Abstract
Genome editing with engineered nucleases (“GEEN”) has emerged as an effective genetic engineering method that uses ‘molecular scissors’—artificially engineered nucleases—to digest DNA at targeted locations in the genome of various organisms including plant species. The DNA binding domains of zinc finger (ZF) proteins were first used as plant genome editing tools via the use of designed ZF nucleases (ZFNs), with TAL-effectors (TALE) and the RNA-DNA recognition system CRISPR/Cas9 now being used as powerful genome editing tools to create targeted gene modifications, not only in model plants but also in crop species. The key to genome editing is the introduction of targeted gene-specific double-stranded DNA breaks (DSBs) using the designed endonucleases, then allowing site-directed mutagenesis via nonhomologous end joining (NHEJ) repair and/or gene targeting via homologous recombination (HR), to occur efficiently at specific sites in the genome. This chapter provides an overview of recent advances in genome editing technologies, giving an insight into current plant molecular biology and breeding techniques.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Historically, natural mutagenesis has contributed to the development of genetic variation in plant breeding programmes aimed at increasing crop quality and yield. In recent decades the increasing use of induced mutagens has required wide-ranging screening to identify superior traits resulting from mutations that have been introduced randomly into the genome. Radiation is often used for this purpose. Physical and chemical mutagens, such as ethyl methanesulfonate (EMS) and radiation using gamma rays and ion beams, which create DNA damage and consequently induce DNA repair systems, are now well established as means of generating new traits by the random introduction of mutations into the genome. In such mutagenesis schemes, the double-stranded DNA break (DSB) genome repair systems that are programmed in the early processes of meiosis are essential to the generation of mutations in plant species as well as in other organisms (Osakabe et al. 2012). To obtain superior plants using such strategies, screening of the mutagenized populations is required to identify individuals possessing the desired phenotypes.
Genome modifications introduced into crop plants by genetic engineering have been utilized recently to create the potential to improve agricultural practices and add nutritional quality to products. For example, "Golden rice" has been engineered to produce beta-carotene to increase its nutritional food value (Gartland et al. 2013). Silencing of a gene of interest at the posttranscriptional stage using short RNA interference (siRNA) has also been utilized effectively; however, gene knockdown by siRNA can sometimes be variable and incomplete (Fei et al. 2013). On the other hand, sequence-specific modifications have also been widely used in genetic analysis; however, there are some technical difficulties with site-specific mutagenesis in several organisms.
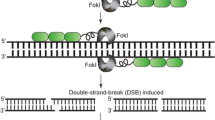
Genome editing with engineered nucleases (GEEN) has been developed as an effective genetic engineering method that uses artificially engineered nucleases to digest DNA at the desired location in the genome of a plant species. An induced DSB introduced at a specific site by the engineered nuclease then undergoes repair by the natural processes of homologous recombination (HR) and nonhomologous end joining (NHEJ). Sequence modifications at the cleaved site, such as deletions or insertions introduced by NHEJ, result in gene disruption and integration of exogenous sequences then occurs in the genome via HR. Currently, three types of engineered nucleases, viz. zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated9), have been developed; of these, TALEN and CRISPR/Cas9 are now especially widely used for genome editing in various plant species (Sander and Joung 2014) (Fig. 13.1).
Current methods in GEEN and an overview of the workflow of plant genome editing. Homing endonucleases/meganucleases (EMN) recognize long (~20 base pair) DNA sequences. The FokI nuclease is the DNA-cleavage domain used in ZFN that binds target DNAs via engineered C2H2-zinc finger (ZF) domains; TALENs recognize targets via the engineered TALE composed of RVD domains derived from the plant pathogen Xanthomonas. The CRISPR/Cas9 system utilizes RNA-guided engineered nucleases (RGNs), which use a short guide RNA (gRNA) to recognize DNA sequences at the target site
In this chapter, we describe recent advances in genome editing in the field of plant genome engineering. We highlight genome editing studies that have led to the modification of various plant genomes. Site-directed sequence modifications using engineered nucleases have been used in studies of both model and crop plant species. We also discuss approaches for the application and future prospects of these technologies in plant molecular breeding and biotechnology.
2 Site-Directed Mutagenesis of Higher Plants Using Genome Editing Tools
2.1 Site-Directed Mutagenesis Using ZFNs
In 2005, Lloyd et al. published the first report of site-directed mutagenesis in plants using ZFN; these authors created a target-specific mutation in Arabidopsis using a synthetic model target (Lloyd et al. 2005). Of the ZFN-induced mutations characterized, 78 % were simple deletions of 1–52 bp, 13 % were simple insertions of 1–4 bp, and 8 % were deletions accompanied by insertions. The results showed that the mutants were present in the subsequent generation in 10 % of induced individuals. Using tobacco plants, Wright et al. (2005) also showed ZFN-stimulated gene targeting (GT) in plants. A modular assembly method for ZFN—oligomerized pool engineering (OPEN) to generate ZF arrays that recognize specific DNA sequences—was developed and used for plant genome modification (Townsend et al. 2009). After establishment of the basic assembling systems, site-directed mutagenesis of Arabidopsis using ZFN driven by an estrogen-inducible system was reported, resulting in highly effective mutation of the alcohol dehydrogenase and chalcone synthase genes (Zhang et al. 2010). In this latter study, the primary transgenic Arabidopsis induced to express ADH1 or TT4 ZFNs exhibited somatic mutation frequencies of 7 % or 16 %, and 69 % or 33 % of the primary lines were transmitted to the next generation (Zhang et al. 2010). We also reported ZFN mutagenesis of the gene ABSISCIC ACID INSENSITIVE 4 in Arabidopsis using the HSP system, and created a new mutant in abi4 (ABA-insensitive) showing the expected response to the plant hormone ABA (Osakabe et al. 2010). Together, looking at these studies and others (Tovkach et al. 2009, 2010), most studies utilizing ZFN have employed the controlled expression of ZFN proteins to create DSBs and induce repair by NHEJ.
2.2 Site-Directed Mutagenesis Using TALENs
With further extended methods using TALENs, several studies have succeeded recently in creating target-specific mutations (Cermak et al. 2011; Christian et al. 2013; Gurushidze et al. 2014; Li et al. 2012; Mahfouz et al. 2011; Zhang et al. 2013; Wendt et al. 2013). Voytas’ group has developed the assembly of custom TALE arrays in a system known as the Golden Gate cloning method, and achieved site-directed mutation of the ADH1 gene in Arabidopsis protoplasts using this system (Cermak et al. 2011). The de novo engineered TALEN derived from Hax3 from the Brassicaceae pathogen X. campestris pv. armoraciae strain 5 was used (Mahfouz et al. 2011). Hax3, which recognizes a 12-bp DNA sequence, was used to construct an engineered nuclease that was then used for targeted mutagenesis in a transient assay using Nicotiana benthamiana (Mahfouz et al. 2011). Targeted TALEN mutagenesis of the Arabidopsis genome using a stable transgenic approach has subsequently been reported (Christian et al. 2013). In this latter study, constitutive TALEN expression in transgenic Arabidopsis was induced to create mutations of several genes and a duplicated gene cluster. The frequency of somatic mutagenesis was 41–73 % in the individual transgenic plant lines, and mutants transmitted to the next generation with a frequency of 1.5–12 %. Because of the relatively low toxicity of TALEN compared with ZFN, TALEN can be considered for use in constitutive expressions systems (Christian et al. 2013).
Very recently, TALEN technology has started to be used in crop species such as rice, barley, and maize (Gurushidze et al. 2014; Li et al. 2012; Liang et al. 2014; Shan et al. 2013; Wendt et al. 2013). The rice disease-susceptibility gene and the sucrose-efflux transporter gene OsSWEET14 were mutagenized by TALEN, resulting in disease-resistant rice with normal phenotypes (Li et al. 2012). Large-scale targeted mutagenesis by TALEN in rice and Brachypodium has been reported by Gao’s group, who generated knockouts of rice genes and eight Brachypodium genes with high efficiency, and also showed large (e.g. 1.3 kb) genomic deletions by simultaneous expression of two pairs of TALENs (Shan et al. 2013). Recently, Gurushidze et al. have developed gene knockout systems based on TALEN in the barley genome using the transformation of embryonic pollen cultures consisting primarily of haploid cells (Gurushidze et al. 2014). These findings have high applicability for detailed studies into gene function and molecular breeding in these various crop species.
2.3 Site-Directed Mutagenesis Using CRISPR/Cas9
In the 2 years from the first publication of this system (in 2012) plant genome editing using the CRISPR/Cas9 system was demonstrated in various plant species, such as Arabidopsis, tobacco, sorghum, rice, wheat, maize, sweet orange, and liverwort (Feng et al. 2013, 2014; Jiang et al. 2013; Jia and Wang 2014; Li et al. 2013; Liang et al. 2014; Mao et al. 2013; Miao et al. 2013; Nekrasov et al. 2013; Shan et al. 2013; Sugano et al. 2014; Upadhyay et al. 2013), suggesting its highly applicability. The first reports of CRISPR/Cas9 in plant genome editing appeared in August 2013, in a report of transient expression of CRISPR/Cas9 in Arabidopsis protoplasts, tobacco cells, and rice plants adapted to create DSBs (Li et al. 2013; Nekrasov et al. 2013; Shan et al. 2013). Li et al., showed that transient expression of a CRISPR/Cas9 vector in Arabidopsis protoplasts induced the mutation of homologous members in a multiplex genome as predicted (Li et al. 2013). Using haploid generation of Marchantia polymorpha L., Sugano et al., demonstrated a simple and rapid genome editing method employing CRISPR/Cas9 using liverwort as a model species in a study on land plant evolution (Sugano et al. 2014). Recently, Zhu’s group reported the multigenerational analysis of CRISPR/Cas9-induced genome editing in Arabidopsis, showing the heritable mutation of a 1-bp insertion and short deletion with high efficiency (Feng et al. 2014). In this study, the mutation frequencies were 71.2 % in the first transgenic lines, 58.3 % in the next generation, and 79.4 % in subsequent generations (Feng et al. 2014). Recently, the same group reported efficient gene modification using CRISPR/Cas9 in rice, suggesting that the CRISPR/Cas9 system will become a powerful tool in crop genome engineering (Zhang et al. 2014).
Since the structure of plant genomes and gene families are highly redundant and overlapping, off-target effects during genome editing are an unavoidable and important issue. Recently, Puchta’s group reported that applying nickase resulted in efficient genome engineering in Arabidopsis (Fauser et al. 2014). Concentrated efforts to extend the above-mentioned findings, together with the precise and detailed evaluation of genome edited-plants to detect off-target effects will be extended to allow further application of genome editing for the crop breeding.
3 Gene Targeting and Targeted Gene Addition in Higher Plants Using Genome Editing Tools
Genome editing tools provide novel strategies for genetic manipulation in plants and are likely to assist engineering of desired plant traits by modifying endogenous genes. For instance, the site-specific addition of genes in major crop species can be used for 'trait stacking', whereby several desired traits are physically linked to ensure their co-segregation during the breeding processes. Gene targeting (GT) is a genome engineering method designed to introduce modifications into endogenous genomic sequences via HR. An exogenous DNA with sequences homologous to the target gene and the modification of interest is used as a template instead of undamaged homologous DNA (Osakabe et al. 2006, 2012). HR is induced by creating DSBs at the target site, e.g. the expression of I-Sce I, a rare-cutting restriction enzyme, has been shown to lead to a significant increase in HR-mediated GT in tobacco cells (Puchta et al. 1993, 1996). Rare-cutting enzymes can also lead to the site-specific integration of foreign DNA molecules and induce site-specific mutagenesis.
To introduce GT in both endogenous and exogenous genes with high efficiency in fruit fly and human genomes, engineered ZFNs have been utilized to induce DSB and GT in the presence of donor DNA (Bibikova et al. 2003; Porteus and Baltimore 2003; Urnov et al 2005). ZFNs have also been reported to increase GT frequency in higher plants. A ZFN designed for maize genes and a heterologous donor molecule introduced into maize cells suggested that the ZFN effectively controlled targeted gene addition at a specific site in the genome and that the change was inheritable and transmitted to the next generation (Shukla et al. 2009). Another study by Voytas’s group demonstrated ZFN-mediated GT in a transient expression system in tobacco (Townsend et al. 2009). Both studies revealed that over 20 % of selected lines showed GT events. These works demonstrate that cleavage of a chromosomal target by ZFNs dramatically stimulates HR-mediated GT in plants, and provides a basis for future experiments with ZFNs directed to any endogenous genomic location.
Recently, highly efficient targeted gene insertion using TALEN in tobacco protoplasts has also been reported (Zhang et al. 2013). The CRISPR/Cas9 system is also useful in HR-mediated targeted gene insertion in tobacco (Li et al. 2013) and rice (Shan et al. 2013). Since a low HR efficiency has been found in several plant species, appropriate optimizations and improvements will be needed to extend plant genome engineering using custom engineered nuclease-mediated targeting.
4 Future Prospects
Current molecular biology techniques allow the direct and effective mutation of particular genes of interest by GEEN. The use of GEEN technology has grown dramatically, the development of CRISPR/Cas9 with simplified methods particularly speeding up the progress of these techniques. Via modification of known genes, these "targetable" nucleases will become a viable alternative to standard breeding methods to identify and introduce novel traits in economically important plants. Genome editing methods now allow us to create newly designed superior traits in various plant species with these applicable and useful methods; however, genome editing will become more effective with further improvements and by conquering several remaining problems such as off-target effects. In particular, a reliable nuclease design, which includes the absence of toxicity and the lack of off-target effects, will be needed for high efficiency and specificity. For example, the double-nicking methods using mutant-types of CAS9 developed by Zhan’s group (Ran et al. 2013) and the double cleavage methods of Joung’s group (Tsai et al. 2014) will also be utilized in the genome editing of higher plants.
One of the most important issues in plant genome editing is how to deliver and express the engineered nucleases in plant cells, since not all useful plant species are amenable to regeneration and transgenic methods. The appropriate choice of plant tissue and optimized methods for transformation and culturing are still major issues to resolve on the way towards the generation of novel useful crops. Thus, efficient systems for the delivery of genome editing tools into plant cells must be developed. Recent studies using viral vectors, such as recombinant adeno-associated virus (Ellis et al. 2013) and geminivirus-based replicons (Baltes et al. 2014), to deliver the DNA of genome editing tools has enabled efficient genome engineering in various plant species. As new plant breeding techniques, these efforts, together with a deeper understanding of whole genome structure and function, promise to deliver future technologies in breeding new and important traits in various plants.
References
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26:151–163
Bibikova M, Beumer K, Trautman JK, Carroll D (2003) Enhancing gene targeting with designed zinc finger nucleases. Science 300:764
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82
Christian M, Qi Y, Zhang Y, Voytas DF (2013) Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 (Bethesda) 3:1697–1705
Ellis BL, Hirsch ML, Porter SN, Samulski RJ, Porteus MH (2013) Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Ther 20:35–42
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359
Fei Q, Xia R, Meyers BC (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25:2400–2415
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas9 system. Cell Res 23:1229–1232
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas9-induced gene modifications in Arabidopsis. Proc Natl Acad Sci U S A 111:4632–4637
Gartland KM, Bruschi F, Dundar M, Gahan PB, Viola Magni M, Akbarova Y (2013) Progress towards the 'Golden Age' of biotechnology. Curr Opin Biotechnol 24(Suppl 1):S6–S13
Gurushidze M, Hensel G, Hiekel S, Schedel S, Valkov V, Kumlehn J (2014) True-breeding targeted gene knock-out in barley using designer TALE-nuclease in haploid cells. PLoS One 9:e92046
Jia H, Wang N (2014) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9:e93806
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392
Li JF, Norville JE, Aach J, McCormack M, Zhang D et al (2013) Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Liang Z, Zhang K, Chen K, Gao C (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas9 system. J Genet Genomics 41:63–68
Lloyd A, Plaisier CL, Carroll D, Drews GN (2005) Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci U S A 102:2232–2237
Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK (2011) De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci U S A 108:2623–2628
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR/Cas9 system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ (2013) Targeted mutagenesis in rice using CRISPR/Cas9 system. Cell Res 23:1233–1236
Nekrasov V, Staskawicz B, Weigel D, Jones JDJ, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNAguided endonuclease. Nat Biotechnol 31:691–693
Osakabe K, Abe K, Yoshioka T, Osakabe Y, Todoriki S, Ichikawa H, Hohn B, Toki S (2006) Isolation and characterization of the RAD54 gene from Arabidopsis thaliana. Plant J 48:827–842
Osakabe K, Osakabe Y, Toki S (2010) Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc Natl Acad Sci U S A 107:12034–12039
Osakabe K, Saika H, Okuzaki A, Toki S (2012) Site-directed mutagenesis in higher plants. In: Shu QY, Forster BP, Nakagawa H (eds) Plant mutation breeding and biotechnology. CAB eBooks, Wallingford, pp 523–533
Porteus MH, Baltimore D (2003) Chimeric nucleases stimulate gene targeting in human cells. Science 300:763
Puchta H, Dujon B, Hohn B (1993) Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res 21:5034–5040
Puchta H, Dujon B, Hohn B (1996) Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci U S A 93:5055–5060
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F (2013) Double nicking by RNA-guided CRISPR/Cas9 for enhanced genome editing specificity. Cell 154:1380–1389
Sander JD, Joung JK (2014) CRISPR/Cas9 systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355
Shan Q, Wang Y, Li J, Zhang Y, Chen K et al (2013) Targeted genome modification of crop plants using a CRISPR/Cas9 system. Nat Biotechnol 31:686–688
Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459:437–441
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55:475–481
Tovkach A, Zeevi V, Tzfira T (2009) A toolbox and procedural notes for characterizing novel zinc finger nucleases for genome editing in plant cells. Plant J 57:747–757
Tovkach A, Zeevi V, Tzfira T (2010) Validation and expression of zinc finger nucleases in plant cells. Methods Mol Biol 649:315–336
Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML et al (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459:442–445
Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK (2014) Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32:569–576
Upadhyay SK, Kumar J, Alok A, Tuli R (2013) RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 3:2233–2238
Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435:646–651
Wendt T, Holm PB, Starker CG, Christian M, Voytas DF, Brinch-Pedersen H, Holme IB (2013) TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol Biol 83:279–285
Wright DA, Townsend JA, Winfrey RJ Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44:693–705
Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D et al (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci U S A 107:12028–12033
Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF (2013) Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161:20–27
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Acknowledgments
The authors are grateful to Dr. S. Toki for his kind support to this work. This work was supported by the Grant-in-Aid for Scientific Research C from the Japan Society for the Promotion of Science (21580125 to Y.O. and 25450002 to K.O.).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Japan
About this chapter
Cite this chapter
Osakabe, Y., Osakabe, K. (2015). Genome Editing in Higher Plants. In: Yamamoto, T. (eds) Targeted Genome Editing Using Site-Specific Nucleases. Springer, Tokyo. https://doi.org/10.1007/978-4-431-55227-7_13
Download citation
DOI: https://doi.org/10.1007/978-4-431-55227-7_13
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-55226-0
Online ISBN: 978-4-431-55227-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)