Abstract
Extracting relations from unstructured Web content is a challenging task and for any new relation a significant effort is required to design, train and tune the extraction models. In this work, we investigate how to obtain suitable results for relation extraction with modest human efforts, relying on a dynamic active learning approach. We propose a method to reliably generate high quality training/test data for relation extraction - for any generic user-demonstrated relation, starting from a few user provided examples and extracting valuable samples from unstructured and unlabeled Web content. To this extent we propose a strategy which learns how to identify the best order to human-annotate data, maximizing learning performance early in the process. We demonstrate the viability of the approach (i) against state of the art datasets for relation extraction as well as (ii) a real case study identifying text expressing a causal relation between a drug and an adverse reaction from user generated Web content.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
1 Introduction
Recent years have seen the rise of neural networks for addressing many Information Extraction tasks. Particular interest is focused on Relation Extraction from unstructured text content. While crafting the right model architecture has gained significant attention, a major and often overlooked challenge is the acquisition of solid training data and reliable gold standard datasets for validation.
Kick-starting a relation extraction process - i.e. acquiring reliable training and testing data - for an arbitrary user-defined relation presents many hurdles. This is especially true when the pool of unannotated data to choose from is virtually infinite, as in the case of Web data and social streams - where one first needs to identify a relevant corpus from which relations should be extracted. Systems extracting relations from open data have been described in the literature. Although they may perform well, they are in general quite expensive: on one hand, supervised methods require an upfront annotation effort for each relation, while on the other hand unsupervised methods present many drawbacks - mainly the reliance on Natural Language Processing (NLP) tools, which might not be available for all languages and which do not perform that well on the ungrammatical text often found in social posts. Moreover for any approach it is desirable to evaluate the performance: this implies the availability of test data, which in general involves an expensive manual annotation process.
In this paper, we address these challenges by presenting a system for extracting relations from unlabeled data which minimizes the required annotations, which needs no NLP tools and performs well with respect to the available state of the art methods. These relations can be fairly well defined (e.g. given a color and an object, does the author imply the object is that color) to somewhat more subjective ones (e.g. detecting asserted causal relations between drugs and adverse events). Here relation detection is defined in a standard way, i.e. determining if a relation is present in a text sample or not, or in Relation Extraction terms, the goal is to recognize whether a predefined set of relations holds between two or more entities in a sentence. We propose an end-to-end system for extracting relations from unstructured Web content. First, the type of entities involved in the relation, e.g. drugs and adverse events, must be specified - this step can be seen as a blackbox component here. Then we obtain a relevant pool of potential examples of the relations from the Web and social media by selecting sentences where the entities co-occur. We discard parts of the corpus which seems to contain highly ambiguous data, while retaining useful data for the task (Sect. 3).
After collecting relevant unlabeled data, we ask a Subject Matter Expert (SME) to annotate the data in small batches (e.g. 100 examples at a time). The selection of examples that are presented to the SME is dependent on the learning model and the active learning strategy which in itself is dependent on the type of relation and data at hand. In this paper, we show how for a given model the system “learns” a quite successful strategy (in general it is not feasible to determine an “optimal” strategy with reasonable effort): we measure the performance of several neural models and several active learning strategies at the end of each batch, devising a method to only promote the most successful strategies for subsequent steps (Sect. 3.2). By optimizing the order of examples to annotate, the work required by the SME is much less than manually creating labeled training data [29] or building/tuning NLP tools for different languages and styles. We show this with several experiments on standard benchmarking datasets.
The contribution of this work is threefold. First, we propose an end-to-end method for relation extraction with a human-in-the-loop. We design a systematic procedure for generating datasets for relation extraction on any domain and any concept that the user is interested in. This is valuable to kick-start arbitrary extraction tasks for which annotated resources are not yet available. Our method does not have to rely on any NLP tools and hence is independent of document style and language. Second, we experiment using a combination of active learning strategies on neural models and devise a method to prune the ones that are not effective for the task at hand. Besides testing the approach on a real use case, we prove its efficacy on publicly available standard datasets for relation extraction and show that by using our pruning technique - and observing the results a posteriori - we achieve similar performance to the “optimal active learning strategy” for the task and the specific dataset. In addition, as one does not know a priori what the optimal strategy is, our system learns which strategy among the available ones to use. The technique works comparably well regardless of the chosen neural architecture. Finally, we present a real use case scenario where we address the challenging task of extracting the causal relation between drugs and their adverse events from user generated content.
The advantage of the proposed approach is the possibility to rapidly deploy a system able to quickly generate high quality train/test data on any relation of interest, regardless of language and text style of the corpus. Given the fact that the method gives feedback on performance after every small annotation step, the user can decide when to stop annotating when she is satisfied with the level of accuracy (e.g. accuracy above 75%) or decide to stop if she understands that the underlying data might not be useful for the task at hand. Substantially, we are able to early identify high quality train/test data for challenging relation extraction tasks while minimizing the user annotation effort.
2 Related Work
One of the key to success for machine learning tasks is the availability of high quality annotated data, which is often costly to acquire. For the relation extraction task, the definition of a relation is highly dependent on the task at hand and on the view of the user, therefore having pre-annotated data available for any specific case is unfeasible. Various approaches have been proposed to minimize the cost of obtaining labelled data, one of the most prominent being distant supervision, which exploits large knowledge bases to automatically label entities in text [5, 12, 16, 26, 27]. Despite being a powerful technique, distant supervision has many drawbacks including poor coverage for tail entities [16], as well as the broad assumption that when two entities co-occur, a certain relation is expressed in the sentence [5]. The latter can be especially misleading for unusual relations, where the entities might co-occur but not fit the desired semantic (e.g. a user wants to classify “positive” or desirable side effects of drugs). One way to tackle the problem is to use targeted human annotations to expand the large pool of examples labelled with distant supervision [3]. This combination approach produced good results in the 2013 KBP English Slot Filling taskFootnote 1. Another way is to address it as a noise reduction problem: e.g. Sterckx et al. [30] exploit hierarchical clustering of the distantly annotated samples to select the most reliable ones, while Fu and Grishman [10] propose to interleave self-training with co-testing. Nonetheless, it is nearly impossible to refrain from manual annotation entirely: at the very least test data (that serves as gold standard) needs to be annotated manually. The question then is how to minimize the human annotation effort.
Active Learning (AL) aims at incorporating targeted human annotations in the process: the learning strategy interactively asks the user to annotate certain specific data points, using several criteria to identify the best data to annotate next. Some of the most commonly used criteria are: (i) uncertainty sampling, which ranks the samples according to the model’s belief it will mislabel them [18]; (ii) density weighted uncertainty sampling, which clusters the unlabeled instances to pick examples that the model is uncertain for, but also are “representative” of the underlying distribution [9, 23]; (iii) QUIRE, which measures each instance’s informativeness and representativeness by its prediction uncertainty [15]; (iv) Bayesian methods such as bald (Bayesian Active Learning by Disagreement) which select examples that maximize the models’s information gain [11]. The effectiveness of these criteria is highly dependent on the underlying data and the relation to extract and it is very difficult to identify strong connections between any of the criteria and the task [14]. The open question is then how to decide which technique to use on a new extraction task. Following [14] we argue that it is best to dynamically decide on the criteria on a task-driven basis. The “active learning by learning” method (albl) [14] has an initial phase where all criteria are tested extensively and one is chosen. Our intuition is that the technique that seems to perform the best at the beginning might not be best one in the long run. Therefore we propose a method that initially distributes the budget of annotation among all considered criteria and discards the worst performing one at each iteration. We argue that keeping a pool of options for a longer number of iterations will maximize performance on average for a larger number of tasks, especially given the very small sample set, and we support the claim with comparative experiments.
For the sake of completeness, it is worth mentioning that in relation extraction, as in many other machine learning tasks, there is no one-fits-all model and many have been proposed ranging from early solutions based on SVMs and tree kernels [7, 8, 20, 32, 34] to most recent ones exploiting neural architectures [24, 31, 33]. Neither the model nor the active learning strategy or any particular combination is universally (on all relations/all data) “the best” performer - hence our proposal of a data driven approach. The aim of this work is to investigate the influence of different active learning strategies on different extraction tasks (regardless of the underlying neural model) and to devise strategies to effectively annotate data, rather than proposing a new neural architecture per-se. Therefore for our experiments we considered several state of the art deep learning models for relation classification, including Convolutional Neural Networks (CNNs) [24, 33], Recurrent Neural Networks (RNNs) [19], such as bi-directional GRUs [35], as well as ensembles [31]. For all models, we do not require any NLP preprocessing (besides tokenization) of the text.
3 Relation Classification
We consider relation extraction as a binary classification task. Given a text snippet s containing one or more target entities \(e_i\)Footnote 2 our goal is to identify if s expresses a certain relation r among the entities \(e_i\). Our goal is two-fold: (i) create a relation classification system that gradually increases accuracy from each recognized relation, as well as (ii) identifying the sentence snippets for which the system is most/least confident about expressing the desired relation. We first obtain a large pool of relevant unlabeled text from a given social media stream (e.g. the Twitter stream, a social forum etc.), applying the following method. We consider the (two) types of entities involved in the relation, for which we construct dictionaries using any off-the-shelf tool (e.g. [2]) and select sentences where the (two) entities co-occur. Note that this will produce lot of noisy data, therefore noise reduction needs to be in place. For this work we treat entity identification in sentences as a blackbox component with various valid available solutions [16, 26, 30].
We then segment the learning process into small steps of b examples at a time (\(b=100\) in this workFootnote 3) and interactively annotate the data as we train the models. Example refers here to a text snippet expressing the relation between the entities and annotation refers to manually assigning a “true/false” label to each example. We select the first batch of b examples with a curriculum learning strategy (details in Sect. 3.2) and manually annotate them. With those we train (i) several neural models, using (ii) two different data representation paradigms and (iii) several active learning strategies to determine the next batch of examples. Our goal is not to specifically improve a particular learning model per-se, but rather (i) identify at an early stage, i.e. with minimal annotation effort, if a specific relation can be learned from the available data and (ii) minimize the labelling effort by using first examples that are more likely to boost the learning performance. As no active learning strategy is universally preferable (we show tests on ready-available gold standard datasets for relation extraction in Sect. 4.2) we propose a pruning method (Sect. 3.2) that dynamically selects the best strategy for a given task.
3.1 Models, Data Representations and Parameter Choices
We employ commonly used neural models for relation extraction, specifically CNNs [24, 33] and bi-directional GRUs [35].
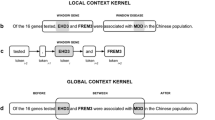
As for data representation, we do not rely on lexical features or any other language-dependent information, but after using a simple tokenizer (white spaces, punctuation) we merely exploit distributional semantics - statistical properties of the text data - to ensure portability to different languages, domains and relation types. We explore two different representations for the text: (i) word sequences concatenated with positional features (as in [33]), i.e. we generate three embedding matrices, one initialized with pre-trained word embeddings and two randomly initialized for the positional features; (ii) a context-wise split of the sentence (as in [1]), i.e. using pre-trained word embeddings and using the two entities in the text as split points to generate three matrices - left, middle and right context.
As for the neural network architectures specifications, following literature, all our models use: 100-dimensional pre-trained GloVe word embeddings [25]; 100-dimensional positional embeddings optimized with Adam [17]; initial learning rate = 0.001; batch size b = 100; validation split = 0.2; early stopping [21] to avoid overfitting (if no improvement happens for 5 consecutive iterations). For the CNNs we use: 100 filters; kernels width = 3; ReLU nonlinearities [22] - for CNNs with multiple filter sizes we set the kernels width from 2 to 5. For the GRU we use: ReLU activations and layer size = 100.
3.2 Active Learning by Pruning
At the bootstrapping phase, we have no information on the performance of each model as all data is unlabeled. We used curriculum learning (CL) strategies [6], where the order of the data is decided in advance - before starting the learning process using several text based criteria. While we tested several criteria, including random as baseline, the best performance was obtained by maximizing dissimilary. Starting from a random example (sentence) we sort the data as to maximize dissimilarity between the sentences. We calculate sentence similarity exploiting GloVe embeddings as proposed by [4].
For all subsequent steps, we can use previously annotated examples to test the performance of the different active learning strategies. We consider a pool-based active learning scenario [28] in which there exists a small set of labeled data \(L={(x_1,y_1),\dots ,(x_{n_l},y_{n_l})}\) (in this case we consider the batch of 100 examples selected by CL) and a large pool of unlabeled data \(U={x_1,\dots ,x_{n_u}}\). The task for the active learner is to draw examples to be labeled from U, so as to maximize the performance of the classifier (the neural net) while limiting the number of required annotations to achieve a certain accuracy. We train the model on the first batch of annotated examples, using 5-fold validation on the batch itself. At each subsequent iteration we select \(\frac{b}{n}\) examples according to each of the n target active learning strategies; after labelling those b examples we calculate the performance for each of them and identify the worst performing AL strategy, which gets discarded in subsequent iterations. After n iterations we remain with one strategy for the particular task. In this particular work, we select \(n=5\) active learning strategies: uncertainty sampling (us), density weighted uncertainty sampling (dwus), bayesian active learning by disagreement (bald), QUIRE and we include as baseline the random selection (rs) of examples. The proposed approach is not limited to those - any other strategy can be added without changing the overall framework.
We perform extensive experiments testing all possible combinations of models, data representations and active learning strategies. Results are summarized in Table 2. We show that our proposed “active learning by pruning” strategy is robust across relation extraction tasks and datasets (Sect. 4).
4 Experiments
The relation extraction task is a challenging one. Especially in the case of developing early prototype systems, little can be done with a traditional neural network in the absence of a significant quantity of hand labeled data. While a task specific labeling system can help [29], it makes sense to consider the “best order” to ask the user for input in the hopes of achieving a sufficiently performant system with minimal human effort.
Assuming the existence of a relevant corpus of unlabelled examples for the relation at hand our aim in this work is to identify the best active learning strategy for each extraction task to prioritize the annotation of examples that have a better impact on the models. We exploit existing benchmark datasets on relation extraction and simulate the human-in-the-loop: we treat all examples as unlabelled and “request” the annotations in small batches from the existing labels, as if they were annotated in real-time by a user. This gives us useful insights, as we can compare partial performance (after any given annotation batch) against the best achievable performance (using the whole dataset), as well as run in parallel all active learning strategies to figure out if any of them is “universally” better for all tasks. A post-hoc analysis reveals that in terms of active learning strategy there is no one-fits-all solution (Sect. 4.2) but that our proposed solution is able to promote good performing ones for the task. We test our pruning technique on all the benchmark relations, as well as on our real case scenario on extracting adverse drug events, for a total of 10 different relations (details on the data in Sect. 4.1).
4.1 Datasets
We test our method in a real case experiment, extracting Adverse Drug Events (ADE) relations from a Web forum (http://www.askapatient.com/). Our human-in-the-loop is a medical doctor using our system to annotate the data. We produced annotations in the same style as CADEC (CSIRO Adverse Drug Event Corpus)Footnote 4, totaling of 646 positive and 774 negative examples of causal relationships between drugs and ADEs. We name this dataset causalADEsFootnote 5. Posts are tagged based on mentions of certain drugs, ADEs, symptoms, findings etc. However, the mere co-occurrence of a drug and an ADE in a sentence does not necessarily imply a causal relation among the two. Figure 1 shows three sentences, one where the drug caused an ADE and others where it did not.
We also test our method on the Semeval2010-Task8 dataset [13], which consists of 8,000 training and 2,717 test examples on nine relation types: Cause-Effect, Component-Whole, Content-Container, Entity-Destination, Entity-Origin, Instrument-Agency, Member-Collection, Message-Topic, and Product-Producer.
4.2 Fixed Active Learning Strategy VS Dynamic Selection
The aim of the experiments is to compare all the considered active learning strategies (as used individually) against dynamic selection, either our proposed pruning strategy or the albl method [14]. We ran experiments using various different configurations of neural networks, data representations and active learning strategies. For the sake of reporting clarity we use CNN with positional features to plot results on the different active learning strategies - but we summarize results for different configurations in Table 2. Figure 2 shows the accuracy on the Semeval extraction tasks for all the strategies as a function of the number of labelled examples (Fig. 2): no single AL strategy is always the best, but we can observe that our pruning strategy has a consistent behavior across all tasks, approximating top performance.
In a real case scenario, where all data is unlabeled and we do not have a designated test set, the feedback that we can provide at each step is the performance calculated with cross-validation on the currently annotated data.
Taking a closer look at individual results (Table 2) one can observe that our proposed pruning strategy (i) obtains top performance - with respect to other strategies - with exhaustive annotation, i.e. when all examples are labelled on most tasks (9 out of 10) and (ii) can consistently “near” top performance (with a loss \(\le \)2% in most cases (7 out of 10) with less than half of the annotated data, for some relations as early as after 400 annotations. For completeness we also compare our pruning strategy to albl [14], which is to the best of our knowledge the best performing method for dynamically selecting active learning strategies. Our pruning method outperforms albl (or has same performance) in all the runs, with a maximal increase of 6% (the Entity-Origin task).
A further observation is about the causalADEs dataset, which is our true real-case experiments (while the others are simulated with existing benchmark datasets). The presence of a causal relation between a Drug and an Adverse Drug Event can be very tricky to identify. Table 1 shows examples of correct and incorrect predictions of our models.
Our overarching goal is to be able to identify a pool of examples for which we are highly confident to have been annotated correctly leading to a reliable training/test dataset to train the extraction of a new relation. This is particularly valuable in situations where the unlabeled sample data is particularly large and we can afford to discard examples, as long as the selected ones are of high quality.
In Table 2 we summarize our results and report top performance for each extraction task. Regarding neural architecture we observe that the simple CNN model performed better in most cases, with a preference for the context-wise split data representation. Regarding active learning strategies we compare our method with (i) each considered state-of-the-art fixed AL strategy (ii) as well as with the albl dynamic AL strategy selection. The experimental results show that our pruning method achieves either better or comparable performance with the best performing AL method, and we surpass the best performance of albl in almost all cases. Our method also has a computational advantage with respect to albl. While we train and test in small batches, albl works in a streaming fashion where a micro-training and performance estimation is done after each new example. While this is affordable in their tested settings (using a Support Vector Machine model) it becomes computationally heavy in neural network settingsFootnote 6. Additionally, our performance using only 500 examples is very close to the best accuracy we can potentially achieve (but do not know a priori) in each task, with comparable results to albl with the same number of examples.
Another observation is that when using the whole available training data, the different active learning strategies tend to converge. On the other hand at the first stages of training some strategies might be “slower” in terms of performance gain. We can observe this in Table 2: after using all available data several AL strategies achieve top performance (as reported in column Best AL), while when using only 500 examples (strategies marked in bold in column Best AL) we have less ties. Regarding our pruning method we report which AL strategy is selected (column selection) after the pruning is completed. It is important to note that this is not equivalent to running the selected strategy alone, because the first stages of training include data selected with various techniques, and this contributes to learning a slightly different model than with a single technique.
5 Conclusions and Future Work
Previous literature on relation extraction has been focusing on improving model performance by either developing new architectures, incorporating additional linguistic features or acquiring additional data. We conjecture that in order to be able to capture any domain specific relation, we need to design models that take into account the effect of the data size and type in addition to the computational cost occurring from training under streamed annotations. To this end, we train neural models with minimal data pre-processing, without using any linguistic knowledge and we propose a novel active learning strategy selection technique. We achieve promising performance on various relation extraction tasks. Moreover, we demonstrate that our method is effective for the rapid generation of train/test data for ambiguous relations and we release a novel dataset for the detection of adverse drug reactions in user generated data. In future work, we will investigate pruning strategies, specifically a hierarchical approach which, given the small amount of data, may result in faster convergence, especially when exploring many AL options.
Notes
- 1.
- 2.
In our experiments we use pairs of entities, however we should note that our models can handle n-ary relations as well. We leave this to future work.
- 3.
The size of the batch is adjustable, the human-in-the-loop can specify it. In our experiments, the involved medical doctor indicated 100 as a good size in terms of keeping focus.
- 4.
- 5.
- 6.
On a Linux server with 48 Intel Xeon CPUs @2.20GHz, 231GBs RAM, NVIDIA GeForce GTX 1080 GPU, on causalADE task albl (the libact implementation https://github.com/ntucllab/libact) took 3hrs-10mins, our pruning method took 7 min.
References
Adel, H., Roth, B., Schütze, H.: Comparing convolutional neural networks to traditional models for slot filling. In: NAACL-HLT (2016)
Alba, A., Coden, A., Gentile, A.L., Gruhl, D., Ristoski, P., Welch, S.: Language agnostic dictionary extraction. In: ISWC (ISWC-PD-Industry). CEUR Workshop Proceedings, vol. 1963 (2017)
Angeli, G., Tibshirani, J., Wu, J., Manning, C.D.: Combining distant and partial supervision for relation extraction. In: EMNLP, pp. 1556–1567 (2014)
Arora, S., Liang, Y., Ma, T.: A simple but tough-to-beat baseline for sentence embeddings. In: ICLR (2017)
Augenstein, I., Maynard, D., Ciravegna, F.: Distantly supervised web relation extraction for knowledge base population. Semant. Web 7(4), 335–349 (2016)
Bengio, Y.: Curriculum learning. In: ICML (2009)
Bunescu, R.C., Mooney, R.J.: A shortest path dependency kernel for relation extraction. In: HLT/EMNLP, pp. 724–731. ACL (2005)
Culotta, A., Sorensen, J.: Dependency tree kernels for relation extraction. In: ACL, p. 423. ACL (2004)
Donmez, P., Carbonell, J.G., Bennett, P.N.: Dual strategy active learning. In: Kok, J.N., Koronacki, J., Mantaras, R.L., Matwin, S., Mladenič, D., Skowron, A. (eds.) ECML 2007. LNCS (LNAI), vol. 4701, pp. 116–127. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-74958-5_14
Fu, L., Grishman, R.: An efficient active learning framework for new relation types. In: IJCNLP, pp. 692–698 (2013)
Gal, Y., Islam, R., Ghahramani, Z.: Deep bayesian active learning with image data. In: ICML (2017)
Gentile, A.L., Zhang, Z., Augenstein, I., Ciravegna, F.: Unsupervised wrapper induction using linked data. In: K-CAP, pp. 41–48. ACM (2013)
Hendrickx, I., Kim, S.N., Kozareva, Z., Nakov, P., Ó Séaghdha, D., Padó, S., Pennacchiotti, M., Romano, L., Szpakowicz, S.: Semeval-2010 task 8: multi-way classification of semantic relations between pairs of nominals. In: DEW Workshop, pp. 94–99. ACL (2009)
Hsu, W., Lin, H.: Active learning by learning. In: Bonet, B., Koenig, S. (eds.) AAAI, pp. 2659–2665. AAAI Press (2015)
Huang, S.J., Jin, R., Zhou, Z.H.: Active learning by querying informative and representative examples. In: NIPS, pp. 892–900 (2010)
Ji, G., Liu, K., He, S., Zhao, J.: Distant supervision for relation extraction with sentence-level attention and entity descriptions. In: AAAI, pp. 3060–3066 (2017)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. In: ICLR (2015)
Lewis, D.D., Catlett, J.: Heterogeneous uncertainty sampling for supervised learning. In: ICML, pp. 148–156 (1994)
Liu, M.X.C.: Semantic relation classification via hierarchical recurrent neural network with attention. In: COLING (2016)
Mooney, R.J., Bunescu, R.C.: Subsequence kernels for relation extraction. In: NIPS, pp. 171–178 (2006)
Morgan, N., Bourlard, H.: Generalization and parameter estimation in feedforward nets: some experiments. In: NIPS, pp. 630–637 (1990)
Nair, V., Hinton, G.E.: Rectified linear units improve restricted boltzmann machines. In: ICML, pp. 807–814 (2010)
Nguyen, H.T., Smeulders, A.: Active learning using pre-clustering. In: ICML. ACM (2004)
Nguyen, T.H., Grishman, R.: Relation extraction: perspective from convolutional neural networks. In: VS@ HLT-NAACL, pp. 39–48 (2015)
Pennington, J., Socher, R., Manning, C.D.: Glove: global vectors for word representation. In: EMNLP, vol. 14, pp. 1532–1543 (2014)
Ratner, A.J., Sa, C.D., Wu, S., Selsam, D., Ré, C.: Data programming: creating large training sets, quickly. In: NIPS, pp. 3567–3575 (2016)
Roth, B., Barth, T., Wiegand, M., Klakow, D.: A survey of noise reduction methods for distant supervision. In: AKBC, pp. 73–78. ACM (2013)
Settles, B.: Active learning literature survey. Univ. Wis. Madison 52(55–66), 11 (2010)
Stanovsky, G., Gruhl, D., Mendes, P.: Recognizing mentions of adverse drug reaction in social media using knowledge-infused recurrent models. In: EACL, pp. 142–151. ACL (2017)
Sterckx, L., Demeester, T., Deleu, J., Develder, C.: Using active learning and semantic clustering for noise reduction in distant supervision. In: AKBC at NIPS, pp. 1–6 (2014)
Vu, N.T., Adel, H., Gupta, P., et al.: Combining recurrent and convolutional neural networks for relation classification. In: NAACL-HLT, pp. 534–539 (2016)
Zelenko, D., Aone, C., Richardella, A.: Kernel methods for relation extraction. J. Mach. Learn. Res. 3, 1083–1106 (2003)
Zeng, D., Liu, K., Lai, S., Zhou, G., Zhao, J., et al.: Relation classification via convolutional deep neural network. In: COLING, pp. 2335–2344 (2014)
Zhao, S., Grishman, R.: Extracting relations with integrated information using kernel methods. In: ACL, pp. 419–426. ACL (2005)
Zhou, P., Shi, W., Tian, J., Qi, Z., Li, B., Hao, H., Xu, B.: Attention-based bidirectional long short-term memory networks for relation classification. In: ACL - Short Papers, vol. 2, pp. 207–212 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Lourentzou, I., Alba, A., Coden, A., Gentile, A.L., Gruhl, D., Welch, S. (2018). Mining Relations from Unstructured Content. In: Phung, D., Tseng, V., Webb, G., Ho, B., Ganji, M., Rashidi, L. (eds) Advances in Knowledge Discovery and Data Mining. PAKDD 2018. Lecture Notes in Computer Science(), vol 10938. Springer, Cham. https://doi.org/10.1007/978-3-319-93037-4_29
Download citation
DOI: https://doi.org/10.1007/978-3-319-93037-4_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-93036-7
Online ISBN: 978-3-319-93037-4
eBook Packages: Computer ScienceComputer Science (R0)