Abstract
Automatic segmentation of brain glioma from multimodal MRI scans plays a key role in clinical trials and practice. Unfortunately, manual segmentation is very challenging, time-consuming, costly, and often inaccurate despite human expertise due to the high variance and high uncertainty in the human annotations. In the present work, we develop an end-to-end deep-learning-based segmentation method using a multi-decoder architecture by jointly learning three separate sub-problems using a partly shared encoder. We also propose to apply smoothing methods to the input images to generate denoised versions as additional inputs to the network. The validation performance indicates an improvement when using the proposed method. The proposed method was ranked 2nd in the task of Quantification of Uncertainty in Segmentation in the Brain Tumors in Multimodal Magnetic Resonance Imaging Challenge 2020.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Glioma is a particular kind of brain tumor that develops from glial cells. It is the most frequently occurring type of brain tumor and the one with the highest mortality rate. Glioma is categorized by the World Health Organization (WHO) into four grades: low-grade glioma (LGG) (class I and II), and high-grade glioma (HGG) (class III and IV), where HGG is being considered a dangerous and life-threatening tumor. Specifically, about 190,000 cases occur annually worldwide [6], and around 90 % [18] of patients die within 24 months of surgical resection. Segmentation of the tumor plays a role both for radiotherapy treatment planning and for diagnostic follow-up of the disease. Manual segmentation is time-consuming, subjective, and associated with uncertainties due to the variation of shape, location, and appearance of the tumors. Hence, decision support or automating the segmentation may improve the treatment quality as well as enhancing the efficiency when handling this patient group.
Inspired by a need of automatic segmentation of brain tumors in multimodal magnetic resonance imaging (MRI) scans, the Brain Tumors in Multimodal Magnetic Resonance Imaging Challenge 2020 (BraTS 2020) [2,3,4,5, 14] is a yearly challenge (associated with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)) that aims to evaluate state-of-the-art methods for brain tumor segmentation. BraTS 2020 provides the participants with images from four structural MRI modalities: post-contrast T1-weighted (T1c), T2-weighted (T2w), T1-weighted (T1w), and T2 Fluid Attenuated Inversion Recovery (FLAIR) for brain tumor analysis and segmentation. Masks were annotated manually by one to four raters followed by improvements by expert raters. The segmentation performances of the participants were evaluated using the Sørensen-Dice coefficient (DSC), sensitivity, specificity, and the \(95^{th}\) percentile of the Hausdorff distance (HD95).
Since the introduction of the U-Net by Ronneberger et al. [16], Convolutional Neural Networks (CNNs) incorporating skip connections have become the baseline architecture for medical image segmentation. Various architectures, often building on or extending this baseline, have been proposed to address the brain tumor segmentation problem. In BraTS 2019, Jiang et al. [11], who was the first-place winner of the challenge, proposed an end-to-end two-stage cascaded U-Net to segment the substructures of brain tumors from coarse (in the first stage) to fine (in the second stage) prediction. In the same challenge, Zhao et al. [21], who won the second place, introduced numerous tricks for 3D MRI brain tumor segmentation including processing methods, model designing methods, and optimizing methods. McKinley et al. [13] proposed DeepSCAN, which is a modification of their previous 3D-to-2D Fully Convolutional Network (FCN), by replacing batch normalization with instance normalization and adding a lightweight local attention mechanism to secure the third place in the BraTS 2019.
The architecture proposed in this work is an extension of the one in [20] from the BraTS 2019 where End-to-end Hierarchical Tumor Segmentation using Cascaded Networks (TuNet) was introduced. Despite achieving a decent performance, the main drawback of TuNet is that it comprises three cascaded networks that make it hard to fit a full volume, with shape \(240 \times 240 \times 155\), into memory on any recent graphics processing units (GPUs). Because of this, the TuNet adapted a patch-based segmentation approach, leading to long training times. In addition to that, the TuNet might suffer from a lack of global information about the image.
Motivated by the successes of the cascaded networks, presented in e.g. [11, 20], the present work proposes a multi-decoder architecture, denoted End-to-end Multi-Decoder Cascaded Network for Tumor Segmentation (MDNet), to separate a complicated problem into simple sub-problems. We also propose to use multiple denoised versions of the original images as inputs to the network. The hypothesis was that this would counteract the salt and pepper noise often seen in MRI scans [1]. To the best of our knowledge, this is the first use of this technique.
The authors hypothesize that the MDNet will reduce overfitting problems by employing a shared encoder between three different decoders, while denoised MRI images will help the network to gain more insight into the multimodal input images with the presence of another two versions of the images: (i) a salt and pepper-free one from the use of a median filter, and (ii) one with reduced high-frequency components by employing a low-pass Gaussian filter.
2 Methods
Inspired by the drawbacks of the method proposed in [20], the authors here also propose an end-to-end framework that separates the complicated multi-class tumor segmentation problem into three simpler binary segmentation problems, but with a major change in the design. The MDNet consumes much less memory compared to the TuNet, which means that whole input volumes can be fit into the GPU memory. Hence, the proposed MDNet can take advantage of global details. In addition to that, the design of MDNet results in shorter training times since it uses whole volumes instead of patches, as was the case with the TuNet.
2.1 Encoder Network
The encoder network consists of conventional convolution blocks [16], where each block includes a convolution layer with batch normalization and a leaky rectified linear unit (LeakyReLU) activation function. Each convolutional block is then followed by a Squeeze-and-Excitation block (SEB) (see Sect. 2.3). Max-pooling layers were used for downsampling. All convolutional filters had the size of \(3\,\times \,3\,\times \,3\), and the initial numbers of filters were set to twelve, which in the proposed architecture is equivalent to three denoising methods applied to the four given modalities (see Sect. 2.4). The encoder output has shape \(96\,\times \,20\,\times \,24\,\times \,16\). The complete architecture of the proposed encoder network is detailed in Table 1.
2.2 Multi-decoder Networks
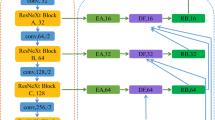
Table 2 illustrates the proposed multi-decoder networks. The decoder networks include three separate paths, where each path is employed to cope with a specific aforementioned tumor region including whole, core, and enhancing, that are denoted by W-Net, C-Net, and E-Net, respectively. Each decoder path comprises skip connections as in U-Net. There was also a SEB after each convolution block and a concatenation operation of the output of the spatial upsampling layers with the feature maps from the encoder at the same level. To enrich the feature maps at the beginning of each level in the C-Net, the feature map at the end of the W-Net on the same level is used. A similar approach is employed in the decoder network of the E-Net and C-Net. By utilizing these, we hypothesize that the W-Net will constrain the C-Net, while the C-Net will constrain the E-Net. Figure 1 illustrates the proposed architecture.
2.3 Squeeze-and-Excitation Block
We added a channel-based SEB as proposed by Hu et al. [8] after each convolution block or concatenation operation. The idea of SEB is to adapt the weight of each channel in a feature map by adding a content-aware mechanism at almost no computational cost. In recent days, SEB has been widely employed to achieve a huge boost in performance. A conventional SEB includes the following layers in sequence: global pooling, fully connected, rectified linear unit (ReLU) activation function, fully connected, and a sigmoid activation function [8].
2.4 Denoising the Inputs
The inputs to the network were the MRI modalities, and also each modality after denoising using two different methods: median denoising and Gaussian smoothing. The authors then concatenated the three versions of the images for each modality (the raw image, and the two denoised versions) to obtain a total of twelve images, that were input as different channels. For the median denoising, we used a \(3\,\times \,3\,\times \,3\) median filter; the Gaussian smoothing used a \(3\,\times \,3\,\times \,3\) Gaussian filter with a standard deviation of 0.5. In this sense, adding a Gaussian smoothed version of the input is similar to adding a down-scaled version of the input image as was proposed for the TuNet [20].
2.5 Preprocessing and Augmentation
All input images were normalized to have a mean zero and unit variance. In order to reduce overfitting and increase the diversity of data available for training models, we used on-the-fly data augmentation [9] comprising: (1) randomly rotating the images in the range \([-1, 1]\) degrees on all three axes, (2) random mirror flipping with a probability of 0.5 on all three axes, (3) elastic transformation with a probability of 0.3, (4) random scaling in the range [0.9, 1.1] with a probability of 0.3, and (5) random cropping with subsequent resizing with a probability of 0.3.
As in [17], the elastic transformations used a random displacement field, \(\varDelta \), such that
where \(\alpha \) is the strength of the displacement, while \(R_w\) and \(R_o\) denote the location of a voxel in the warped and original image, respectively. For each axis, a random number was drawn uniformly in \([-1, 1]\) such that \(\varDelta _x \sim \mathcal {U}(-1, 1)\), \(\varDelta _y \sim \mathcal {U}(-1, 1)\), and \(\varDelta _z \sim \mathcal {U}(-1, 1)\). The displacement field was finally convolved with a Gaussian kernel having standard deviation \(\sigma \). In the present case, \(\alpha =1\) and \(\sigma =0.25\).
2.6 Post-processing
The most challenging task of BraTS 2020 specifically, and BraTS challenges in general, is to distinguish between LGG and HGG patients by labeling small vessels lying in the tumor core as edema or necrosis. In order to tackle this problem, we used the same strategy as proposed in our previous work [20]. In specific, we labeled all small enhancing tumor region with less than 500 connected voxels as necrosis. The proposed post-processing step aims to handle a few cases where the proposed networks fail to differentiate between the whole and core tumor regions.
2.7 Task 3: Quantification of Uncertainty in Segmentation
The organizers of the BraTS challenge introduced the task of “Quantification of Uncertainty in Segmentation” in BraTS 2019 and was held again in BraTS 2020. This task is aimed to measure the uncertainty in the context of glioma region segmentation by rewarding predictions that are (a) confident when correct and (b) uncertain when incorrect. Participants were expected to generate uncertainty maps in the range of [0, 100], where 0 represents the most certain and 100 represents the most uncertain. The performance was evaluated based on three metrics: Dice Area Under Curve (DAUC), Ratio of Filtered True Positives (RFTPs), and Ratio of Filtered True Negatives (RFTNs).
Similar to [20], the proposed network, MDNet, predicts the probability of three tumor regions, it thus benefits from this task. Following [20], an uncertainty score, \(u^r_{i,j,k}\), at voxel (i, j, k) is defined by
where \(u^r_{i,j,k} \in [0, 100]^{|\mathcal {R}|}\) and \(p^{r}_{i,j,k} \in [0, 1]^{|\mathcal {R}|}\) are the uncertainty score map and probability map, respectively. Here, \(r \in \mathcal {R}\), where \(\mathcal {R}\) is the set of tumor regions, i.e. whole, core, and enhancing region.
3 Experiments
3.1 Implementation Details and Training
The proposed method was implemented in Keras 2.2.4Footnote 1 with TensorFlow 1.12.0Footnote 2 as the backend. The experiments were trained on NVIDIA Tesla V100 GPUs from the High Performance Computer Center North (HPC2N) at Umeå University, Sweden. Seven models were trained from scratch for \(N_e=200\) epochs, with a mini-batch size of one. The training time for a single model was about six days.
3.2 Loss
For evaluation of the segmentation performance, we used a combination of the DSC loss and categorical cross–entropy (CE) as the loss function. The DSC is defined as [19, 20]
where u and v are the output segmentation and its corresponding ground truth, respectively. To include the the DSC in the loss function, we employed the soft DSC loss, which is defined as [10, 19, 20]
where for each label i, the \(u_i\) is the softmax output of the proposed network for label i, v is a one-hot encoding of the ground truth labels (segmentation maps in this case), and \(\epsilon = 1 \cdot 10^{-5}\) is a small constant added to avoid division by zero.
Following [10, 19], for unbalanced data sets with small structures like in the BraTS 2020 data, we added the CE term to our loss function to make the loss surface smoother. The CE is defined as
The combination of the DSC loss and CE (denoted a hybrid loss) is simply defined as the sum of the two losses, as
The final loss function that was used for training contained one hybrid loss for each tumor region, and was thus
where \(\mathcal {R}\) again is the set of tumor regions (the whole, core, and enhancing regions) and \(\mathcal {L}_{\mathrm {hybrid}}(u_r, v_r)\) is the hybrid loss for a particular tumor region.
The segmentation performance was also evaluated using the HD95, a common metric for evaluating segmentation performances. The Hausdorff distance (HD) is defined as [7]
where
in which \(\Vert u_i - v_i\Vert _2\) is the spatial Euclidean distance between points \(u_i\) and \(v_i\) on the boundaries of output segmentation u and ground truth v.
3.3 Optimization
The authors used the Adam optimizer [12] with an initial learning rate of \(\alpha _0 = 1 \cdot 10^{-4}\) and momentum parameters of \(\beta _1=0.9\) and \(\beta _2=0.999\). Following Myronenko et al. in [15], the learning rate was decayed as
where e and \(N_e=200\) are epoch counter and total number of epochs, respectively.
The authors also used \(L_2\) regularization with a penalty parameter of \(1 \cdot 10^{-5}\), which was applied to the kernel weight matrices, for all convolutional layers to counter overfitting. The activation function of the final layer was the logistic sigmoid function.
4 Results and Discussion
Table 3 shows the mean DSC and HD95 scores and standard deviations (SDs) computed from the five-folds of cross-validation on 369 cases of the training set. From Table 3 we see that: (i) the U-Net with denoised input improved the DSC and HD95 on all tumor regions, and (ii) the proposed model with denoising boosted the performance in both metrics (DSC and HD95) by a large margin.
Table 4 shows the mean DSC and HD95 scores on the validation set, computed on the predicted masks by the evaluation serverFootnote 3 (team name UmU). The BraTS 2020 final validation dataset results were 90.55, 82.67 and 77.17 for the average DSC, and 4.99, 8.63 and 27.04 for the average HD95, for whole tumor, tumor core and enhanced tumor core, respectively. These results were slightly lower than the top-ranking teams.
Table 5 provides the mean DAUC, RFTPs, and RFTNs scores on the validation set obtained after uploading the predicted masks and corresponding uncertainty maps to the evaluation serverFootnote 4. As can be seen from Table 5, the RFTNs scores were the best amongst the best-ranking participants.
Table 6 and Table 7 show the mean DSC and HD95, and the mean DAUC, RFTPs, and RFTNs scores on the test set, respectively. In the task of Quantification of Uncertainty in Segmentation, our proposed method was ranked 2nd.
5 Conclusion
In this work, we proposed a multi-decoder network for segmenting tumor substructures from multimodal brain MRI images by separating a complex problem into simpler sub-tasks. The proposed network adopted a U-Net-like structure with Squeeze-and-Excitation blocks after each convolution and concatenation operation. We also proposed to stack original images with their denoised versions to enrich the input and demonstrated that the performance was boosted in both DSC and HD95 metrics by a large margin. The results on the test set indicated that: (i) the proposed method performed competitively in the task of Segmentation, with DSC scores of 88.26/82.49/80.84 and HD95 scores of 6.30/22.27/20.06 for the whole tumor, tumor core, and enhancing tumor core, respectively, (ii) the proposed method was top 2 performing ones in the task of Quantification of Uncertainty in Segmentation.
References
Ali, H.M.: A new method to remove salt pepper noise in magnetic resonance images. In: 2016 11th International Conference on Computer Engineering Systems (ICCES), pp. 155–160 (2016)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The cancer imaging archive (2017) (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. Cancer Imaging Archive 286 (2017)
Bakas, S., et al.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4, 170117 (2017)
Bakas, S., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629 (2018)
Castells, X., et al.: Automated brain tumor biopsy prediction using single-labeling CDNA microarrays-based gene expression profiling. Diagn. Mol. Pathol. 18, 206–218 (2009)
Hausdorff, F.: Erweiterung einer stetigen Abbildung, pp. 555–568. Springer, Heidelberg (2008). https://doi.org/10.1007/978-3-540-76807-4_16
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7132–7141 (2018)
Isensee, F., et al.: batchgenerators–a python framework for data augmentation, January 2020
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: No new-net. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 234–244. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_21
Jiang, Z., Ding, C., Liu, M., Tao, D.: Two-stage cascaded U-Net: 1st place solution to BraTS challenge 2019 segmentation task. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 231–241. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_22
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
McKinley, R., Rebsamen, M., Meier, R., Wiest, R.: Triplanar ensemble of 3D-to-2D CNNs with label-uncertainty for brain tumor segmentation. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 379–387. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_36
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE TRans. Med. Imaging 34(10), 1993–2024 (2014)
Myronenko, A.: 3D MRI brain tumor segmentation using autoencoder regularization (2018). http://arxiv.org/abs/1810.11654
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Simard, P.Y., Steinkraus, D., Platt, J.C.: Best practices for convolutional neural networks applied to visual document analysis. In: Seventh International Conference on Document Analysis and Recognition, 2003. Proceedings, pp. 958–963 (2003)
Thurnher, M.: The 2007 WHO classification of tumors of the central nervous system–what has changed? Am. J. Neuroradiol. (2012)
Vu, M.H., Grimbergen, G., Nyholm, T., Löfstedt, T.: Evaluation of multi-slice inputs to convolutional neural networks for medical image segmentation. arXiv preprint arXiv:1912.09287 (2019)
Vu, M.H., Nyholm, T., Löfstedt, T.: TuNet: end-to-end hierarchical brain tumor segmentation using cascaded networks. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 174–186. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_17
Zhao, Y.-X., Zhang, Y.-M., Liu, C.-L.: Bag of tricks for 3D MRI brain tumor segmentation. In: Crimi, A., Bakas, S. (eds.) BrainLes 2019. LNCS, vol. 11992, pp. 210–220. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-46640-4_20
Acknowledgement
The computations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at the HPC2N in Umeå, Sweden. We are grateful for the financial support obtained from the Cancer Research Fund in Northern Sweden, Karin and Krister Olsson, Umeå University, The Västerbotten regional county, and Vinnova, the Swedish innovation agency.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Vu, M.H., Nyholm, T., Löfstedt, T. (2021). Multi-decoder Networks with Multi-denoising Inputs for Tumor Segmentation. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science(), vol 12658. Springer, Cham. https://doi.org/10.1007/978-3-030-72084-1_37
Download citation
DOI: https://doi.org/10.1007/978-3-030-72084-1_37
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72083-4
Online ISBN: 978-3-030-72084-1
eBook Packages: Computer ScienceComputer Science (R0)