Abstract
Sebacinales form mycorrhizae with a variety of plant families including Ericaceae and Orchidaceae. Structurally, these mutual associations are quite distinct, with specific structures largely depending on the individual plant family. Many plants that are associated with Sebacinales occur in the same habitats and it has been questioned whether they share a common pool of mycobionts or are associated with different Sebacinales guilds.
This chapter presents a study of Sebacinales associated with coexisting Ericaceae and Orchidaceae from two different habitats; pristine forest and regenerating landslides in a tropical mountain rain forest in South Ecuador. The structural distinctness of mycorrhizal associations in Ericaceae and Orchidaceae formed by Sebacinales is presented, and evidence that different guilds are associated with both plant families is given. Phylogenetic relationships of these Sebacinales on higher and lower taxonomic levels are illustrated and discussed in an evolutionary context. In addition, this chapter addresses community analyses based on a phylogenetic concept, a promising approach used in the field of ecology and evolution of microorganisms.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Since the first description of Sebacina vermifera as intrahymenial fungus by Oberwinkler (1964) and the first identification of an orchid-mycorrhiza isolate as S. vermifera Oberwinkler by Warcup and Talbot (1967), Sebacinales were repeatedly retrieved as important mycorrhiza-forming fungi with members of diverse plant families (Warcup 1988; Glen et al. 2002; Selosse et al. 2002; Urban et al. 2003; Kottke et al. 2003; Allen et al. 2003; Setaro et al. 2006a, b; Suárez et al. 2008). Structurally, mycorrhizae involving Sebacinales are quite distinct, with specific structures largely depending on the individual plant family. Sebacinales form ectomycorrhizae with members of Fagales and Myrtales (Glen et al. 2002; Selosse et al. 2002; Urban et al. 2003), ericoid and cavendishioid mycorrhizae with Ericaceae (Allen et al. 2003; Setaro et al. 2006a, b), orchid mycorrhizae with Orchidaceae (Suárez et al. 2008), and jungermannioid mycorrhizae with liverworts (Kottke et al. 2003; Newsham and Bridge 2010). A molecular phylogeny revealed ectomycorrhiza-forming Sebacinales in a distinct clade together with Sebacina incrustans, separated from the other mycorrhiza-forming Sebacinales that cluster with S. vermifera (Weiß et al. 2004).
In a previous small-scale investigation comparing Sebacinales mycobionts of Ericaceae and Orchidaceae from the same sampling area in the tropical mountain rain forest of Southern Ecuador, we found molecular indications for a distinction between these associations (Kottke et al. 2008). These results were based on sampling of roots from epiphytic orchids on stems of standing trees, but of hemiepiphytic Ericaceae growing in the humus fraction of soil and mineral soil (Kottke et al. 2008). Therefore, it was unclear whether the distinctiveness of Sebacinales clades was due to habitat differences or whether it was determined by the association with a distinct plant family. For the present study, a larger sample size was collected at the same locality. Sampling included epiphytic and terrestrial orchids on humus and mineral soil as well as terrestrial and hemiepiphytic ericads from pristine forest and disturbed habitats. In contrast to the 2008 study by Kottke et al., we sequenced not only the 28S of the rDNA but also included the more variable ITS (Nilsson et al. 2008) in order to get higher resolution on subgeneric levels. Phylogenetic inference, cluster analyses, and a phylogenetic-Bray-Curtis (PBC) transformation approach were used to address the following questions: (1) Is it possible to delimitate distinct clades of Sebacinales in the host plants under investigation? (2) Do Sebacinales show preference for either one of the two plant families independent of habitat and substrate? (3) In addition, since Sebacinales have been isolated from diverse geographic areas and display a worldwide distribution, the phylogenetic position of Sebacinales from the Neotropics was also of interest.
2 Materials and Methods
2.1 Study Sites and Sampling
Mycorrhizae from coexisting Ericaceae and Orchidaceae were collected in the Reserva Biológica San Francisco (3°58′S, 79º4′W), part of the Biosphere Reserve Podocarpus—El Condor, Southern Ecuador. The protected area contains pristine rain and cloud forest and sites that are disturbed due to anthropogenic impacts. Five different sites were selected for sampling of mycorrhizae of Ericaceae and Orchidaceae. Two sites were located inside undisturbed forest at 2,000 m (site 1) and 2,170 m (site 4), one in a 40-year-old regenerating part of the pristine forest at 2,170 m (site 3) and two on a 55-year-old anthropogenic landslide surrounded by mountain rain forest at 1,900 m (site 2 and 5). Detailed information on the area is given in Beck et al. (2008).
Roots from Ericaceae were collected in March 2007 and from Orchidaceae in February to May 2008. In total, 103 Orchidaceae, 17 of which were shown to be associated with Sebacinales and 19 Ericaceae were sampled. Mycorrhizae of Ericaceae were sampled from flowering individuals selected randomly by excavating the entire length of a root, beginning at the stem base and leading out to the finest rootlets. The rootlets of each host individual were pooled, cleaned under tap water, and freed from organic debris within 2 days after sampling. Fresh mycorrhizae were selected using a dissection microscope. A root cluster consisting of two to eight rootlets was dried and stored in 1.5-ml microcentrifuge tubes with silica gel for DNA extraction.
Roots from orchids were collected from 2 or 3 orchid individuals in 56 permanently established plots of 1 m2, 8 terrestrial or epiphytic plots per respective site. Only roots in contact with the tree bark were collected from epiphytic orchids. The roots of terrestrial orchids in the forest were sampled from the pure humus layer while on the landslide mycorrhizal roots were sampled from the mineral soil. Preliminary observations had shown the presence of orchid mycorrhizae in these microhabitats (Kottke et al. 2010). Roots were screened for fungal colonization of the cortical tissue the day of sampling by microscopic observation of freehand sections stained using methyl blue (0.05 % in lactic acid, Merck C.I. 42780). Well-colonized parts of roots were selected for DNA extraction and ultrastructural examination. DNA was conserved at −20 °C.
Ericaceae were identified to species level with the Flora of Ecuador (Luteyn 1996). In case where specimens could not be certainly identified, we consulted the experts of Neotropical Ericaceae, Jim Luteyn and Paola Pedraza. Orchid specimens were not sent to experts because of conservational issues and lack of flowering material available, so identification of Orchidaceae is on the level of morphospecies only. DNA barcoding of Orchidaceae allowed further identification on genus level.
2.2 Light and Transmission Electron Microscopy
Mycorrhizae of Orchidaceae were fixed in 2.5 % glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) on the day of sampling. Samples were postfixed in 1 % osmium tetroxide and embedded in Spurr’s plastic according to Bauer et al. (2006), but with prolonged infiltration steps of up to 12 h. Semi-thin sections were stained by crystal violet and observed for fungal colonization of the cortical tissue by use of light microscopy. Healthy looking colonized parts were selected for ultra-thin sectioning. Serial ultra-thin sections were prepared and examined using a ZEISS transmission electron microscope at 80 kV. Vouchers of the plastic embedded samples were deposited in the Herbarium Tubingense, Eberhard-Karls-University Tübingen.
Mycorrhizae of Ericaceae had already been collected and processed in the case of the Setaro et al. study 2006a, b in the study area. The preparation was analogous to those of Orchidaceae; for details see Setaro et al. (2006a, b).
2.3 Processing of Fungal DNA Sequences
For roots of Ericaceae, genomic DNA was isolated from dried mycorrhiza samples (~5 mm root length per host individual) using the DNAeasy Plant Mini Kit (Qiagen), according to the manufacturer’s instructions but without the use of RNase A. To amplify part of the 18S, the ITS and portions of the 28S genes, we used the universal forward primers NS23 (Gargas and Taylor 1992) and ITS1F (Gardes and Bruns 1993) as well as the Sebacinales-specific reverse primer MWS1 (kindly provided by Michael Weiß), in polymerase chain reactions (PCRs). In some cases, a nested PCR was required to obtain products suitable for cloning. The first amplification was conducted with the primers NS23/ITS1F and MWS1. This product was used as a template with final concentrations of 1:50, 1:500, or 1:5,000 for a second PCR with the primers NS23/ITS1F and the Sebacinales-specific primer MWS2 (kindly provided by Michael Weiß). We used the Phusion Master Mix with HF buffer (Finnzymes) as PCR reagents with a final volume of 20 μl. The template for the PCR reaction was the genomic DNA extract with a final concentration of 1:10. The PCR protocol was conducted according to the manufacturer’s instructions, but with 30 cycles and an annealing temperature of either 60 °C, 63 °C, or 66.5 °C.
All positive PCR products were cloned with the Zero Blunt Topo PCR Cloning Kit (Invitrogen, Life Technologies) using a PCR product volume of 0.5 μl for the cloning reaction. Clones were checked for positive inserts by picking up to eight bacterial clones with a toothpick and placing them directly into a PCR reaction mixture. The reaction volume was 50 μl, with concentrations of 1.5 mM MgCl2, 200 μM of each dNTP (Life Technologies, Invitrogen), 1 U of Taq polymerase (Life Technologies, Invitrogen), an amplification buffer (Life Technologies, Invitrogen), bovine serum albumin (0.004 %), and 0.5 μM of each primer (Biomers). We used M13F and M13R as the forward and reverse primer for this PCR reaction. One to ten inserts were amplified by rolling circle amplification with the TempliPhi Amplification Kit (GE Healthcare) and sequenced without prior purification on an ABI 3730 sequencing machine (GATC Biotech, Germany). Aside from M13F and M13R, we used several insert primers (NS23; ITS3Seb; kindly provided by M. Berbee; LR3: (Hopple and Vilgalys 1994); LR0R: (Hopple and Vilgalys 1994); SSS1: 5′-GTGAACCTGCGGAAGGATCATTA-3′; MWS1; ITS1F; NL4 reverse complement: (O’Donnell 1993); SSS2: 5′-TAGATG TTCTGGGCCGCACGC-3′; SSS3: 5′-GGAATAGGGAGAATCTGC-3′) to obtain the full sequence length in good quality.
For roots of Orchidaceae, DNA isolation, polymerase chain reaction, cloning and sequencing, a 1–2 cm long piece of root was cut for one DNA extraction per plant individual. The pieces were rinsed in sterile water and freed from the velamen. Genomic DNA was recovered using the DNAeasy Plant Mini Kit (Qiagen) according to the manufacturers’ instructions. The whole ITS1-5.8S- ITS2 region and part of the 28S rDNA were amplified with the universal primers ITS1 (White et al. 1990) and TW14 (Cullings 1994) using the Phusion High-Fidelity PCR Mastermix (Finnzymes). PCR products were cloned with the Zero Blunt TOPO PCR Cloning Kit (Invitrogen) according to the manufacturer’s protocol (Stockinger et al. 2009). Twelve colonies per individual were selected for PCR amplification using modified M13F and M13R primers (Krüger et al. 2009). Success of PCR was tested in 1 % agarose gel. Eight colonies per orchid individual showing correct fragment size were grown in liquid LB Broth, MILLER (Difco) and plasmids were isolated with the S.N.A.P. miniprep kit (Invitrogen) according to manufacturers’ instructions. Plasmids were sequenced on an ABI 3730xl sequencing machine (Macrogen, Korea) using universal primers M13F and M13R.
All sequences were edited and congruent strands were combined using Sequencher 4.6 (Gene Codes, Ann Arbor, MI, USA). BLAST (Altschul et al. 1990) against the NCBI nucleotide database (GenBank; http://www.ncbi.nlm.nih.gov/) was used to find published sequences with high similarity. Sequences that matched Sebacinales were compiled in a data set and checked for potential chimeric sequences. To do this, sequences were roughly aligned with Poa (Grasso and Lee 2004) and analyzed with Bellerophon (Huber et al. 2004) and a window width of 200 base pairs. In addition, the alignment was partitioned with a window width of 400 base pairs and each partition was blasted separately. All sequences have been submitted to Genbank, the accession numbers are given in Table 5.1 and Fig. 5.2.
2.4 DNA Bar Coding of Orchidaceae
Genomic DNA of fresh or dried leaves was isolated using the DNAeasy Plant Mini Kit (Qiagen) according to the manufacturers’ instructions. To amplify the chloroplast gene matK, we used the primer combinations 19 F (Whitten et al. 2000)—881R (Pridgeon et al. 2001) and 731 F (Pridgeon et al. 2001)—trnK-2R (Johnson and Soltis 1995) in PCRs. We used the Phusion High-Fidelity Polymerase (Finnzyme) as PCR reagent in a final volume of 25 μl according to the manufacturers’ instructions but with an annealing step for 20 s and annealing temperatures of 52 °C for the primers 19 F-881R and 58 °C for the primers 731 F-trnK2R. We added Bovine Serum Albumin (BSA-SIGMA) with a final concentration of 0.8 μg μl−1 to the reaction mix (Iotti and Zambonelli 2006). Every PCR was run with a negative control.
All positive PCR products were sequenced on an ABI 3730xl sequencing machine (Macrogen, Korea) using the same primers as for DNA amplification. The sequences were edited and congruent strands were combined using Sequencher 4.6 (Gene Codes, Ann Arbor, MI, USA). BLAST (Altschul et al. 1990) against the NCBI nucleotide database (GenBank; http://www.ncbi.nlm.nih.gov/) was used to find published sequences with high similarity to identify the Orchidaceae to genus level.
2.5 Phylogenetic Analyses of Sebacinales
Phylogenetic analyses were performed with additional sequences of Sebacinales from NCBI in order to ascertain the phylogenetic position of Sebacinales from the sampling area in relation to Sebacinales from other hosts and locations all over the world. NCBI was searched for Sebacinales including ITS data and a minimum length of 400 base pairs in order to exclude excessively short sequences. The NCBI sequences that met these criteria were added to our data set. Sequences were roughly aligned with Poa and alignment columns upstream the 5′ direction of the ITS1F primer region were excluded from the data set. Furthermore, all sequences that were not aligned across the 5.8S region were excluded as well as were all sequences with more than 65 % of leading and trailing gaps. Because of these preconditions, not all Sebacinales from Genbank could be included in the data set.
To prune the data set, single linkage clustering with a threshold of 0.5 % was performed with Optsil(Göker et al. 2009). All Sebacinales with identical names or isolated from the same host plant, and affiliated to the same cluster, were excluded with the exception of one representative. The remaining sequences were joined with our data set of Sebacinales from Ecuadorian Ericaceae and Orchidaceae.
All gaps and missing data were removed and the sequences were aligned with Mafft v6.602b (Katoh et al. 2002) and the option linsi as well as with Poa (Grasso and Lee 2004). The most reliable of both alignments was chosen with Trimal and the option comparesets (Capella-Gutiérrez et al. 2009). The best alignment was used to perform a Maximum Likelihood analysis with 1,000 rapid bootstrap replicates (Stamatakis et al. 2008) in Raxml 7.0.4 sequential version using the GTRmix model (Stamatakis 2006).
After the analysis, the ML tree was rooted with the midpoint method in FigTree (Rambaut 2009). To enhance visualization of the tree, a large clade (in total 100 sequences) containing sequences of Sebacinales group A (Weiß et al. 2004) and related taxa was collapsed. None of the Sebacinales associated with Ericaceae and Orchidaceae from this study were in the collapsed clade.
2.6 Cluster Analysis of Ericaceae and Orchidaceae
We used the UniFrac metric (Lozupone and Knight 2005) in combination with hierarchical clustering to determine whether Ericaceae and Orchidaceae are significantly associated with different guilds of Sebacinales. The UniFrac metric measures the difference between two environments (in our case host species) in terms of the branch length that is unique to one host or the other (Lozupone and Knight 2005).
All sequences of Sebacinales were aligned with Mafft v6.602b (Katoh et al. 2002) and the option linsi as well as with Poa (Grasso and Lee 2004). The most reliable of both alignments was chosen with Trimal and the option comparesets (Capella-Gutiérrez et al. 2009). A Maximum Likelihood (ML) tree was reconstructed in Raxml 7.0.4 sequential version using the GTRmix model (Stamatakis 2006). The ML tree was rooted using the midpoint method in FigTree (Rambaut 2009) and analyzed with FastUnifrac (Hamady et al. 2010) clustering and 1,000 jackknife replications.
2.7 Distance Transformation and Neighbor-Net Analysis of Ericaceae and Orchidaceae
Another approach to determine whether Ericaceae and Orchidaceae are associated with different guilds of Sebacinales is the PBC transformation approach (Göker and Grimm 2008) in combination with Neighbor-Net analysis. This novel approach transforms phylogenetic distance matrices of the “associates” to distance matrices of the “host” using Sørensen’s distance coefficient. In contrast to the Sørensen distances, PBC does not require knowledge of the taxonomic affiliation of the sampled species in advance. Rather, it determines the most similar specimens between two hosts automatically when computing pair-wise distances from the reduced matrix (Göker and Grimm 2008).
The same alignment was used as for the UniFrac distance measure to compute a sequence distance matrix (GTR model of evolution) with Paup 4.0b10 (Swofford 2002). The distance matrix was used as “associate” distances and the respective host species as “hosts.” The distance transformation was calculated with Pbc (Göker and Grimm 2008) and the option—minimum one associate per host. A Neighbor-Net analysis of the PBC distance matrix was conducted with Splitstree v4 (Huson and Bryant 2006) to visualize the outcome of the distance transformation. To test the treelikeness of the PBC network, delta scores were computed in Splitstree v4. Delta scores are based on quartets of taxa and range between zero (maximum treelikeness) to one and are a reliable predictor of phylogenetic accuracy (Holland et al. 2002; Auch et al. 2006).
3 Results
3.1 Structural Differences in Colonization of Roots of Neotropical Ericaceae and Orchidaceae
Light and transmission electron microscopic observations revealed Sebacinales as the dominant mycobionts of Ericaceae in the area under investigation. Hyphae of Sebacinales were recognized by the typical septal pore structure (Fig. 5.1b). While species belonging to the Andean-clade of Ericaceae (Kron et al. 2002) displayed ectendomycorrhizas (cavendishioid mycorrhizae - Setaro et al. 2006a, b) with a hyphal sheath, Hartig net, and intracellular colonization (Fig. 5.1c), the non-Andean clade ericads showed only intracellular colonization (Fig. 5.1a) characterizing ericoid mycorrhizae (Smith and Read 2008). Sebacinales in orchid roots formed the typical pelotons in the multilayered cortical tissue that are encased by cell wall material after degeneration (Fig. 5.1d). Hyphae were also found in the dead cells of the velamen entering through passage cells of the exodermal layer into the cortical layer (Kottke and Suárez 2009). Sebacinales were only rarely observed in the tissue of the orchids under investigation. The most frequent fungi of orchids observed in this study were Tulasnellales, followed by Atractiellomycetes (Kottke et al. 2010).
Structural differences of the mycorrhizal types (a) Light microscopic micrograph of a square section through the ericoid mycorrhiza of a species of Sebacinales in Gaultheria erecta; hyphae colonizing the single layer of epidermal cells that surrounds the endodermis and stele; scale bar 6.3 μm. (b) Transmission electron micrograph of a hyphal septum of a species of Sebacinales in epidermal cells of Ericaceae displaying dolipore structure and continuous parenthesome; scale bar 0.15 μm. First published in Setaro et al. (2006a) (c) Light micrograph of tangential section through the epidermal cells of a mycorrhiza of Semiramisia speciosa and a species of Sebacinales (cavendishioid mycorrhiza); hyphal sheath, Hartig net, and enlarged intracellular hyphae visible; scale bar 4 μm. (d) Light micrograph of a square section through the orchid mycorrhiza of Sobralia rosea and a species of Sebacinales; velamen, exodermal layer with passage cell containing hyphae and multilayered cortical cells with hyphal pelotons and degenerating pelotons are displayed; scale bar 20 μm. Abbreviations: cc cortical cell, ep epidermis, dp degenerating pelotons, en endodermis, ex exodermis, ihy intracellular hyphae, Hn Hartig net, hs hyphal sheath, pc passage cell, pe pelotons, hc hyphal coils, ve velamen
3.2 Phylogeny of Sebacinales Associated with Neotropical Ericaceae and Orchidaceae
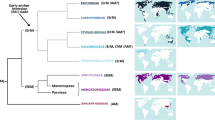
Detailed information of Sebacinales with respect to hosts, sampling localities, and life-forms is given in Table 5.1. Sebacinales associated with Neotropical Ericaceae and Orchidaceae from the present study fall in seven major clades (clades I, II, III, IV, V, VI, VII) with high bootstrap support (Figs. 5.2a, b). All of these clades contain Sebacinales either associated with Ericaceae or Orchidaceae, but not with both host families. Clade I and VI contain Sebacinales from tropical and temperate Ericaceae as well as undefined Sebacinales from soil (Fig. 5.2a, b). The remaining clades (II, III, IV, V, and VII) only contain Sebacinales associated with Neotropical Orchidaceae from the present study. Orchid clade III is closely related (96 % bootstrap support) to S. vermifera isolated from an Australian orchid, Eriochilus cucullatus. The Orchidaceae clades II, III, IV, and V are closely related (100 % bootstrap support) to clade I containing Sebacinales associated with Ericaceae (Fig. 5.2a). A sequence of Sebacinales associated with Phleum pratense (EU910901) falls within that group. Sebacinales in clade VI are related to S. vermifera (DQ520096, EU625995) and to Sebacinales associated with grasses, herbs, trees, and an arbutoid mycorrhizal Ericaceae (Pyrola rotundifolia EU668934) (Fig. 5.2b). Clade VII is rather isolated, as its close relationship to clade VI is not supported. However, all seven clades are more closely related to each other than to Sebacinales of group A sensu Weiß et al. (2004) as they cluster together with 100 % bootstrap support (Fig. 5.2a, b).
Maximum Likelihood tree of Sebacinales associated with Ericaceae and Orchidaceae inferred from ITS and 28S data. (a) Upper part of the tree, (b) lower part of the tree. Maximum Likelihood analysis was performed with RAxML and 500 rapid bootstrap replicates. The tree was midpoint rooted. Sequences marked in bold are from the present data set. The labels correspond to Genbank number | host species | host family | sampling locality. The remaining sequences are from Genbank and the labels indicate Genbank number and fungal species (if provided on NCBI) | host species (if provided on NCBI) | host plant family. Roman numbers indicate clade names
3.3 UNIFRAC Cluster and PBC-Neighbor-Net Analyses
The Unifrac clustering (UC) of host individuals based on phylogenetic distances of their associates (Sebacinales) shows two major clades and one isolated sequence (Sphyrospermum cordifolium—Fig. 5.3). Clade 2 has 83 % jackknife support, clade 1 is not supported. Clade 2 contains all hosts of Orchidaceae with the exception of Orchid sp. 12 and Orchid sp. 2 that fall in a supported subclade (subclade 2) of clade 1. In clade 2 there are three subclades (# 3, 4, 5) that each consist of epiphytic as well as terrestrial Orchidaceae from different sampling localities (Table 5.1). The positions of the species of Ericaceae are mainly unsupported, with the exception of the two terrestrial species, Gaultheria erecta and Gaultheria reticulata that form a subclade (# 2) with moderate support (72 %) (Fig. 5.3).
UPGMA cluster of Ericaceae and Orchidaceae associated with Sebacinales. Analysis was performed with FastUnifrac and 1,000 jackknife replicates. Labels indicate names of host species, if known. For more information about hosts, please see Table 5.1
The PBC-Neighbor-Net analysis (PBC) also shows a general division of Sebacinales from Ericaceae and Orchidaceae and the distinction of clade 2 from clade 1 (Fig. 5.4). The delta score of the PBC-Neighbor-Net is 0.31, indicating rather high treelikeness of the network. The unsupported clade 1 from the UC analysis appear also in the PBC analysis but large box-like structures between the hosts indicate a high amount of reticulation between host species of clade 1. All subclades in the UC analysis appear also in the PBC analysis (Figs. 5.3 and 5.4). The orchid species of sublcade 2 and the isolated ericad species S. cordifolium are intersectional species as they are in between ericad and orchid species in the PBC network (Fig. 5.4). Orchid species of subclade 2 are closer to ericad species than to orchid species, whereas S. cordifolium is in between both clades (# 1 and 2).
PBC-Neighbor-Net Analysis of Ericaceae and Orchidaceae associated with Sebacinales. Analysis was performed with PBC and Neighbor-Net construction with Splitstree. Delta value is 0.31. Labels indicate names of host species, if known. Large box-like structures indicate reticulation between respective hosts. For more information about hosts, please see Table 5.1
4 Discussion
4.1 Distinct guilds of Sebacinales in Neotropical Ericaceae and Orchidaceae
Comparative investigations of mycobionts of Ericaceae and Orchidaceae were carried out over several years (2003–2009) in the tropical mountain rain forest of the Northern Andes in South Ecuador. We showed that these two plant families harbor Sebacinales as mycobionts. The involved Sebacinales appeared to be closely related but are probably not identical species (Kottke et al. 2008; Suárez et al. 2008). This behavior distinguishes Sebacinales from other fungal orders, since Sebacinales are the only order that contains fungi forming mycorrhiza with chlorophyllous species of both host families (Taylor et al. 2003). Data of the 28S rDNA (Kottke et al. 2008; Suárez et al. 2008) and 5.8S-ITS2 (Suárez et al. 2008) were used in previous investigations to compare Sebacinales mycobionts of epiphytic Orchidaceae and mostly hemiepiphytic Ericaceae. We found that the investigated Orchidaceae and Ericaceae were associated with distinct guilds of Sebacinales, inferred from the absence of well-supported clades on a lower taxonomic level. The overall support values of the 28S rDNA and 5.8S-ITS2 analyses were moderate, with good support values only for some ericad and orchid clades (Kottke et al. 2008; Suárez et al. 2008). The phylogenetic relationships of Sebacinales in the unsupported groups were unclear (Kottke et al. 2008; Suárez et al. 2008).
The present study is more extensive, including Sebacinales associated with epiphytic and terrestrial Orchidaceae and terrestrial and hemiepiphytic Ericaceae from the pristine forest and disturbed habitats. Furthermore, we sequenced ITS (ITS1-5.8S-ITS2) and the D1/D2 region of the 28S rDNA in order to infer phylogenetic relationships of Sebacinales based on a combined dataset and to test the hypothesis that Ericaceae and Orchidaceae are associated with distinct guilds of Sebacinales.
The results of the ML (Fig. 5.2a, b), UN (Fig. 5.3), and PBC analyses (Fig. 5.4) presented here give further indication that the coexisting Ericaceae and Orchidaceae in the tropical mountain rain forest in Southern Ecuador form mycorrhizae with distinct guilds of Sebacinales. The ML analysis shows that Sebacinales in Ericaceae and Orchidaceae form seven distinct clades, which are all well supported (Fig. 5.2a, b). None of the seven clades contain Sebacinales sequences from both host families. The orchid clades (# II, III, IV, V, VII) contain closely related Sebacinales associated with epiphytic as well as terrestrial hosts and from different sites. The same is true for Sebacinales associated with Ericaceae (clades I, VI). This suggests that the association with either Ericaceae or Orchidaceae is more important than the host’s life-form or the sampling site of Sebacinales in the study area.
Ericaceae and Orchidaceae have different mycorrhizal types (Smith and Read 2008), with Orchidaceae forming orchid mycorrhizae (Fig. 5.1d, Burgeff 1909) and Ericaceae forming either monotropoid (Duddridge and Read 1982), arbutoid (Molinia and Trappe 1982), ericoid (Fig. 5.1a, Pearson and Read 1973; Read 1996), or cavendishioid (Fig. 5.1c; Setaro et al. 2006a, b) mycorrhizae. In contrast to orchid mycorrhiza, ericoid and cavendishioid mycorrhizae can be formed by the same Sebacinales phylotype, indicating that the mycorrhizal features are induced by signals from the plant rather than the fungi (Kottke et al. 2008). The finding that the same fungi are able to form ericoid and cavendishioid mycorrhizae corresponds with the similarity of both mycorrhizal associations (Setaro et al. 2006a, b). The root anatomy of both ericoid and cavendishioid mycorrhizae display a small stele, the endodermis, and one further cell layer, the epidermis (identified by some authors as epidermis e.g., Peterson and Massicotte 2004 and by others as cortical layer e.g., Bonfante-Fasolo and Gianinazzi-Pearson 1979). The host plants forming ericoid and cavendishioid mycorrhizae are closely related and the associated fungi in both mycorrhizal associations are Sebacinales and ascomycetes, mainly Helotiales (Allen et al. 2003; Hambleton and Sigler 2005; Setaro et al. 2006a, b). However, the UN and PBC analysis both indicate that ericoid and cavendishioid mycorrhizal Ericaceae in this study are associated with distinct fungal guilds, although the fungi are closely related (Figs. 5.3 and 5.4).
Orchid mycorrhiza is distinct from ericoid and cavendishioid mycorrhizae and also from other mycorrhizal associations regarding the anatomy of the mycorrhizal interaction and the fungal taxonomy (Taylor et al. 2003). Pelotons are formed in the cortical cells of the orchid root that are digested by the plant shortly after their formation (Smith and Read 2008). In addition to Sebacinales, basidiomycetes belonging to Tulasnellales (Warcup 1981; Suárez et al. 2006), Ceratobasidiales (Warcup 1981) and Atractiellomycetes (Kottke et al. 2010) are orchid-mycorrhiza-forming fungi. The differences between orchid mycorrhiza and ericoid/cavendishioid mycorrhiza corresponds to the finding that Neotropical Orchidaceae and Ericaceae are associated with distinct Sebacinales.
4.2 Phylogenetic Position and Biogeography of Sebacinales Associated with Neotropical Ericaceae and Orchidaceae
In general, Sebacinales have a broad mycorrhizal potential as exemplified in the introduction, but include also species of supposedly endophytic lifestyle. In our present phylogenetic analysis, we included all currently available sequences for Sebacinales that contain ITS data and that were at least 400 base pairs long. Many of these Sebacinales were isolated from roots of herbaceous plants for which an endophytic lifestyle is suggested (Selosse et al. 2009) but with no indication of mutualisms so far. The ML analysis (Fig. 5.2a,b) shows that Neotropical Sebacinales do not have a monophyletic origin and have close relatives in the Northern hemisphere (e.g., FM997953—clade I, EU910901—clade IV, FM997954—clade VI) and Australia (EU625991—clade III). Close relatives of the Neotropical Sebacinales are species belonging to the Sebacina vermifera complex. This corroborates previous studies of Sebacinales that were based on a different taxon sampling and on 28S rDNA data (Weiß et al. 2004; Setaro et al. 2006a; Selosse et al. 2007; Suárez et al. 2008; Kottke et al. 2008).
The phylogenetic relationships of the seven clades of Sebacinales show that orchid clades II, III, and IV are more closely related to ericad clade I than to orchid clades V and VII (Fig. 5.2a, b) and thus indicate that the mycorrhizal association with Neotropical Ericaceae and Orchidaceae evolved several times in Sebacinales. In the close relationship of clades I, II, III, IV, and V are relatively few sequences from other places besides the Neotropics (Fig. 5.2a, b). However, clades VI and VII are closely related to Sebacinales associated with many different hosts (grasses, trees, Fabaceae, Ericaceae, and Orchidaceae) from several regions of the world. This indicates that Sebacinales of clades I, II, III, IV and V have a different evolutionary history than those of clade VI and VII (Fig. 5.2a, b). Clade I and II have relatively short branch lengths indicating that these clades are rather young, which corresponds to the relatively recent uplift of the Andes (Gregory-Wodzicki 2000) and the subsequent diversification of Orchidaceae (Gustafsson et al. 2010) and Neotropical Ericaceae due to adaptive radiation (Luteyn 2002; Kron and Luteyn 2005). However, shorter branch lengths could also be the result of lower substitution rates (Takezaki et al. 1995). In general, Sebacinales are considered as rather old because of their relatively basal position in the Agaricomycotina (Weiß et al. 2004; Hibbett and Matheny 2009), but so far there are no detailed studies about age estimation in Sebacinales.
Knowledge of the ecology of the ancestors of Sebacinales from Neotropical Ericaceae and Orchidaceae would be important to elucidate whether the different mycorrhizal states evolved from endophytic/saprophytic or mycorrhizal precursors. Furthermore, the ancestors of Sebacinales associated with Neotropical Ericaceae and Orchidaceae could have been introduced to the area together with their host plants or could have been adapted from the local Sebacinales community. Hibbet and Matheny (2009) performed Bayesian relaxed molecular clock analyses in order to infer the ecological lifestyle of the ancestors of ectomycorrhizal basidiomycetes with the result that the ectomycorrhizal state in basidiomycetes evolved independently several times from saprophytic ancestors (Hibbett and Matheny 2009). This approach is also conceivable for Neotropical Sebacinales in the future but a more extensive sampling of Sebacinales from other hosts, especially herbaceous plants, or soil of the same area would be crucial.
4.3 Suitability of Unifrac and PBC-Neighbor-Net Analyses for Ecological Analyses of Sebacinales
Most of the Sebacinales lack distinguishing morphological characters to identify them to species level (Weiß et al. 2004). Therefore, conventional ecological approaches based on species entities (e.g., cluster analysis and ordination techniques—Legendre and Legendre 1998) are not feasible to characterize communities of Sebacinales. The Unifrac and PBC approaches are a useful alternative, since they both use phylogenetic data without the need to delimitate fungal sequences to species or molecular operational taxonomic units (Lozupone and Knight 2005; Göker and Grimm 2008). Especially the Unifrac approach has been used for a variety of organisms for which the same species delimitation problems exist as for Sebacinales (e.g., Schloss 2008; Chu et al. 2010; Dimitriu and Grayston 2010; Doherty et al. 2010). The Unifrac metric measures the difference between two environments (in our case host species) in terms of the branch length that is unique to one host or the other (Lozupone and Knight 2005). The PBC approach was published in 2008 (Göker and Grimm 2008) and used to detect hybrids in Acer spp. and also discussed in the context of a broad applicability to ecological questions where “host” and “associate” data are available (Göker and Grimm 2008). This novel approach transforms phylogenetic distance matrices of the “associates” to distance matrices of the “host” using Sørensen’s distance coefficient and determines the most similar specimens between two hosts automatically when computing pair-wise distances from the reduced matrix (Göker and Grimm 2008). Subsequently, phylogenetic analyses like Neighbor-Net analysis allows visualization of the transformed distance matrix and Delta values can be computed to statistically evaluate phylogenetic accuracy (Göker and Grimm 2008). For Sebacinales, this is the first study that uses the PBC approach to investigate fungal communities in different host species.
Both analyses showed the general division of Sebacinales associated with Ericaceae or Orchidaceae since almost all orchid species sampled from the study site group together in a well-supported clade in the UN cluster analysis (Fig. 5.3) and appear together in the PBC-Neighbor-Net analysis (Fig. 5.4). Clade 1 has no jackknife support in the Unifrac (Fig. 5.3) and a relatively high occurrence of reticulation in the PBC analysis (Fig. 5.4). The jackknife statistic implemented in FastUnifrac indicates whether the result of the analysis is robust in terms of sample size and evenness (Hamady et al. 2010). Thus, the missing support values for most of the Ericaceae indicate that species of Ericaceae are associated with a more diverse community of Sebacinales than are Orchidaceae.
Findings from both ecological analyses, UN (Fig. 5.3) and PBC (Fig. 5.4), are congruent and correspond with results from the phylogenetic ML analysis of the associated Sebacinales (Fig. 5.2a, b). The results show that both analyses performed well. The UN approach has the advantage that it is already well established in ecology as exemplified above, and resampling statistics that support the outcome of the analyses are integrated in the program (Lozupone and Knight 2005; Hamady et al. 2010). However, the PBC approach gives the user more freedom how to analyze and visualize the data so that even networks can be computed (Göker and Grimm 2008). Networks have the advantage that data can be displayed that are not necessarily treelike (Huson and Bryant 2006), what might be the case for many host species and was true for most of the Ericaceae in this study. We think that both approaches are a suitable tool for dealing with ecological questions in Sebacinales, especially when many hosts and localities are to be investigated.
References
Allen T, Millar T, Berch S, Berbee M (2003) Culturing and direct DNA extraction find different fungi from the same ericoid mycorrhizal roots. New Phytol 174:864–878
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Auch A, Henz S, Holland B, Göker M (2006) Genome BLAST distance phylogenies inferred from whole plastid and whole mitochondrion genome sequences. BMC Bioinformatics 7:350
Bauer R, Begerow D, Sampaio JP, Weiß M, Oberwinkler F (2006) The simple-septate basidiomycetes: a synopsis. Mycol Progr 5:41–66
Beck E, Bendix J, Kottke I, Makeschin F, Mosandl R (2008) Gradients in a tropical mountain ecosystem of Ecuador. Ecological Studies 198. Springer
Burgeff H (1909) Die Wurzelpilze der Orchideen. Gustav Fischer, Jena, Germany
Bonfante-Fasolo P, Gianinazzi-Pearson V (1979) Ultrastructural aspects of Endomycorrhiza in the Ericaceae I. Naturally infected hair roots of Calluna vulgaris L. Hull. New Phytol 83:739–744
Capella-Gutiérrez S, Silla-Martinez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Chu H, Fierer N, Lauber C, Caporaso J, Knight R, Grogan P (2010) Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol. doi:10.1111/j.1462-2920.2010.02277.x
Cullings K (1994) Molecular phylogeny of the Monotropoideae (Ericaceae) with a note on the placement of the Pyroloideae. J Evolut Biol 7:501–516
Dimitriu PA, Grayston SJ (2010) Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and natural boreal forest soils. Microb Ecol 59:563–573
Doherty M, Tamura M, Costas BA, Ritchie ME, McManus GB, Katz LA (2010) Ciliate diversity and distribution across an environmental and depth gradient in Long Island Sound, USA. Environ Microbiol 12:886–898
Duddridge JA, Read D (1982) An ultrastructural analysis of the development of mycorrhizas in Monotropa hypopitys L. New Phytol 92:203–214
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Gargas A, Taylor JW (1992) Polymerase Chain Reaction (PCR) primers for amplifying and sequencing nuclear 18S rDNA from lichenized fungi. Mycologia 84:589–592
Glen M, Tommercup IC, Bougher NL, O’Brien PA (2002) Are Sebacinaceae common and widespread ectomycorrthizal associates of Eucalyptus species in Australian forests? Mycorrhiza 12:243–247
Göker M, Grimm G (2008) General functions to transform associate data to host data, and their use in phylogenetic inference from sequences with intra-individual variability. BMC Evol Biol 8:86
Göker M, García-Blázquez G, Voglmayr H (2009) Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One 4:e6319
Grasso C, Lee C (2004) Combining partial order alignment and progressive multiple sequence alignment increases alignment speed and scalability to very large alignment problems. Bioinformatics 20:1546–1556
Gregory-Wodzicki K (2000) Uplift history of the Central and Northern Andes: a review. GSA Bulletin 112:1091–1105
Gustafsson AL, Verola CF, Antonelli A (2010) Reassessing the temporal evolution of orchids with new fossils and a Bayesian relaxed clock, with implications for the diversification of the rare South American genus Hoffmannseggella (Orchidaceae: Epidendroideae). BMC Evol Biol 10:177
Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27
Hambleton S, Sigler L (2005) Meliniomyces, a new anamorph genus for root-associated fungi with phylogenetic affinities to Rhizoscyphus ericae (≡ Hymenoscyphus ericae), Leotiomycetes. Stud Mycol 53:1–27
Hibbett D, Matheny P (2009) The relative ages of ectomycorrhizal mushrooms and their plant hosts estimated using Bayesian relaxed molecular clock analyses. BMC Biol 7:13
Holland B, Huber K, Dress A, Moulton V (2002) ∂ Plots: a tool for analyzing phylogenetic distance data. Mol Biol Evol 19:2051
Hopple J, Vilgalys R (1994) Phylogenetic relationships among coprinoid taxa and allies based on data from restriction site mapping of nuclear rDNA. Mycologia 86:96–107
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Iotti M, Zambonelli A (2006) A quick and precise technique for identifying ectomycorrhizas by PCR. Mycol Res 110:60–65
Johnson LA, Soltis DE (1995) Phylogenetic inference in Saxifragaceae sensu stricto and Gilia (Polemoniaceae) using matK sequences. Ann Missouri Bot Gard 82:149–175
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kottke I, Beiter A, Weiß M, Haug I, Oberwinkler F, Nebel M (2003) Heterobasidiomycetes form symbiotic associations with hepatics: Jungermanniales have sebacinoid mycobionts while Aneura pinguis (Metzgeriales) is associated with a Tulasnella species. Mycol Res 107:957–968
Kottke I, Haug I, Setaro S, Suárez J, Weiß M, Preussing M, Nebel M, Oberwinkler F (2008) Guilds of mycorrhizal fungi and their relation to trees, ericads, orchids and liverworts in a neotropical mountain rain forest. Basic Appl Ecol 9:13–23
Kottke I, Suárez JP (2009) Mutualistic, root-inhabiting fungi of orchids – identification and functional types. In: Pridgeon AM, Suárez JP (eds) Proceedings of the Second Scientific Conference on Andean Orchids. Universidad Técnica Particular de Loja, Loja, Ecuador, pp 84–99. ISBN 978-9942-00-502-1
Kottke I, Suárez J, Herrera P, Cruz D, Bauer R, Haug I, Garnica S (2010) Atractiellomycetes belonging to the ‘rust’ lineage (Pucciniomycotina) form mycorrhizae with terrestrial and epiphytic neotropical orchids. Proc R Soc B 277:1289–1298
Kron KA, Judd WS, Stevens PF, Crayn DM, Anderberg AA, Gadek PA, Quinn CJ, Luteyn JL (2002) Phylogenetic classification of Ericaceae: molecular and morphological evidence. Bot Rev 68:335–423
Kron KA, Luteyn JL (2005) Origins and biogeographic patterns in Ericaceae: new insights from recent phylogenetic analyses. Biol Skr 55:479–500
Krüger M, Stockinger H, Krüger C, Schüßler A (2009) DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol 183:212–223
Legendre P, Legendre L (1998) Numerical ecology, 2nd edn. Elsevier Science B. V, Amsterdam
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Luteyn JL (1996) Ericaceae. In: Flora of Ecuador, Vol. 54. Department of Systematic Botany, University of Göteborg, Stockholm, Sweden, pp 1–404
Luteyn JL (2002) Diversity, Adaptation, and Endemism in Neotropical Ericaceae: Biogeographical Patterns in the Vaccinieae. Bot Rev 68:55–87
Molinia R, Trappe JM (1982) Lack of mycorrhizal specificity by the ericaceous hosts Arbutus menziesii and Arctostaphylos uva-ursi. New Phytol 90:495–509
Newsham KK, Bridge PB (2010) Sebacinales are associates of the leafy liverwort Lophozia excisa in the southern maritime Antarctic. Mycorrhiza 20:307–313
Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH (2008) Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinform 4:193–201
Oberwinkler F (1964) Intrahymeniale Heterobasidiomyceten. Fruchtkörperlose Sebacina-Sippen und ihre systematische Stellung. Nova Hedwigia 7:489–498
O’Donnell K (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Washington, DC, USA, pp 225–233
Pearson V, Read DJ (1973) The biology of mycorrhiza in the Ericaceae: I. The isolation of the endophyte and synthesis of mycorrhizas in aseptic culture. New Phytol 72:371–379
Peterson RL, Massicotte HB (2004) Exploring structural definitions of mycorrhizas, with emphasis on nutrient-exchange interfaces. Can J Bot 82:1074–1088
Pridgeon AM, Solano R, Chase MW (2001) Phylogenetic relationships in Pleurothallidinae (Orchidaceae): combined evidence from nuclear and plastid DNA sequences. Am J Bot 88:2286–2308
Rambaut A (2009) FigTree Tree Figure Drawing Tool Version 1.3.1. http://tree.bio.ed.ac.uk/
Read DJ (1996) The structure and function of the Ericoid mycorrhizal root. Ann Bot 77:365–374
Schloss P (2008) Evaluating different approaches that test whether microbial communities have the same structure. ISME J 2:265–275
Selosse MA, Bauer R, Moyersoen B (2002) Basal hymenomycetes belonging to the Sebacinaceae are ectomycorrhizal on temperate deciduous trees. New Phytol 155:183–195
Selosse MA, Dubois MP, Alvarez N (2009) Do Sebacinales commonly associate with plant roots as endophytes? Mycol Res 113:1062–1069
Selosse MA, Setaro S, Glatard F, Richard F, Urcelay C, Weiß M (2007) Sebacinales are common mycorrhizal associates of Ericaceae. New Phytol 174:864–878
Setaro S, Weiß M, Oberwinkler F, Kottke I (2006a) Sebacinales form ectendomycorrhizae with Cavendishia nobilis, a member of the Andean clade of Ericaceae, in the mountain rain forest of southern Ecuador. New Phytol 169:355–365
Setaro S, Kottke I, Oberwinkler F (2006b) Anatomy and ultrastructure of mycorrhizal associations of neotropical Ericaceae. Mycol Prog 5:243–254
Smith SE, Read DJ (2008) Mycorrhizal symbiosis, 3rd edn. Elsevier, Great Britain
Stamatakis A (2006) RaxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stamatakis A, Hoover P, Rougemont J (2008) A rapid Bootstrap algorithm for the RAxML Web Servers. Syst Biol 57:758
Stockinger H, Walker C, Schüßler A (2009) 'Glomus intraradices DAOM197198', a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol 183:1176–1187
Suárez JP, Weiß M, Abele A, Garnica S, Oberwinkler F, Kottke I (2006) Diverse tulasnelloid fungi form mycorrhizas with epiphytic orchids in an Andean cloud forest. Mycol Res 110:1257–1270
Suárez JP, Weiß M, Abele A, Oberwinkler F, Kottke I (2008) Members of Sebacinales subgroup B form mycorrhizae with epiphytic orchids in a neotropical mountain rain forest. Mycol Prog 7:75–85
Swofford DL (2002) Paup* Phylogenetic Analysis Using Parsimony (* and other Methods). Version 4. Sinauer Associates, Sunerland, Massachusetts
Takezaki N, Rzhetsky A, Nei M (1995) Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol 12:823–833
Taylor DL, Bruns TD, Leaky JR, Read DJ (2003) Mycorrhizal specificity and function in Myco-heterotrophic plants. In: Heijden MGA, Sanders IR (eds) Mycorrhizal ecology. Springer, Berlin, pp 375–414
Urban A, Weiß M, Bauer R (2003) Ectomycorrhizae involving sebacinoid mycobionts. Mycol Res 107:3–14
Warcup J, Talbot P (1967) Perfect states of rhizoctonias associated with orchids. New Phytol 66:631–641
Warcup J (1981) The mycorrhizal relationships of Australian orchids. New Phytol 87:371–381
Warcup JH (1988) Mycorrhizal associations of isolates of Sebacina vermifera. New Phytol 110:227–231
Weiß M, Selosse MA, Rexer KH, Urban A, Oberwinkler F (2004) Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential. Mycol Res 108:1003–1010
White TJ, Bruns TD, Lee SB, Taylor JW (1990) Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal RNA genes. In: PCR protocols: a guide to method and applications, pp 315–322
Whitten WM, Williams NH, Chase MW (2000) Subtribal and generic relationships of Maxillarieae (Orchidaceae) with emphasis on Stanhopeinae: combined molecular evidence. Am J Bot 87:1842–1856
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Setaro, S., Suárez, J.P., Herrera, P., Cruz, D., Kottke, I. (2013). Distinct but Closely Related Sebacinales form Mycorrhizae with Coexisting Ericaceae and Orchidaceae in a Neotropical Mountain Area. In: Varma, A., Kost, G., Oelmüller, R. (eds) Piriformospora indica. Soil Biology, vol 33. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-33802-1_5
Download citation
DOI: https://doi.org/10.1007/978-3-642-33802-1_5
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-33801-4
Online ISBN: 978-3-642-33802-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)