Abstract
A deep learning approach to glioma segmentation is presented. An encoder and decoder pair deep learning network is designed which takes T1, T2, T1-CE (contrast enhanced) and T2-Flair (fluid attenuation inversion recovery) images as input and outputs the segmented labels. The encoder is a 49 layer deep residual learning architecture that encodes the \(240\,\times \,240\,\times \,4\) input images into \(8\,\times \,8\,\times \,2048\) feature maps. The decoder network takes these feature maps and extract the segmented labels. The decoder network is fully convolutional network consisting of convolutional and upsampling layers. Additionally, the input images are downsampled using bilinear interpolation and are inserted into the decoder network through concatenation. This concatenation step provides spatial information of the tumor to the decoder, which was lost due to pooling/downlsampling during encoding. The network is trained on the BRATS-17 training dataset and validated on the validation dataset. The dice score, sensitivity and specificity of the segmented whole tumor, core tumor and enhancing tumor is computed on validation dataset. The mean dice score for whole tumor, core tumor and enhancing tumor for validation dataset were 0.824, 0.627 and 0.575, respectively.
Caffe model and code available at: https://github.com/kamleshpawar17/BratsNet-2017.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Gliomas are the tumors of the central nervous system which arises from glial cells. The gliomas are classified into two types depending on the aggressiveness of the tumor: high grade (HGG) and low grade (LGG) gliomas, both types of tumors are malignant and need treatment [1]. The accurate segmentation of gliomas is important in grading, treating and monitoring tumor progression. Multiple magnetic resonance (MR) image contrasts are used to evaluate the type and extent of tumors. The different contrasts T1, T2, T1-CE and T2-Flair are analysed by a radiologist and tumor regions are manually segmented. Segmenting brain tumor is a comprehensive task, and large intra-rater variability is often reported, e.g. 20% [2]. Thus it is imperative to have a reliable automatic segmentation algorithm that standardizes the process of segmentation, resulting in more precise planning, treatment and monitoring.
Manual segmentation by an expert is a time consuming and expensive process, thus computer assisted tumor segmentation is imperative to the problem of brain tumor segmentation. Computer assisted methods [3,4,5,6,7,8] can be broadly classified into two categories: semiautomatic brain tumor segmentation and automatic brain tumor segmentation. Semiautomatic approach involves seeding some initial information by an expert such as a location of the tumor and the fine delineation and computational task is offloaded to a computer. This approach minimises the time spent by human experts. However large number of MR images are generated routinely in clinics, demanding fully automatic segmentation methods. The automatic segmentation methods require no human intervention and can segment the brain tumor into different classes such as necrotic tumor, enhancing tumor, tumor core and edema.
In this work, we present a deep learning based approach to brain tumor segmentation on the BRATS-17 dataset [9,10,11,12]. Deep learning methods [13, 14] based on convolutional neural networks (CNN) [15, 16] have demonstrated highly accurate results in image classification [17, 18] and segmentation [19, 20]. However, selection of the number of CNN layers is a complex task. On one hand, increasing the number of layers improves complexity of the network and leads to more accurate results. On the other hand, designing deeper CNN may result in performance degradation due to exploding/vanishing gradients. This problem is partially solved by the batch normalization layers [21] that minimize the chances of exploding/vanishing gradients. Another limitation of designing deep neural network is that the training process becomes difficult and, after a certain depth the network ceases to converge. However recently introduced residual networks [22], consisting of short cut connections, can be trained to the larger depths. In this paper we present an encoder-decoder based CNN architecture to solve the tumor segmentation problem.
If there exist a nonlinear function H(x) between point A and B. (a): a network with no shortcut connection, here the layers Conv1 and Conv2 approximate the non linear function H(x); (b): a network with shortcut connection, here the layers Conv1 and Conv2 approximate the nonlinear residual function \({f(x)=H(x)-x}\). Both (a) and (b) approximate the same non-linear function H(x) between A and B, however individual layers learn different nonlinear functions.
2 Deep Residual Learning Networks
Deep learning CNN uses the layers of convolution as a feature extractor. The initial layers extract the basic features such as horizontal, vertical and slanted edges, the later layers extract more complicated features by combining the basic features. Thus increasing the depth results in more complicated features being extracted, hence better performance in classification/segmentation. However, it is found that increasing the network depth does not always increase the accuracy. The training error reduces with the depth of the network to a certain level but the error increasing as further layers are added to the network [23, 24]. This behaviour is counter intuitive, one may expect the error to decrease or stay constant after certain depth. The network may just learn identity mapping after certain depth and keep the error constant with even further increasing the depth of the network. Deep learning residual networks overcome this problem, by inserting the bottleneck units to increase the depth of the network. The bottleneck units learn the residual function that minimises the error.
Residual functions are realised with shortcut connection (Fig. 1(b)), a shortcut connection is a direct connection between two layers in a network skipping one or few layers. Consider that there exists a nonlinear relationship between the two points A and B in the network given by H(x) as shown in Fig. 1. The conventional CNN without the shortcut connection (Fig. 1(a)), would train the layers (Conv1 and Conv2) such that the combined effect of the learning represents H(x). However in case of the network with shortcut connection (Fig. 1(b)), the same layers (Conv1 and Conv2) would learn a residual function given by:
In both the cases the relationship between the point A and B remains the same, however the convolution layers have learned different functions. The advantage of using a residual network is that it may approximate an identity relationship between the point A and B, if the convolution layers Conv1 and Conv2 are deemed to be unnecessary. Thus the residual learning in effect avoids the performance degradation on going to arbitrarily large depths.
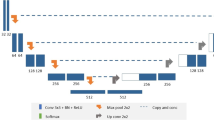
Residual encoder and convolutional decoder network; the encoder is a 49 layer deep residual network and the decoder is a 10 layer deep fully convolutional network with bilinear upsampling layers. The input data is also downsampled using bilinear interpolation and is inserted back into the decoder through concatenation.
3 Proposed Segmentation Method
The method presented here is based on residual learning convolutional neural network [22] and the design is similar to the Unet encoder-decoder architecture [25]. The network is a 2D CNN, which performs segmentation on individual slices taken one by one from the full 3D dataset. The network is designed as an encoder-decoder pair, the four input images of size \(240\,\times \,240\) are given as input to the encoder that encodes them into \(8\,\times \,8\times 2048\) data. This encoded data are provided to a fully convolutional decoder network that predict the labels for the glioma segmentation. The Resent-50 [22] which was the winner of ILSRVC 2015 image classification challenge is used as an encoder network followed by fully convolutional layers of decoder network.
3.1 Deep Learning Network Architecture
Fig. 2, shows the network architecture; it consists of the first 49 layers of Resnet-50 as the encoder. Each convolutional layer in the encoder is followed by a batch normalization and scaling layers [21], which avoids vanishing/exploding gradients. The first layer of the encoder consists of a larger kernel size \(7\,\times \,7\) with a stride of 2, and this reduces the input \(240\,\times \,240\) image to \(120\,\times \,120\). The first layer is followed by a max-pooling layer that further reduces the size of the data to \(60\,\times \,60\). The use of larger kernel increases the field of view, at the same time the stride of 2 and max pooling reduces the size of the data, which results in reduction on memory requirement and reduces the training time. The layer parameters for encoder network, which is derived from first 49 layers of Resnet-50 are presented in [22].
The decoder network consists of upsampling layers that enlarges the dimension of image by a factor of 2. The weights of upsampling layer are fixed to bilinear upsampling and were not learned. During the encoding process the spatial information is lost due to pooling/downsampling, therefore the spatial information is reintroduced into the decoding network by concating the original images scaled by bilinear interpolation after each upsampling layer. After each convolutional layer, the batch normalization and scaling is performed followed by an ReLU non linear activation function. The last layer in the network is a multinomial logistic layer that predicts the probability of a given pixel being either normal (label 0), necrotic/non-enhancing tumor (NCR/NET, label 1), edema (label 2) or enhancing tumor (label 4). The decoder networks’ input and output dimensions of the feature maps and convolutional kernel sizes are shown in the Table 1. The input to the decoder network is 2048 features of size \(8\,\times \,8\), which are then passed successively through a series of convolution and upsampling layers, eventually generating a probability maps for the segmentation labels.
3.2 Dataset
The training dataset [9,10,11,12] used to train the network was provided by organisers of BRATS-17 challenge. The data consisted of 3D brain images from 285 subjects with four different contrast: T1, T1-CE, T2 and T2-Flair. There were 210 HGG subjects and 75 LGG subjects. The manually segmented labels were also provided for each subject. We divided the dataset into two groups:
-
Train dataset: 259 subjects consisting of 192 HGG and 67 HGG subjects, used to train the network
-
Test dataset: 26 subjects consisting of 18 HGG and 8 LGG subjects, used to evaluate the generalisation of the network.
Another two datasets for which the ground truth was not known were provided: one for validation consisting of 46 subjects and other for testing consisting of 147 subjects.
3.3 Pre-processing
The Caffe [26] deep learning library was used to train the network. The images provided by the organisers were in the NIFTI format, which were first converted to HDF5 file format, so that it can be read by Caffe. All the 3D volumes were normalised using histogram matching [27], the reference histogram for matching was obtained by averaging histograms of all the training dataset. Only the voxels with signal were used for computing the reference histogram and matching the histogram.
3.4 Training
The training was performed on the train dataset using stochastic gradient descent with momentum in Caffe. The training parameters were: base learning rate \((lr_{base})\) = 0.01, momentum = 0.9. The weights were decayed using the formula:
where, \(lr_{iter}\) is learning rate during \(iter^{th}\) iteration, \(\gamma = 0.0001\), and \(power = 0.75\). The network was trained for 45 K iterations with the batch size of 8. The weights were regularized with \(l_2\) regularization of 0.0005 during the training. Different base learning rate were experimented and the one that provided minimum loss was used for the final training.
3.5 Evaluation Method
The results of segmentation were evaluated using the dice score, sensitivity (true positive rate) and specificity (true negative rate). All the evaluation matrices were computed locally to test the local dataset of 26 subjects. The evaluation on the validation and test dataset was calculated on an online web-portal provided by the BRATS-17 organisers.
4 Results
The trained network was tested on the validation data and the results of segmentation were uploaded on the computing portal provided by the organisers. The dice score, sensitivity, specificity and Hausdorff distance were computed on the segmented labels. The Tables 2 and 3 show the result of segmentation of 46 subjects on the validation dataset and 147 subjects on the test dataset, respectively. The results of the segmentation from 2 different subjects on the local testing dataset are shown in Fig. 3. The whole tumor is defined as union of label 1, 2, and 4; the tumor core is defined as the union of label 1 and 4; and enhancing tumor is defined as label 4.
Segmentation results on the test dataset for 2 different subjects; GT: represents the ground truth segmentation; P: represents the segmentation predicted by the proposed residual encoder-decoder network. Colour coding scheme: magenta (label 0, NCR/NET), yellow (label 4, enhancing tumor) and cyan (label 2, edema). (Color figure online)
5 Discussion
The median of dice score, sensitivity and specificity are all greater than the mean, which indicates that the proposed method performed well for most of the dataset but did not performed well for a few, that reduces the overall mean. The boundaries of the labels segmented by the proposed algorithm are smooth compared to the ground truth (Fig. 3), and this may be due to the fact that some of the ground truth were created using automated algorithms rather than a human rater.
The network was trained only on the 2D axial slices of the 3D volume, thus it considered each slice as a separate instance and does not learn any correlation across the slices. This is evident from the predicted labels in the sagittal and coronal slices (Fig. 3), where there are few isolated false positive prediction. These false positive predictions can be suppressed by incorporating 3D information during training. One way to achieve this is to design a 3D residual network which takes a 3D volume and predicts a 3D label. This approach however would require large internal memory to store the network and 3D data/feature maps, which may not be feasible for a large depth. A moderate approach would be to use more than one slices at a time instead of full 3D volume for training. This approach would not increase the memory requirement and at the same time provide 3D information to the network. The number of slices to be used for this moderate approach would be determined by the available memory of the computing hardware. Ideally, all the slices should be used for the 3D architecture, but given the limitation of GPU internal memory a trade of would be required between accuracy and memory.
6 Conclusion
In this paper, we developed a 59-layer deep residual encoder-decoder convolutional neural network that takes 2D slices of 3D MRI images as input and outputs the segmented labels. The bottleneck residual units make it feasible to train a 49-layer deep encoder. The bilinear scaling of input images serves as a guidance for the decoder network to spatially locate the tumor within a image.
Supporting information
The trained caffe model and all the scrips to train/finetune the network presented in this work are available at: https://github.com/kamleshpawar17/BratsNet-2017
References
Louis, D.N., Ohgaki, H., Wiestler, O.D., Cavenee, W.K., Burger, P.C., Jouvet, A., Scheithauer, B.W., Kleihues, P.: The 2007 who classification of tumours of the central nervous system. Acta Neuropathol. 114(2), 97–109 (2007)
Mazzara, G.P., Velthuizen, R.P., Pearlman, J.L., Greenberg, H.M., Wagner, H.: Brain tumor target volume determination for radiation treatment planning through automated MRI segmentation. Int. J. Radiat. Oncol. Biol. Phys. 59(1), 300–312 (2004)
Liu, J., Li, M., Wang, J., Wu, F., Liu, T., Pan, Y.: A survey of MRI-based brain tumor segmentation methods. Tsinghua Sci. Technol. 19(6), 578–595 (2014)
Angelini, E.D., Clatz, O., Mandonnet, E., Konukoglu, E., Capelle, L., Duffau, H.: Glioma dynamics and computational models: a review of segmentation, registration, and in silico growth algorithms and their clinical applications. Current Med. Imaging Rev. 3(4), 262–276 (2007)
Gupta, M.P., Shringirishi, M.M., et al.: Implementation of brain tumor segmentation in brain MR images using K-means clustering and fuzzy C-means algorithm. Int. J. Comput. Technol. 5(1), 54–59 (2013)
Corso, J.J., Sharon, E., Dube, S., El-Saden, S., Sinha, U., Yuille, A.: Efficient multilevel brain tumor segmentation with integrated Bayesian model classification. IEEE Trans. Med. Imaging 27(5), 629–640 (2008)
Sharma, N., Aggarwal, L.M.: Automated medical image segmentation techniques. J. Med. Phys./Assoc. Med. Physicists India 35(1), 3 (2010)
Pham, D.L., Xu, C., Prince, J.L.: Current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2(1), 315–337 (2000)
Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, R., Lanczi, L., Gerstner, E., Weber, M.-A., Arbel, T., Avants, B.B., Ayache, N., Buendia, P., Collins, D.L., Cordier, N., Corso, J.J., Criminisi, A., Das, T., Delingette, H., Demiralp, C., Durst, C.R., Dojat, M., Doyle, S., Festa, J., Forbes, F., Geremia, E., Glocker, B., Golland, P., Guo, X., Hamamci, A., Iftekharuddin, K.M., Jena, R., John, N.M., Konukoglu, E., Lashkari, D., Mariz, J.A., Meier, R., Pereira, S., Precup, D., Price, S.J., Raviv, T.R., Reza, S.M.S., Ryan, M., Sarikaya, D., Schwartz, L., Shin, H.-C., Shotton, J., Silva, C.A., Sousa, N., Subbanna, N.K., Szekely, G., Taylor, T.J., Thomas, O.M., Tustison, N.J., Unal, G., Vasseur, F., Wintermark, M., Ye, D.H., Zhao, L., Zhao, B., Zikic, D., Prastawa, M., Reyes, M., Leemput, K.V.: The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015)
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J., Freymann, J., Farahani, K., Davatzikos, C.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat. Sci. Data 4 (2017)
Bakas, S., Sotiras, H., Bilello, M., Kirby, J., Freymann, J., Farahani, K., Davatzikos, C.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive (2017). https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q
Bakas, S., Sotiras, H., Bilello, M., Kirby, J., Freymann, J., Farahani, K., Davatzikos, C.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive (2017). https://doi.org/10.7937/K9/TCIA.2017.GJQ7R0EF
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521(7553), 436–444 (2015)
Schmidhuber, J.: Deep learning in neural networks: an overview. Neural Networks 61, 85–117 (2015)
LeCun, Y., Boser, B.E., Denker, J.S., Henderson, D., Howard, R.E., Hubbard, W.E., Jackel, L.D.: Handwritten digit recognition with a back-propagation network. In: Advances in Neural Information Processing Systems, pp. 396–404 (1990)
LeCun, Y., Bottou, L., Bengio, Y., Haffner, P.: Gradient-based learning applied to document recognition. Proc. IEEE 86(11), 2278–2324 (1998)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Ciregan, D., Meier, U., Schmidhuber, J.: Multi-column deep neural networks for image classification. In: 2012 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3642–3649. IEEE (2012)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Tajbakhsh, N., Shin, J.Y., Gurudu, S.R., Hurst, R.T., Kendall, C.B., Gotway, M.B., Liang, J.: Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imaging 35(5), 1299–1312 (2016)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. In International Conference on Machine Learning, pp. 448–456 (2015)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
He, K., Sun, J.: Convolutional neural networks at constrained time cost. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 5353–5360 (2015)
Srivastava, R.K., Greff, K., Schmidhuber, J.: Training very deep networks. In: Advances in Neural Information Processing Systems, pp. 2377–2385 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Jia, Y., Shelhamer, E., Donahue, J., Karayev, S., Long, J., Girshick, R., Guadarrama, S., Darrell, T.: Caffe: convolutional architecture for fast feature embedding, arXiv preprint arXiv:1408.5093 (2014)
Shapira, D., Avidan, S., Hel-Or, Y.: Multiple histogram matching. In: 2013 20th IEEE International Conference on Image Processing (ICIP), pp. 2269–2273. IEEE (2013)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Pawar, K., Chen, Z., Shah, N.J., Egan, G. (2018). Residual Encoder and Convolutional Decoder Neural Network for Glioma Segmentation. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2017. Lecture Notes in Computer Science(), vol 10670. Springer, Cham. https://doi.org/10.1007/978-3-319-75238-9_23
Download citation
DOI: https://doi.org/10.1007/978-3-319-75238-9_23
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-75237-2
Online ISBN: 978-3-319-75238-9
eBook Packages: Computer ScienceComputer Science (R0)