Abstract
Estimating health care cost of patients provides promising opportunities for better management and treatment to medical providers and patients. Existing clinical approaches only focus on patient’s demographics and historical diagnoses but ignore ample information from clinical records. In this paper, we formulate the problem of patient’s cost profile estimation and use Electronic Medical Records (EMRs) to model patient visit for better estimating future health care cost. The performance of traditional learning based methods suffered from the sparseness and high dimensionality of EMR dataset. To address these challenges, we propose Patient Visit Probabilistic Generative Model (PVPGM) to describe a patient’s historical visits in EMR. With the help of PVPGM, we can not only learn a latent patient condition in a low dimensional space from sparse and missing data but also hierarchically organize the high dimensional EMR features. The model finally estimates the patient’s health care cost through combining the effects learned both from the latent patient condition and the generative process of medical procedure. We evaluate the proposed model on a large collection of real-world EMR dataset with 836,033 medical visits from over 50,000 patients. Experimental results demonstrate the effectiveness of our model.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Electronic medical records
- Cost profile estimation
- Health care data mining
- Probabilistic generative model
1 Introduction
The growth rate of health care cost is an alarming problem in many countries all over the world. The cost of health care in the United States is steadily rising, making it the most expensive in the worldFootnote 1. Studies show that advanced analytics can help slow this upward trend [12], among which accurate estimation of health care cost serves as the first step. In clinical practice, a more expensive treatment regimen at the onset of diseases could lower the patient’s total cost in the next 10 years. Doctors need to predict the future cost of a patient in order to give a more reasonable and economical treatment. For health insurance companies, there is a significant need to look beyond applicants’ future cost, especially for those with chronic diseases.
Therefore, it is important to estimate patient’s health care cost aiming at better management and treatment for both medical providers and patients. Traditionally, previous approaches in this field are mainly based on demographics and diagnosis data. The Diagnosis-Related Groups (DRGs) classifies patients into groups by their age, sex and history diagnosis to infer patient’s current problem and their potential health care cost [1, 7, 10]. Moturu et al. [21] utilized different classification algorithms to predict future high cost patients. Recently, with the development of health informatics, a large amount of data from medical institute become available. These Electronic Medical Records (EMR) document patient visits, including demographic information (birth date, gender, etc.), diagnosis (historic and current), treatment (medication and surgery), laboratory results and clinical notes, providing an opportunity for researchers to develop data-driven models for health care data analysis.
In this paper, we study the problem of estimating future health care cost of a patient from his history data in EMR. More specifically, given a series of hospital visit data of a patient, the goal of this study is to estimate the high cost risk of future visits. This approach could help doctors identify potential high cost patients in the near future, who could be enrolled in case-management or enroll-management program and get early intervention. The health insurance companies can also benefit from them to develop personalized contracts. However, there are some obstacles for making analysis on EMR due to its intricate nature. Generally speaking, there are two aspects of challenges:
Sparse and Missing Data. There are thousands of possible items in the clinical laboratory, only a few of them could be done in each patient visit. Even for a certain kind of disease, a patient can take a small bundle of medical tests in hundreds of related tests, leading to a highly sparse dataset. There are 566 different medical test features in our dataset, but each patient visit only contains 13.92 medical tests (2.46%) on average. 70.37% of the 836,033 patient visits have less than 10 medical tests. The sparse dataset will make it difficult in constructing the model and definitely deteriorate learning performance. In clinical trails, Wood et al. [26] proposed multiple imputation methods to handle missing data, but little statistical imputation could handle our highly sparse dataset with guaranteed bias.
Confounding Effect and High Dimensionality. Confounding arises when a variable (confounder) is not connecting the exposure with the outcome but associated with them [22]. Using EMRs often has confounding effects [20]. For medical cost prediction problems, risk score features based on DRGs and indicators of chronic conditions can be extracted [8]. However, a spurious association may be generated when we study on a small bundle of risk factors and the high cost risk of patients. On the other hand, there are 746 different features in our EMR dataset containing detailed information of patient visits. But modeling of the high dimensional features without domain knowledge is still an open question.
To address these challenges, we propose the Patient Visit Probabilistic Generative Model (PVPGM) to describe the generative process of the high dimensional features of a patient visit. For handling sparse and missing data, PVPGM learns a latent patient condition in a low dimensional space from the sparse and missing medical test data. For handling the confounding effect and high dimensionality, PVPGM hierarchically organizes the high dimensional features of a patient visit including demographic information, diagnosis, treatment and laboratory results, and then describes the generative process by logistic functions. With the help of such a generative model, we can predict the high cost risk of a patient as well as expose the risk factors. To the best of our knowledge, it is the first paper to study the cost prediction problem in EMR data mining. We evaluate the proposed model on a large collection of the real-world electronic medical records from a famous cardiovascular hospital in China and the experiment results demonstrate the effectiveness of our model.
To summarize, our work contributes on the following aspects:
-
We study the problem of estimating patients’ cost, which has been found widespread application prospect. To the best of our knowledge, no previous work has studied this problem in machine learning perspective.
-
We propose a probabilistic generative model, PVPGM, to solve this problem. Our model aims at addressing two common challenges in EMR data mining: it first projects the high dimensional sparse data into a low dimensional space and then models the features of a patient visit from electronic medical records to estimate the patient’s cost profile.
-
We evaluate our proposed model on a large scale real-world dataset, including 836,033 medical visits from over 50,000 patients. Experimental results show that our model outperforms baseline methods significantly.
2 Problem Definition
In this section, we introduce and define related concepts and formulate our cost profile estimation problem.
2.1 Definition
As we mentioned in the above section, our model jointly describes patient visits in the generative process. Formally, we define a patient visit for a patient first.
Definition 1
(Patient Visit). A patient visit n for a patient is defined as a set \(s_n=\{x, \iota , d, t, c, \tau \}\), where \(x=\{(m_l, r_l)\}\) is the medical test set of (item, result) pairs, demographical information \(\iota \), diagnosis d, medical treatment t, health care cost c and time stamp \(\tau \).
A patient visit is the behavior and corresponding record of the inpatient or outpatient documented by hospital information system. We organize all possible information in a clinical record that EMR provides to us. A patient may visit a hospital multiple times, so we use a patient visit sequence \(S=(s_i)\) to represent a patient where \(\tau _i<\tau _j\) if \(i<j\). Then we define the concept of patient condition as below.
Definition 2
(Patient Condition). The patient condition \(\psi =(\iota , \theta )\) denotes one particular patient’s current condition, which is defined as a pair of demographical information \(\iota \) and intrinsic condition \(\theta \).
The patient condition for one patient visit includes the extrinsic condition (the demographic information such as age, gender) and intrinsic one. \(\theta \) is a latent low dimensional probability vector learned from patient’s medical test. Each dimension of the latent intrinsic condition denotes the weight of the generative distribution since we adopt a mixture of the generative process to the medical tests. Similar to the above, we further define a condition sequence \(\varPsi =(\psi _i)\) for a particular patient to describe the development of his/her disease, where \(\psi _i\) is earlier than \(\psi _j\) if \(i<j\). Furthermore, we define the concept related to our object problem as follows.
Definition 3
(Cost Profile). The cost profile is the description of one patient’s health care cost sequence \(C=(c_i)\). For a particular patient visit i, the m-period cost profile is the average cost of the future m patient visits:
The cost profile of patient visit i includes two aspects: short-term description \(STCP=CP(i, 1)\) and m periods long-term description \(LTCP(m)=CP(i, m)\). Based on the above definitions, we further define our prediction task as follows.
2.2 Cost Profile Estimation Problem
The problem we address is to forecast cost profile based on the patient visit sequence. The input of cost profile estimation includes a set of patients \(V=\{v_n\}\) and a patient visit sequence \(S_n(\tau _0)=(..., s_{i_0-1}, s_{i_0})\) for each patient \(v_n\) where \(\tau _{i_0} \le \tau _0 < \tau _{i_0+1}\). Our goal of cost profile estimation is for each patient \(v_n\) and \(i_0+1\) patient visit, determining if \(STCP_n\) is in the high group. The threshold will be tuned to verify the robustness of our approach.
3 Model
In this section, we describe our proposed Patient Visit Probabilistic Generative Model, PVPGM to estimate the cost profile of a patient. The model includes the generative process of patient visit and patient condition so as to produce the unknown condition of next period and corresponding cost profile.
3.1 Assumptions
With the support of our clinical experts, we have the following assumptions based on the medical insights.
Assumption 1
The health care cost of patients with non-communicable disease (NCD) mainly depends on the progression of disease.
Non-communicable diseases (NCDs) tend to be of long duration, generally slow progression such as cardiovascular disease (CVD) and diabetes mellitusFootnote 2. Our problem focuses on NCDs whose health care cost of inpatient and outpatient is more predictable than other diseases such as surgery injuries. We can learn patient condition from medical tests to infer the progression of disease and make a more accurate prediction about patient’s future cost.
Assumption 2
A diagnosis is made by obtaining patient condition and physicians give out the treatment based on the diagnosis.
In order to make a diagnose, physicians analyze patient’s extrinsic and intrinsic condition from inquiry and medical tests based on their own experience. Then, the corresponding treatment is given and the health care cost is incurred.
3.2 Patient Visit Probabilistic Generative Model
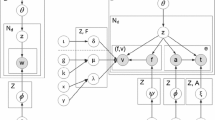
Based on the assumptions, we propose our Patient Visit Probabilistic Generative Model, PVPGM, as shown in Fig. 1. Table 1 describes the variables of our model.
According to our problem definition, we have a patient visit sequence \(S(\tau _0)=(..., s_{i_0-1}, s_{i_0})\) for each patient. We use \(c_n\) to denote whether STCP of patient visit n is in the high group. \(c_n=1\) represents the positive result and \(c_n=-1\) is the negative result. Our target is to map the input visit sequence to the label of patient visit \(i_0+1\), i.e., \(f:S(\tau _0) \mapsto c_{i_0+1}\). We can build a classification model to map the sequence to a target label. In order to handle the challenges of our dataset, in each time slice, PVPGM models the medical procedure and extracts low dimensional features from the sparse medical tests.
Medical Procedure. The right part of each time slice in Fig. 1 represents the generative process of medical procedure. Assumption 2 describes our intuitive insights. According to the model, for each patient visit n, the diagnose \(\varvec{d_n}\) is generated from a multinomial logistic regression \(\varOmega \) based on the patient condition \(\psi _n=(\varvec{\iota _n, \theta _n})\). We concatenate \(\psi _n\) to a condition vector
Then the treatment \(\varvec{t}\) is generated from a \(\varvec{d_n}\)-specific multinomial distribution \(\varvec{\lambda _{d_n}}\). Finally c is generated from logistic regression based on the treatment.
Mixture Generative Process for Medical Tests. To extract low dimensional latent feature \(\theta \) from the sparse medical tests dataset, we use the hierarchical generative architecture which is similar to the Latent Dirichlet Allocation (LDA) model [2]. Left part of our model shown in Fig. 1 denotes the design. Marginalized out z, we have

Unlike LDA, there are two kinds of generative target r: numerical and categorical. Similar to Liu et at. [19], we generate numerical variables from Gaussian distributions and categorical variable from multinomial distributions. Thus the k-specific distribution \(\varPi _{l,k}\) is defined as
The whole generative process is shown in Algorithm 1. We define
as our parameter configuration. By adopting the chain rule of probability, the log-likelihood objective function can be obtained as follows:
where \(l_1\) denotes the continuous medical tests and \(l_2\) denotes the categorical tests.
is an indicator function of our dataset.

3.3 Model Learning
From the above, we show the maximum-likelihood estimation problem of PVPGM as follows:
In this paper, we use iterative approximation method to learn the parameters because Eq. (5) does not have the closed-form solution. This algorithm separates the objective function into two parts and maximizes them iteratively. For the first part, we fix the parameters of medical procedure \(\{\varOmega , \varvec{\lambda , \eta }\}\) and learn the remaining mixture generative process. We have
We can adopt Eq. (7) to get the lower bound of Eq. (6) by Jensen’s inequality, then maximize and update the lower bound iteratively to generate the global maximum. We have the following update functions.
where \(1_x(y)\) indicates whether x is equal to y.
For the second part, we fix the parameters of mixture generative process \(\{\varvec{\theta ,} {\varphi }, \mu , \sigma \}\) and learn the medical procedure. We have
We have closed-form solution for \(\lambda _{d,t}\):
We adopt gradient ascent algorithm to update the remaining parameters of \(O_2\) iteratively. Following are the update functions.
where \(\alpha \) is the learning rate with the gradient and \(\alpha '\) is the weight decay term. Algorithm 2 summarizes the learning algorithm.
3.4 Cost Profile Estimation
With the generative model above, a patient’s diagnose code \(\varvec{d}\), treatment \(\varvec{t}\) and health care cost c can be generated from his condition \(\psi \) in a certain patient visit n. In order to estimate cost profile of this patient, we learn the sequence of his history condition. Figure 2 shows the procedure of our cost estimation. Instead of directly producing \(c_{i_0+1}\) (or equivalently \(s_{i_0+1}\)), we first learn a probabilistic graphical model which hierarchically organizes patients’ medical features. Then the condition vectors for different time slices of one patient can be used to predict the condition of next visit. Finally \(c_{i_0+1}\) can be drawn from the inference of probabilistic graph.
With the model parameters and patient condition vector \(\varvec{\vartheta _n}\) for each time slice, we assume the patient condition sequence \(\varPsi =(\psi _i)\) has Markov property. So we can learn the transition matrix by solving the following optimization problem
where \(\tau _{i_0} \le \tau _0\) is the last patient visit before \(\tau _0\).
Then we can calculate next condition vector by \(\psi _{i_0+1}=X \psi _{i_0}\) and generate the corresponding cost profile.
4 Experiments
In this section, we comprehensively evaluate our PVPGM model on a real-world dataset and compared it with multiple state-of-the-art methods. Firstly, we introduce the experiment setup, including the description of our EMR dataset, evaluation metrics and baseline models. Then we present the experimental result and illustrate the effectiveness of our proposed approach. We also discuss the effect of some important parameter values in this section.
4.1 Experiment Setup
We use a collection of real medical records from a leading hospital in cardiovascular medicine, cardiovascular surgery and geriatrics in Beijing. Our dataset is described in Table 2. It contains over 836,033 patient visits from over 50,624 patients. Each record consists of 566 medical tests with 96 demographical information and 102 treatment columns. On average each EMR record contains 13.92 different medical tests (2.46% of all medical tests), which indicates our dataset has a serious problem with feature sparsity.
We randomly picked 80% of the patients as training set and the rest for testing, then adopt 5-fold cross validation during training. We evaluate the proposed model in terms of precision, recall and F-Measure. In order to better evaluate the classifier on imbalanced data, we also compare the ROC curve with different baseline methods to validate its effectiveness. These metrics are widely used in data mining studies. Receiver Operating Characteristic (ROC) curve is extensively adopted to evaluate the imbalanced dataset especially in clinical practice [9].
4.2 Baseline Methods
We select the following methods as baselines for the cost estimation problem and compare our proposed method with them:
-
DTC. Decision Tree Classifier (DTC) [3] learns simple decision rules inferred from the data features, which will generate a data-driven model similar to the rule-based DRGs models. Diagnosis-Related Group (DRG) is a widely used classification model to estimate patient’s health care cost [1, 7] as we mentioned in Sect. 1. We adopt DTC to illustrate the effectiveness of DRGs.
-
NI+SVM. Normal Imputation (NI) is employed on our dataset. In medical tests, it is reasonable that doctors would not check the irrelevant or unnecessary medical lab tests when they are in the reference range. Normal imputation is a widely used method in clinical practice [26]. We treat medical records as features and LIBSVM [5] is employed as the classification model for health care cost estimation.
-
ALS+SVM. Alternative Least Square (ALS) collaborative filtering [13] is an algorithm based imputation method. ASL features are employed as the input of SVM classification model.
-
PGM. A traditional probabilistic generative model is used as classification model. It is a part of PVPGM and has the same architecture with the medical procedure mentioned in Sect. 3.2. We employ gradient descent algorithm to learn the parameters in PGM [16] and set the learning rate parameter as 0.1.
-
PGM+PCA. PCA [11] converts high dimensional data into a set of principal components. We adopt PCA to solve the data sparsity problem and employ it as the input of our PGM classifier.
4.3 Performance Comparison
We now compare the performance with baseline models to evaluate the effectiveness of our model in the health care cost estimation. We set the threshold of the high group for patient’s health care cost to the top 10%, 15% and 20%. We also empirically set the compression parameter of latent variables (dimension of latent variable/origin) as 0.05.
Figure 3 shows the results of precision, recall and F-measure of different models with the cost threshold as 10%. Figure 4 demonstrates the ROC curves on different cost thresholds of health care cost estimation task. From these results, we can see that PVPGM achieves the best aggregate performance in all the three thresholds. Generally, PVPGM models the medical procedure and lab tests of a patient visit at the same time, which captures the dependence and constraint between medical features and latent condition variable of patients. It improves the precision without lowering recall, thus results in the best \(F_1\) score.
DTC is used to simulate DRGs model to learn decision rules from patient’s demographics and diagnosis. NI+SVM and ALS+SVM adopt the rule-based or statistical method to impute missing data in medical test. Compared with DTC, NI+SVM and ALS+SVM include patient’s condition information and get a better performance. However, noise is introduced in such imputation methods and they also suffer from high dimensionality and feature sparsity.
Unlike above methods, PGM models the medical procedure of patient visit with a graph-based method. Such traditional probabilistic graphical model adopts domain knowledge from the medical field to reduce the parameter space, which improves the precision without hurting the recall substantially compared with DTC. However, PGM+PCA does not improve the overall performance because the sparse modeling and classification process are separated into two steps. While our PVPGM learns the patient’s hidden condition and features from the medical process at the same time. Such integrating methods estimate the parameters better and outperform all the baselines.
4.4 Sensitivity Analysis
We also investigate the impact of the latent variable dimensionality. In this study, our PVPGM embeds high dimensional and sparse medical tests into a low dimensional space representing patient’s hidden condition. So we focus on the compression parameter of latent variable dimensionality (latent variables/origin). Figure 5 displays the F-measure of PVPGM to the compression parameter. We can see that the overall performance of our model does not change much in terms of the dimensionality of latent variables when the cost threshold is set as 15% or 20%. Lower dimension is better for 10% cost probably because a larger latent space will increase the number of training parameters and thus hurt the generalization of the classifier. There is also a trade-off between effectiveness and training time. So in our experiment, we choose the compression parameter as 0.05.
5 Related Work
In this section, we review related work about cost estimation problem using EMR dataset in health care data mining and sparse modeling problem using probabilistic graphical models.
Cost Estimation Problem in EMR Data Mining. Recently modeling electronic medical records for prediction has attracted the interests of researchers from various areas. There is a long stream of studies for clinical problems in machine learning perspective [23, 27]. Various medical models are developed based on the specific clinical problems such as patient phenotype identification [18, 24], potential complications of diseases [6, 29] and risk profiling [17]. For health care cost estimation problem, current studies mainly focus on statistical and economic models [1, 21] but few researchers in this domain adopt medical test features from EMR dataset. To the best of our knowledge, none of the previous work estimates cost profile of a patient from a medical model.
Probabilistic Graphical Models in Sparse Modeling. Due to the interpretability and good generalization ability, lots of papers in health care adopt probabilistic generative models to analyze EMR dataset. Liu et al. [18] employ the generative model to capture the relationship of medical events. Some studies [4, 28] design dynamic latent variables to model patient’s behavior over time. Sparse modeling is an important problem in machine learning tasks. Lee et al. [15] and Zhang et al. [37] extensively study the sparse coding problem. Probabilistic graphical models are widely adopted in sparse modeling. The most important applications include probabilistic topic models [30,31,32,33,34,35]. Sparse conditional random field [39], sparse generalized linear models [14] and sparse factor models [29, 36] utilize probabilistic graphical structure to solve the sparse problem. CNN based solution [25] and considering syntactic similarity issues [38] are also good ways to address data sparsity. Inspired by the medical assumptions, we describe a distribution over the observed patient visit by recovering latent random variables from a probabilistic graphical structure.
6 Conclusion
In this paper, we study the problem of using a large volume of electronic medical records to estimate the cost profile of a patient. We propose Patient Visit Probabilistic Generative Model (PVPGM) to model patient visit for better estimating future health care cost. Our model learns the latent patient condition from the sparse dataset and integrates it into the generative process of medical procedure. We validate our model on a large collection of real-world electronic medical records from a famous cardiovascular hospital in China and the experiment results show the effectiveness of our model.
For the future work, we will continuously work with our medical group to reveal the explanations of our model. The mixture generative process of high dimensional medical tests can be regarded as clusters of lab items, which may help mine the possible medical phenotypes. The sensitivity of demographics can reveal a lot of risk factors on health economics. Our estimation architecture can also be used for the intelligent diagnosis or personalized treatment.
Notes
References
Ash, A.S., Ellis, R.P., Pope, G.C., Ayanian, J.Z., Bates, D.W., Burstin, H., Iezzoni, L.I., MacKay, E., Yu, W.: Using diagnoses to describe populations and predict costs. Health Care Financ. Rev. 21(3), 7 (2000)
Blei, D.M., Ng, A.Y., Jordan, M.I.: Latent dirichlet allocation. J. Mach. Learn. Res. 3(Jan), 993–1022 (2003)
Breiman, L., Friedman, J., Stone, C.J., Olshen, R.A.: Classification and Regression Trees. CRC Press, Boca Raton (1984)
Caballero Barajas, K.L., Akella, R.: Dynamically modeling patient’s health state from electronic medical records: a time series approach. In: KDD, pp. 69–78 (2015)
Chang, C., Lin, C.: LIBSVM: a library for support vector machines. ACM TIST 2(3), 27:1–27:27 (2011)
Feld, S.I., Cobian, A.G., Tevis, S.E., Kennedy, G.D., Craven, M.: Modeling the temporal evolution of postoperative complications. In: AMIA (2016)
Fetter, R.B., Shin, Y., Freeman, J.L., Averill, R.F., Thompson, J.D.: Case mix definition by diagnosis-related groups. Med. Care 18(2), i–53 (1980)
Fleishman, J.A., Cohen, J.W.: Using information on clinical conditions to predict high-cost patients. Health Serv. Res. 45(2), 532–552 (2010)
Hajian-Tilaki, K.: Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Casp. J. Internal Med. 4(2), 627 (2013)
Horn, S.D., Bulkley, G., Sharkey, P.D., Chambers, A.F., Horn, R.A., Schramm, C.J.: Interhospital differences in severity of illness: problems for prospective payment based on diagnosis-related groups (DRGs). N. Engl. J. Med. 313(1), 20–24 (1985)
Jolliffe, I.T.: Principal component analysis and factor analysis. In: Jolliffe, I.T. (ed.) Principal Component Analysis, pp. 115–128. Springer, New York (1986). https://doi.org/10.1007/978-1-4757-1904-8_7
Koh, H.C., Tan, G., et al.: Data mining applications in healthcare. J. Healthc. Inf. Manag. 19(2), 65 (2011)
Koren, Y., Bell, R.M., Volinsky, C.: Matrix factorization techniques for recommender systems. IEEE Comput. 42(8), 30–37 (2009)
Krishnapuram, B., Carin, L., Figueiredo, M.A.T., Hartemink, A.J.: Sparse multinomial logistic regression: fast algorithms and generalization bounds. IEEE Trans. Pattern Anal. Mach. Intell. 27(6), 957–968 (2005)
Lee, H., Battle, A., Raina, R., Ng, A.Y.: Efficient sparse coding algorithms. In: NIPS, pp. 801–808 (2006)
Lee, J.D., Hastie, T.J.: Learning the structure of mixed graphical models. J. Comput. Graph. Stat. 24(1), 230–253 (2015)
Lin, Y.K., Chen, H., Brown, R.A., Li, S.H., Yang, H.J.: Healthcare predictive analytics for risk profiling in chronic care: a Bayesian multitask learning approach. MIS Q. 41(2), 473–495 (2017)
Liu, C., Wang, F., Hu, J., Xiong, H.: Temporal phenotyping from longitudinal electronic health records: a graph based framework. In: KDD, pp. 705–714 (2015)
Liu, L., Tang, J., Cheng, Y., Agrawal, A., Liao, W.K., Choudhary, A.: Mining diabetes complication and treatment patterns for clinical decision support. In: CIKM, pp. 279–288 (2013)
Moher, D., Jones, A., Cook, D.J., Jadad, A.R., Moher, M., Tugwell, P., Klassen, T.P., et al.: Does quality of reports of randomised trials affect estimates of intervention efficacy reported in meta-analyses? Lancet 352(9128), 609–613 (1998)
Moturu, S.T., Johnson, W.G., Liu, H.: Predicting future high-cost patients: a real-world risk modeling application. In: BIBM, pp. 202–208. IEEE (2007)
Pearl, J.: Causality. Cambridge University Press, Cambridge (2009)
Shickel, B., Tighe, P., Bihorac, A., Rashidi, P.: Deep EHR: a survey of recent advances on deep learning techniques for electronic health record (EHR) analysis. arXiv preprint arXiv:1706.03446 (2017)
Shivade, C., Raghavan, P., Fosler-Lussier, E., Embi, P.J., Elhadad, N., Johnson, S.B., Lai, A.M.: A review of approaches to identifying patient phenotype cohorts using electronic health records. J. Am. Med. Inform. Assoc. 21(2), 221–230 (2013)
Wang, J., Wang, Z., Zhang, D., Yan, J.: Combining knowledge with deep convolutional neural networks for short text classification. In: IJCAI, pp. 2915–2921 (2017)
Wood, A.M., White, I.R., Thompson, S.G.: Are missing outcome data adequately handled? A review of published randomized controlled trials in major medical journals. Clin. Trials 1(4), 368–376 (2004)
Yadav, P., Steinbach, M., Kumar, V., Simon, G.: Mining electronic health records: a survey. arXiv preprint arXiv:1702.03222 (2017)
Yang, S., Khot, T., Kersting, K., Natarajan, S.: Learning continuous-time Bayesian networks in relational domains: a non-parametric approach. In: AAAI, pp. 2265–2271 (2016)
Yang, Y., Luyten, W., Liu, L., Moens, M.F., Tang, J., Li, J.: Forecasting potential diabetes complications. In: AAAI, pp. 313–319 (2014)
Yin, H., Cui, B.: Spatio-Temporal Recommendation in Social Media. Springer Briefs in Computer Science. Springer, Singapore (2016). https://doi.org/10.1007/978-981-10-0748-4
Yin, H., Cui, B., Zhou, X., Wang, W., Huang, Z., Sadiq, S.W.: Joint modeling of user check-in behaviors for real-time point-of-interest recommendation. ACM Trans. Inf. Syst. 35(2), 11:1–11:44 (2016)
Yin, H., Hu, Z., Zhou, X., Wang, H., Zheng, K., Hung, N.Q.V., Sadiq, S.W.: Discovering interpretable geo-social communities for user behavior prediction. In: ICDE, pp. 942–953. IEEE Computer Society (2016)
Yin, H., Wang, W., Wang, H., Chen, L., Zhou, X.: Spatial-aware hierarchical collaborative deep learning for POI recommendation. IEEE Trans. Knowl. Data Eng. 29(11), 2537–2551 (2017)
Yin, H., Zhou, X., Cui, B., Wang, H., Zheng, K., Hung, N.Q.V.: Adapting to user interest drift for POI recommendation. IEEE Trans. Knowl. Data Eng. 28(10), 2566–2581 (2016)
Yin, H., Zhou, X., Shao, Y., Wang, H., Sadiq, S.W.: Joint modeling of user check-in behaviors for point-of-interest recommendation. In: CIKM, pp. 1631–1640. ACM (2015)
Yoshida, R., West, M.: Bayesian learning in sparse graphical factor models via variational mean-field annealing. J. Mach. Learn. Res. 11, 1771–1798 (2010)
Zhang, X., Yu, Y., White, M., Huang, R., Schuurmans, D.: Convex sparse coding, subspace learning, and semi-supervised extensions. In: AAAI (2011)
Zhang, Y., Li, X., Wang, J., Zhang, Y., Xing, C., Yuan, X.: An efficient framework for exact set similarity search using tree structure indexes. In: ICDE, pp. 759–770 (2017)
Zhong, P., Wang, R.: Learning sparse crfs for feature selection and classification of hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 46(12), 4186–4197 (2008)
Acknowledgment
This work was supported by NSFC (91646202), the National High-tech R&D Program of China (SS2015AA020102), NSSFC (15CTQ028), Research/Project 2017YB142 supported by Ministry of Education of The People’s Republic of China, the 1000-Talent program and Tsinghua Fudaoyuan Research Fund.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Zhao, K. et al. (2018). Modeling Patient Visit Using Electronic Medical Records for Cost Profile Estimation. In: Pei, J., Manolopoulos, Y., Sadiq, S., Li, J. (eds) Database Systems for Advanced Applications. DASFAA 2018. Lecture Notes in Computer Science(), vol 10828. Springer, Cham. https://doi.org/10.1007/978-3-319-91458-9_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-91458-9_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-91457-2
Online ISBN: 978-3-319-91458-9
eBook Packages: Computer ScienceComputer Science (R0)