strives to maintain robust homeostasis. Under this assumption, a modest modulation executed in a combinatorial mode can be manifested by a substantial change in cell physiology and phenotype. We illustrate the combinatorial regulation concept at the level of the individual target (Sect. 16.4.4), at the level of a set of genes whose expression were moderately changed (Sect. 16.4.3), and finally at the level of human pathway integrity (Sect. 16.5). While many questions remain to be solved, we expect the ensemble-oriented tools will prove essential to the biological interpretation of miRNA data.
Abbreviations
- AGO:
-
Argonaute
- DB:
-
database
- DIS:
-
disconnecting score
- GO:
-
gene ontology
- HITS-CLIP:
-
high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation
- PAR-CLIP:
-
photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation
- miRNA (miR):
-
microRNA
- ML:
-
machine learning
- MS:
-
mass spectrometry
- ncRNA:
-
non-coding RNA RISCRNA-induced silencing complex
- SILAC:
-
stable isotope labeling by amino acids in cell culture
- UTR:
-
untranslated region.
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Landgraf P, Rusu M, Sheridan R et al (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414
Iorio MV, Ferracin M, Liu CG et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070
Grad Y, Aach J, Hayes GD et al (2003) Computational and experimental identification of C. elegansmicroRNAs. Mol Cell 11:1253–1263
Friedlander MR, Chen W, Adamidi C et al (2008) Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26:407–415
Bushati N, Cohen SM (2007) microRNA functions. Annu Rev Cell Dev Biol 23:175–205
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Brodersen P, Voinnet O (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10:141–148
Gregory RI, Chendrimada TP, Cooch N et al (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123:631–640
Pillai RS, Bhattacharyya SN, Filipowicz W (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17:118–126
Baek D, Villen J, Shin C et al (2008) The impact of microRNAs on protein output. Nature 455:64–71
Bandyopadhyay S, Mitra R (2009) TargetMiner: MicroRNA target prediction with systematic identification of tissue specific negative examples. Bioinformatics 25:2625–2631
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114
Cullen BR (2009) Viral and cellular messenger RNA targets of viral microRNAs. Nature 457:421–425
Alvarez-Garcia I, Miska EA (2005) MicroRNA functions in animal development and human disease. Development 132:4653–4662
Liang R, Bates DJ, Wang E (2009) Epigenetic control of MicroRNA expression and aging. Curr Genomics 10:184–193
Schroen B, Heymans S (2012) Small but smart-microRNAs in the centre of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and ageing. Cardiovasc Res 93:605–613
Zhang B, Pan X, Anderson TA (2006) MicroRNA: a new player in stem cells. J Cell Physiol 209:266–269
Xiao C, Rajewsky K (2009) MicroRNA control in the immune system: basic principles. Cell 136:26–36
Zhao Y, Samal E, Srivastava D (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436:214–220
Liao R, Sun J, Zhang L et al (2008) MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem 104:805–817
Cheng AM, Byrom MW, Shelton J et al (2005) Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33:1290–1297
Kumar MS, Lu J, Mercer KL et al (2007) Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39:673–677
Volinia S, Calin GA, Liu CG et al (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103:2257–2261
He L, Thomson JM, Hemann MT et al (2005) A microRNA polycistron as a potential human oncogene. Nature 435:828–833
Calin GA, Ferracin M, Cimmino A et al (2005) A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353:1793–1801
Lee YS, Dutta A (2007) The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21:1025–1030
Calin GA, Sevignani C, Dumitru CD et al (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101:2999–3004
Griffiths-Jones S, Grocock RJ, van Dongen S et al (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Cullen BR (2004) Transcription and processing of human microRNA precursors. Mol Cell 16:861–865
Fernandez-Valverde SL, Taft RJ, Mattick JS (2010) Dynamic isomiR regulation in Drosophila development. RNA 16:1881–1888
Berezikov E, Robine N, Samsonova A et al (2010) Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res 21:203–215
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157
Rodriguez A, Griffiths-Jones S, Ashurst JL et al (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14:1902–1910
Barik S (2008) An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res 36:5232–5241
Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448:83–86
Berezikov E, Thuemmler F, van Laake LW et al (2006) Diversity of microRNAs in human and chimpanzee brain. Nat Genet 38:1375–1377
Liu N, Okamura K, Tyler DM et al (2008) The evolution and functional diversification of animal microRNA genes. Cell Res 18:985–996
Betel D, Wilson M, Gabow A et al (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36:D149–D153
Krek A, Grun D, Poy MN et al (2005) Combinatorial microRNA target predictions. Nat Genet 37:495–500
Wang X (2008) miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA 14:1012–1017
Lim LP, Lau NC, Garrett-Engele P et al (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773
Doench JG, Sharp PA (2004) Specificity of microRNA target selection in translational repression. Genes Dev 18:504–511
Enright AJ, John B, Gaul U et al (2003) MicroRNA targets in Drosophila. Genome Biol 5:R1
Chi SW, Zang JB, Mele A et al (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460:479–486
Bentwich I (2005) Prediction and validation of microRNAs and their targets. FEBS Lett 579:5904–5910
Sethupathy P, Megraw M, Hatzigeorgiou AG (2006) A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods 3:881–886
Rajewsky N (2006) microRNA target predictions in animals. Nat Genet 38(Suppl):S8–S13
Sethupathy P, Corda B, Hatzigeorgiou AG (2006) TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA 12:192–197
Lewis BP, I-h S, Jones-Rhoades MW et al (2003) Prediction of mammalian microRNA targets. Cell 115:787–798
John B, Enright AJ, Aravin A et al (2004) Human MicroRNA targets. PLoS Biol 2:e363
Maragkakis M, Reczko M, Simossis VA et al (2009) DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 37:W273–W276
Kertesz M, Iovino N, Unnerstall U et al (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39:1278–1284
Hausser J, Berninger P, Rodak C et al (2009) MirZ: an integrated microRNA expression atlas and target prediction resource. Nucleic Acids Res 37:W266–W272
Nielsen CB, Shomron N, Sandberg R et al (2007) Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 13:1894–1910
Hsu S-D, Chu C-H, Tsou A-P et al (2008) miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res 36:D165–D169
Long D, Lee R, Williams P et al (2007) Potent effect of target structure on microRNA function. Nat Struct Mol Biol 14:287–294
Alexiou P, Maragkakis M, Papadopoulos GL et al (2009) Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics 25:3049–3055
Martin RC, Liu PP, Goloviznina NA et al (2010) microRNA, seeds, and Darwin?: diverse function of miRNA in seed biology and plant responses to stress. J Exp Bot 61:2229–2234
Orom UA, Lund AH (2009) Experimental identification of microRNA targets. Gene 451:1–5
Thomson DW, Bracken CP, Goodall GJ (2011) Experimental strategies for microRNA target identification. Nucleic Acids Res 39:6845–6853
Barrett T, Edgar R (2006) Mining microarray data at NCBI’s Gene Expression Omnibus (GEO)*. Methods Mol Biol 338:175–190
Parkinson H, Sarkans U, Kolesnikov N et al (2011) ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res 39:D1002–D1004
van Dongen S, Abreu-Goodger C, Enright AJ (2008) Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods 5:1023–1025
Creighton CJ, Reid JG, Gunaratne PH (2009) Expression profiling of microRNAs by deep sequencing. Brief Bioinform 10:490–497
Witten D, Tibshirani R, Gu SG et al (2010) Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol 8:58
Stark MS, Tyagi S, Nancarrow DJ et al (2010) Characterization of the melanoma miRNAome by deep sequencing. PLoS One 5:e9685
Bar M, Wyman SK, Fritz BR et al (2008) MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26:2496–2505
Goff LA, Davila J, Swerdel MR et al (2009) Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS One 4:e7192
Easow G, Teleman AA, Cohen SM (2007) Isolation of microRNA targets by miRNP immunopurification. RNA 13:1198–1204
Zhang L, Ding L, Cheung TH et al (2007) Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell 28:598–613
Selbach M, Schwanhausser B, Thierfelder N et al (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63
Papadopoulos GL, Reczko M, Simossis VA et al (2009) The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res 37:D155–D158
Hua Y-J, Tang Z-Y, Tu K et al (2009) Identification and target prediction of miRNAs specifically expressed in rat neural tissue. BMC Genomics 10:214
Mendes ND, Freitas AT, Sagot MF (2009) Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res 37:2419–2433
Hafner M, Landthaler M, Burger L et al (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141
Jiang Q, Wang Y, Hao Y et al (2009) miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 37:D98–D104
Backes C, Meese E, Lenhof HP et al (2010) A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res 38:4476–4486
Papadopoulos GL, Alexiou P, Maragkakis M et al (2009) DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics 25:1991–1993
Saj A, Lai EC (2011) Control of microRNA biogenesis and transcription by cell signaling pathways. Curr Opin Genet Dev 21:504–510
Winter J, Jung S, Keller S et al (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234
Chatterjee S, Grosshans H (2009) Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461:546–549
Khan AA, Betel D, Miller ML et al (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27:549–555
Beitzinger M, Peters L, Zhu JY et al (2007) Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol 4:76–84
Linsley PS, Schelter J, Burchard J et al (2007) Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27:2240–2252
Arvey A, Larsson E, Sander C et al (2010) Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol 6:363
Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4:721–726
Seitz H (2009) Redefining microRNA targets. Curr Biol 19:870–873
Liu J, Valencia-Sanchez MA, Hannon GJ et al (2005) MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7:719–723
Salmena L, Poliseno L, Tay Y et al (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358
Hon LS, Zhang Z (2007) The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol 8:R166
Brennecke J, Stark A, Russell RB et al (2005) Principles of microRNA-target recognition. PLoS Biol 3:e85
Tu K, Yu H, Hua YJ et al (2009) Combinatorial network of primary and secondary microRNA-driven regulatory mechanisms. Nucleic Acids Res 37:5969–5980
Du L, Schageman JJ, Subauste MC et al (2009) miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res 7:1234–1243
Ivanovska I, Cleary MA (2008) Combinatorial microRNAs working together to make a difference. Cell Cycle 7:3137–3142
Mu P, Han YC, Betel D et al (2009) Genetic dissection of the miR-17 92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 23:2806–2811
Zhou YM, Ferguson J, Chang JT et al (2007) Inter-and intra-combinatorial regulation by transcription factors and microRNAs. BMC Genomics 8:396
Wu S, Huang S, Ding J et al (2010) Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene 29:2302–2308
Jiang Q, Feng MG, Mo YY (2009) Systematic validation of predicted microRNAs for cyclin D1. BMC Cancer 9:194
Le Brigand K, Robbe-Sermesant K, Mari B et al (2010) MiRonTop: mining microRNAs targets across large scale gene expression studies. Bioinformatics 26:3131–3132
Alexiou P, Maragkakis M, Papadopoulos GL et al (2010) The DIANA-mirExTra web server: from gene expression data to microRNA function. PLoS One 5:e9171
Antonov AV, Dietmann S, Wong P et al (2009) GeneSet2miRNA: finding the signature of cooperative miRNA activities in the gene lists. Nucleic Acids Res 37:W323–W328
Friedman Y, Naamati G, Linial M (2010) MiRror: a combinatorial analysis web tool for ensembles of microRNAs and their targets. Bioinformatics 26:1920–1921
Altschul SF, Madden TL, Schaffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Darnell RB (2011) HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA 1:266–286
Wen J, Parker BJ, Jacobsen A et al (2011) MicroRNA transfection and AGO-bound CLIP-seq data sets reveal distinct determinants of miRNA action. RNA 17:820–834
Alves L, Niemeier S, Hauenschild A et al (2009) Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Res 37:4010–4021
Yang JH, Li JH, Shao P et al (2011) starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 39:D202–D209
Jensen LJ, Kuhn M, Stark M et al (2009) STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–D416
Vastrik I, D’Eustachio P, Schmidt E et al (2007) Reactome: a knowledge base of biologic pathways and processes. Genome Biol 8:R39
Rappoport N, Fromer M, Schweiger R et al (2010) PANDORA: analysis of protein and peptide sets through the hierarchical integration of annotations. Nucleic Acids Res 38:W84–W89
da Huang W, Sherman BT, Tan Q et al (2007) The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8:R183
Kanehisa M (2002) The KEGG database. Novartis Found Symp 247:91–101, discussion 101–103, 119–128, 244–152
Lall S, Grun D, Krek A et al (2006) A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16:460–471
Schaefer CF, Anthony K, Krupa S et al (2009) PID: the Pathway Interaction Database. Nucleic Acids Res 37:D674–D679
Ogata H, Goto S, Fujibuchi W et al (1998) Computation with the KEGG pathway database. Biosystems 47:119–128
D’Eustachio P (2010) Reactome knowledgebase of human biological pathways and processes. Methods Mol Biol 694:49–61
Chowbina SR, Wu X, Zhang F et al (2009) HPD: an online integrated human pathway database enabling systems biology studies. BMC Bioinform 10(Suppl 11):S5
Ideker T, Sharan R (2008) Protein networks in disease. Genome Res 18:644–652
Stelling J, Sauer U, Szallasi Z et al (2004) Robustness of cellular functions. Cell 118:675–685
Cui Q, Yu Z, Purisima EO et al (2006) Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol 2:46
Gusev Y (2008) Computational methods for analysis of cellular functions and pathways collectively targeted by differentially expressed microRNA. Methods 44:61–72
Peter ME (2010) Targeting of mRNAs by multiple miRNAs: the next step. Oncogene 29:2161–2164
Balaga O, Friedman Y, Linial M (2012) Toward a combinatorial nature of microRNA regulation in human cells Nucl. Acids Res 40:9404–9416
Acknowledgements
We thank Guy Naamati whose contribution to the miRror platform development was seminal. We thank Nati Linial for insightful ideas. We thank Solange Karsenty for maintenance of the miRror website. We thank Manor Askenazi for a critical reading and editing of the manuscript. A student fellowship (O.B) is awarded by the SCCB, the Sudarsky Center for Computational Biology. We apologize to those that we could not cite. In numerous instances we replace the primary citations by review articles due to space constraints. This study is partially supported by the ISF 592/07, the BSF 2007219 and the EU Framework VII of Prospects.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Friedman, Y., Balaga, O., Linial, M. (2013). Working Together: Combinatorial Regulation by microRNAs. In: Schmitz, U., Wolkenhauer, O., Vera, J. (eds) MicroRNA Cancer Regulation. Advances in Experimental Medicine and Biology, vol 774. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-5590-1_16
Download citation
DOI: https://doi.org/10.1007/978-94-007-5590-1_16
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-5589-5
Online ISBN: 978-94-007-5590-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)